Assessing Genomic Mutations in SARS-CoV-2: Potential Resistance to Antiviral Drugs in Viral Populations from Untreated COVID-19 Patients
et al., Microorganisms, doi:10.3390/microorganisms12010002, Dec 2023
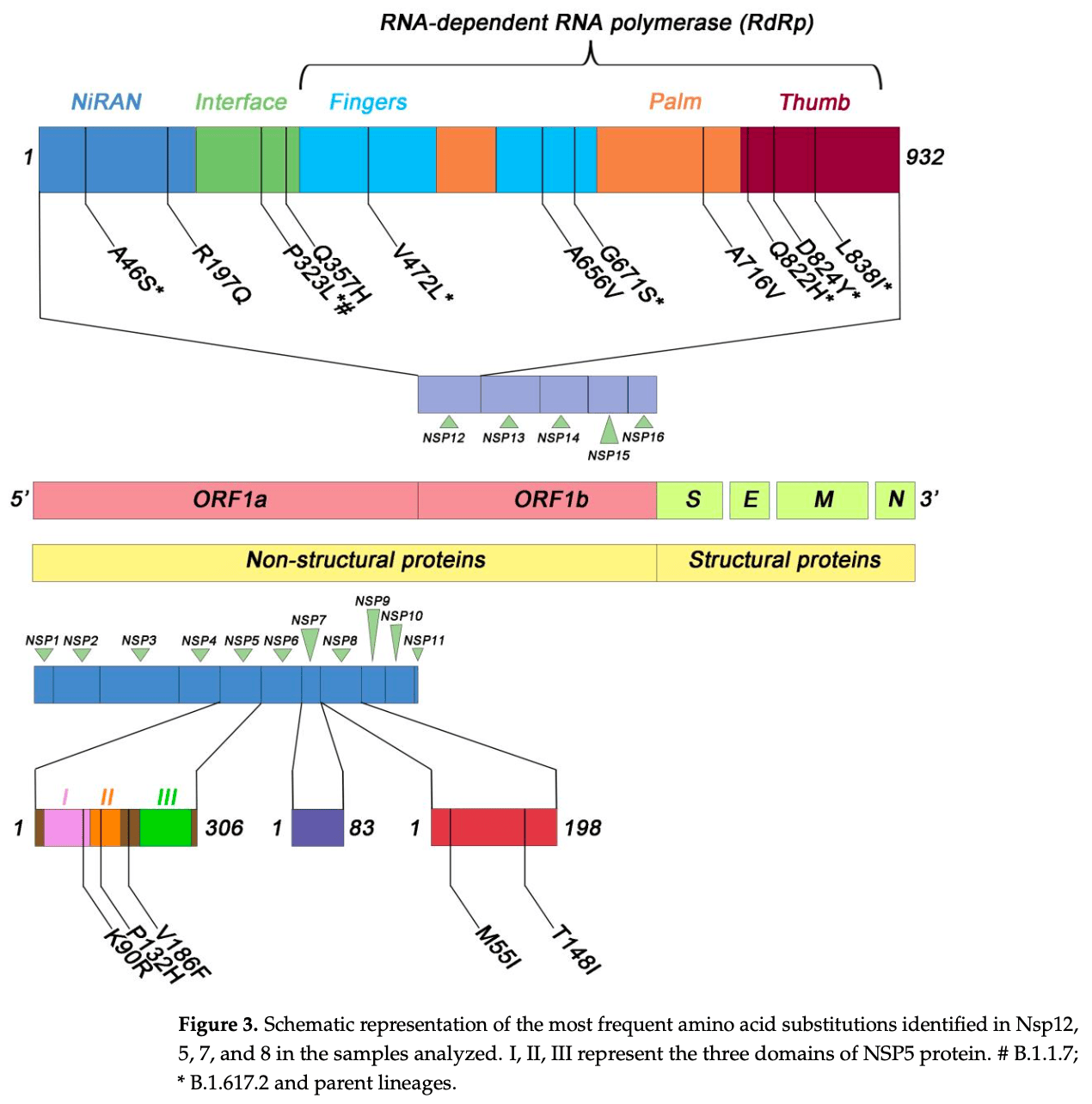
Analysis of naturally occurring SARS-CoV-2 mutations in genomic regions targeted by remdesivir, molnupiravir, and paxlovid in 4,155 antiviral-naive COVID-19 patients. Authors identified 84 amino-acid substitutions in Nsp12 (RdRp; target of remdesivir/molnupiravir) and 28 in Nsp5 (main protease; target of paxlovid), with additional changes in Nsp7 (14) and Nsp8 (24). The findings show that drug-target regions accumulate natural variation in untreated populations, indicating a risk to future antiviral efficacy.
Gérard, Zhou, Wu, Kamo, Choi, Kim show increased risk of acute kidney injury, Leo, Briciu, Muntean, Petrov show increased risk of liver injury, and Negru, Cheng, Mohammed, Kwok show increased risk of cardiac disorders with remdesivir.
Study covers remdesivir, molnupiravir, and paxlovid.
1.
Gérard et al., Remdesivir and Acute Renal Failure: A Potential Safety Signal From Disproportionality Analysis of the WHO Safety Database, Clinical Pharmacology & Therapeutics, doi:10.1002/cpt.2145.
2.
Zhou et al., Acute Kidney Injury and Drugs Prescribed for COVID-19 in Diabetes Patients: A Real-World Disproportionality Analysis, Frontiers in Pharmacology, doi:10.3389/fphar.2022.833679.
3.
Wu et al., Acute Kidney Injury Associated With Remdesivir: A Comprehensive Pharmacovigilance Analysis of COVID-19 Reports in FAERS, Frontiers in Pharmacology, doi:10.3389/fphar.2022.692828.
4.
Kamo et al., Association of Antiviral Drugs for the Treatment of COVID-19 With Acute Renal Failure, In Vivo, doi:10.21873/invivo.13637.
5.
Choi et al., Comparative effectiveness of combination therapy with nirmatrelvir–ritonavir and remdesivir versus monotherapy with remdesivir or nirmatrelvir–ritonavir in patients hospitalised with COVID-19: a target trial emulation study, The Lancet Infectious Diseases, doi:10.1016/S1473-3099(24)00353-0.
6.
Kim et al., Investigating the Safety Profile of Fast‐Track COVID‐19 Drugs Using the FDA Adverse Event Reporting System Database: A Comparative Observational Study, Pharmacoepidemiology and Drug Safety, doi:10.1002/pds.70043.
7.
Leo et al., Hepatocellular liver injury in hospitalized patients affected by COVID-19: Presence of different risk factors at different time points, Digestive and Liver Disease, doi:10.1016/j.dld.2021.12.014.
8.
Briciu et al., Evolving Clinical Manifestations and Outcomes in COVID-19 Patients: A Comparative Analysis of SARS-CoV-2 Variant Waves in a Romanian Hospital Setting, Pathogens, doi:10.3390/pathogens12121453.
9.
Muntean et al., Effects of COVID-19 on the Liver and Mortality in Patients with SARS-CoV-2 Pneumonia Caused by Delta and Non-Delta Variants: An Analysis in a Single Centre, Pharmaceuticals, doi:10.3390/ph17010003.
10.
Petrov et al., The Effect of Potentially Hepatotoxic Medicinal Products on Alanine Transaminase Levels in COVID-19 Patients: A Case–Control Study, Safety and Risk of Pharmacotherapy, doi:10.30895/2312-7821-2025-458.
11.
Negru et al., Comparative Pharmacovigilance Analysis of Approved and Repurposed Antivirals for COVID-19: Insights from EudraVigilance Data, Biomedicines, doi:10.3390/biomedicines13061387.
12.
Cheng et al., Cardiovascular Safety of COVID-19 Treatments: A Disproportionality Analysis of Adverse Event Reports from the WHO VigiBase, Infectious Diseases and Therapy, doi:10.1007/s40121-025-01225-z.
Lombardo et al., 19 Dec 2023, retrospective, Italy, peer-reviewed, 8 authors, study period April 2021 - October 2022.
Contact: teresa.pollicino@unime.it (corresponding author), daniele.lombardo@unime.it, valeria.chines@unime.it, giuseppe.caminiti@unime.it, claudia_palermo@icloud.com, irene.cacciola@unime.it, giuseppina.raffa@unime.it, cristina.musolino@unime.it.
Assessing Genomic Mutations in SARS-CoV-2: Potential Resistance to Antiviral Drugs in Viral Populations from Untreated COVID-19 Patients
Microorganisms, doi:10.3390/microorganisms12010002
Naturally occurring SARS-CoV-2 variants mutated in genomic regions targeted by antiviral drugs have not been extensively studied. This study investigated the potential of the RNA-dependent RNA polymerase (RdRp) complex subunits and non-structural protein (Nsp)5 of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) to accumulate natural mutations that could affect the efficacy of antiviral drugs. To this aim, SARS-CoV-2 genomic sequences isolated from 4155 drug-naive individuals from southern Italy were analyzed using the Illumina MiSeq platform. Sequencing of the 4155 samples showed the following viral variant distribution: 71.2% Delta, 22.2% Omicron, and 6.4% Alpha. In the Nsp12 sequences, we found 84 amino acid substitutions. The most common one was P323L, detected in 3777/4155 (91%) samples, with 2906/3777 (69.9%) also showing the G671S substitution in combination. Additionally, we identified 28, 14, and 24 different amino acid substitutions in the Nsp5, Nsp7, and Nsp8 genomic regions, respectively. Of note, the V186F and A191V substitutions, affecting residues adjacent to the active site of Nsp5 (the target of the antiviral drug Paxlovid), were found in 157/4155 (3.8%) and 3/4155 (0.07%) samples, respectively. In conclusion, the RdRp complex subunits and the Nsp5 genomic region exhibit susceptibility to accumulating natural mutations. This susceptibility poses a potential risk to the efficacy of antiviral drugs, as these mutations may compromise the drug ability to inhibit viral replication
potential limitations, phylogenetic trees have played a crucial role in tracking the spread of the virus, identifying potential sources of outbreaks, and improving our understanding of the evolutionary dynamics of SARS-CoV-2. The results of our study highlight the dynamic genetic diversity of SARS-CoV-2, with variants such as Delta and Omicron exhibiting distinct patterns of amino acid substitutions in key genomic regions and proteins. Although the scope of our study is limited to eastern Sicily, it has shown that even within this restricted area, conserved genomic regions of SARS-CoV-2, such as Nsp5, Nsp7, Nsp8, and Nsp12, are prone to accumulate spontaneous mutations in individuals who have not been exposed to antiviral treatments. Some of these mutations, might compromise the efficacy of antiviral drugs, which are of fundamental importance for patients at risk of severe COVID-19. While this study provides valuable insights into the evolutionary landscape of SARS-CoV-2, it is not without limitations. Notably, this study did not delve into the functional consequences of the identified mutations, leaving unanswered questions regarding their impact on viral fitness and transmission. To address these limitations and gain a more comprehensive understanding of the evolutionary trajectory of SARS-CoV-2, future research should focus on conducting functional studies to evaluate the impact of observed mutations on viral replication, infectivity, and immune evasion, and investigating..
References
Bravo, Dangerfield, Taylor, Johnson, Remdesivir is a delayed translocation inhibitor of SARS-CoV-2 replication, Mol. Cell, doi:10.1016/j.molcel.2021.01.035
Cao, Wang, Yin, Gao, Xia, Human microbiota is a reservoir of SARS-CoV-2 advantageous mutations, bioRxiv, doi:10.1101/2023.11.16.567485
Chand, Banerjee, Azad, Identification of novel mutations in RNA-dependent RNA polymerases of SARS-CoV-2 and their implications on its protein structure, PeerJ, doi:10.7717/peerj.9492
De Sabato, Vaccari, Knijn, Ianiro, Di Bartolo et al., SARS-CoV-2 RECoVERY: A multi-platform open-source bioinformatic pipeline for the automatic construction and analysis of SARS-CoV-2 genomes from NGS sequencing data, bioRxiv, doi:10.1101/2021.01.16.425365
De, Dutta, Role of the Microbiome in the Pathogenesis of COVID-19, Front. Cell. Infect. Microbiol, doi:10.3389/fcimb.2022.736397
Dragelj, Mroginski, Ebrahimi, Hidden in Plain Sight: Natural Products of Commensal Microbiota as an Environmental Selection Pressure for the Rise of New Variants of SARS-CoV-2, Chembiochem, doi:10.1002/cbic.202100346
Eskier, Suner, Karakülah, Oktay, Mutation density changes in SARS-CoV-2 are related to the pandemic stage but to a lesser extent in the dominant strain with mutations in spike and RdRp, PeerJ, doi:10.7717/peerj.9703
Felsenstein, Confidence limits on phylogenies: An approach using the bootstrap, Evolution, doi:10.2307/2408678
Gao, Yan, Huang, Liu, Zhao et al., Structure of the RNA-dependent RNA polymerase from COVID-19 virus, Science, doi:10.1126/science.abb7498
Hillen, Kokic, Farnung, Dienemann, Tegunov et al., Structure of replicating SARS-CoV-2 polymerase, Nature, doi:10.1038/s41586-020-2368-8
Hodge, Field, General Mechanisms of Antiviral Resistance, doi:10.1016/B978-0-12-384890-1.00013-3
Ilmjärv, Abdul, Acosta-Gutiérrez, Estarellas, Galdadas et al., Concurrent mutations in RNA-dependent RNA polymerase and spike protein emerged as the epidemiologically most successful SARS-CoV-2 variant, Sci. Rep, doi:10.1038/s41598-021-91662-w
Ip, Chu, Chan, Leung, Abdullah et al., Global prevalence of SARS-CoV-2 3CL protease mutations associated with nirmatrelvir or ensitrelvir resistance, EBioMedicine, doi:10.1016/j.ebiom.2023.104559
Jin, Du, Xu, Deng, Liu et al., Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors, Nature, doi:10.1038/s41586-020-2223-y
Kabinger, Stiller, Schmitzová, Dienemann, Kokic et al., Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis, Nat. Struct. Mol. Biol, doi:10.1038/s41594-021-00651-0
Kim, Kim, Casel, Kim, Sun et al., SARS-CoV-2 variants with NSP12 P323L/G671S mutations display enhanced virus replication in ferret upper airways and higher transmissibility, Cell Rep, doi:10.1016/j.celrep.2023.113077
Kneller, Phillips, O'neill, Jedrzejczak, Stols et al., Structural plasticity of SARS-CoV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography, Nat. Commun, doi:10.1038/s41467-020-16954-7
Kokic, Hillen, Tegunov, Dienemann, Seitz et al., Mechanism of SARS-CoV-2 polymerase stalling by remdesivir, Nat. Commun, doi:10.1038/s41467-020-20542-0
Koyama, Platt, Parida, Variant analysis of SARS-CoV-2 genomes, Bull. World Health Organ, doi:10.2471/BLT.20.253591
Kumar, Stecher, Li, Knyaz, Tamura et al., Molecular Evolutionary Genetics Analysis across Computing Platforms, Mol. Biol. Evol, doi:10.1093/molbev/msy096
Lee, Worrall, Vuckovic, Rosell, Gentile et al., Crystallographic structure of wild-type SARS-CoV-2 main protease acyl-enzyme intermediate with physiological C-terminal autoprocessing site, Nat. Commun, doi:10.1038/s41467-020-19662-4
Letunic, Bork, Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation, Nucleic Acids Res, doi:10.1093/nar/gkab301
Lou, Rao, The Life of SARS-CoV-2 Inside Cells: Replication-Transcription Complex Assembly and Function, Annu. Rev. Biochem, doi:10.1146/annurev-biochem-052521-115653
Mahase, COVID-19: Pfizer's paxlovid is 89% effective in patients at risk of serious illness, company reports, BMJ, doi:10.1136/bmj.n2713
Mahase, Molnupiravir reduces risk of hospital admission or death by 50% in patients at risk, MSD reports, BMJ, doi:10.1136/bmj.n2422
Malone, Campbell, Molnupiravir, Coding for catastrophe, Nat. Struct. Mol. Biol, doi:10.1038/s41594-021-00657-8
Malone, Urakova, Snijder, Campbell, Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design, Nat. Rev. Mol. Cell Biol, doi:10.1038/s41580-021-00432-z
Martinot, Jary, Fafi-Kremer, Leducq, Delagreverie et al., Emerging RNA-Dependent RNA Polymerase Mutation in a Remdesivir-Treated B-cell Immunodeficient Patient with Protracted Coronavirus Disease, Clin. Infect. Dis, doi:10.1093/cid/ciaa1474
Mason, Devincenzo, Toovey, Wu, Whitley, Comparison of antiviral resistance across acute and chronic viral infections, Antivir. Res, doi:10.1016/j.antiviral.2018.07.020
Mohammad, Al-Mulla, Wei, Abubaker, Remdesivir, Simulations Suggest a More Favourable Binding to SARS-CoV-2 RNA Dependent RNA Polymerase Mutant P323L Than Wild-Type, Biomolecules, doi:10.3390/biom11070919
Pachetti, Marini, Benedetti, Giudici, Mauro et al., Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant, J. Transl. Med, doi:10.1186/s12967-020-02344-6
Patel, Ono, Bassit, Verma, Amblard et al., Assessment of a Computational Approach to Predict Drug Resistance Mutations for HIV, HBV and SARS-CoV-2, Molecules, doi:10.3390/molecules27175413
Peloquin, Dimaio, Bierer, Barnes, Disruptive and avoidable: GDPR challenges to secondary research uses of data, Eur. J. Hum. Genet, doi:10.1038/s41431-020-0596-x
Peng, Peng, Yuan, Zhao, Wang et al., Structural and Biochemical Characterization of the Nsp12-Nsp7-Nsp8 Core Polymerase Complex from SARS-CoV-2, Cell Rep, doi:10.1016/j.celrep.2020.107774
Reshamwala, Likhite, Degani, Deb, Noronha, Mutations in SARS-CoV-2 Nsp7 and Nsp8 proteins and their predicted impact on replication/transcription complex structure, J. Med. Virol, doi:10.1002/jmv.26791
Rubin, Chan-Tack, Farley, Sherwat, FDA Approval of Remdesivir-A Step in the Right Direction, N. Engl. J. Med, doi:10.1056/NEJMp2032369
Szemiel, Merits, Orton, Maclean, Pinto et al., In vitro selection of Remdesivir resistance suggests evolutionary predictability of SARS-CoV-2, PLoS Pathog, doi:10.1371/journal.ppat.1009929
Tamura, Nei, Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees, Mol. Biol. Evol, doi:10.1093/oxfordjournals.molbev.a040023
Tao, Tzou, Nouhin, Bonilla, Jagannathan et al., SARS-CoV-2 Antiviral Therapy, Clin. Microbiol. Rev, doi:10.1128/CMR.00109-21
Trypsteen, Van Cleemput, Snippenberg, Gerlo Svandekerckhove, On the whereabouts of SARS-CoV-2 in the human body: A systematic review, PLoS Pathog, doi:10.1371/journal.ppat.1009037
Van Cleemput, Van Snippenberg, Lambrechts, Dendooven, D'onofrio et al., Organ-specific genome diversity of replication-competent SARS-CoV-2, Nat. Commun, doi:10.1038/s41467-021-26884-7
Vicenti, Zazzi, Saladini, SARS-CoV-2 RNA-dependent RNA polymerase as a therapeutic target for COVID-19, Expert Opin. Ther. Pat, doi:10.1080/13543776.2021.1880568
Wise, COVID-19: Remdesivir is recommended for authorisation by European Medicines Agency, BMJ, doi:10.1136/bmj.m2610
DOI record:
{
"DOI": "10.3390/microorganisms12010002",
"ISSN": [
"2076-2607"
],
"URL": "http://dx.doi.org/10.3390/microorganisms12010002",
"abstract": "<jats:p>Naturally occurring SARS-CoV-2 variants mutated in genomic regions targeted by antiviral drugs have not been extensively studied. This study investigated the potential of the RNA-dependent RNA polymerase (RdRp) complex subunits and non-structural protein (Nsp)5 of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) to accumulate natural mutations that could affect the efficacy of antiviral drugs. To this aim, SARS-CoV-2 genomic sequences isolated from 4155 drug-naive individuals from southern Italy were analyzed using the Illumina MiSeq platform. Sequencing of the 4155 samples showed the following viral variant distribution: 71.2% Delta, 22.2% Omicron, and 6.4% Alpha. In the Nsp12 sequences, we found 84 amino acid substitutions. The most common one was P323L, detected in 3777/4155 (91%) samples, with 2906/3777 (69.9%) also showing the G671S substitution in combination. Additionally, we identified 28, 14, and 24 different amino acid substitutions in the Nsp5, Nsp7, and Nsp8 genomic regions, respectively. Of note, the V186F and A191V substitutions, affecting residues adjacent to the active site of Nsp5 (the target of the antiviral drug Paxlovid), were found in 157/4155 (3.8%) and 3/4155 (0.07%) samples, respectively. In conclusion, the RdRp complex subunits and the Nsp5 genomic region exhibit susceptibility to accumulating natural mutations. This susceptibility poses a potential risk to the efficacy of antiviral drugs, as these mutations may compromise the drug ability to inhibit viral replication</jats:p>",
"alternative-id": [
"microorganisms12010002"
],
"author": [
{
"affiliation": [
{
"name": "Department of Clinical and Experimental Medicine, University Hospital of Messina, 98124 Messina, Italy"
}
],
"family": "Lombardo",
"given": "Daniele",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Department of Human Pathology, University Hospital of Messina, 98124 Messina, Italy"
}
],
"family": "Musolino",
"given": "Cristina",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Clinical and Experimental Medicine, University Hospital of Messina, 98124 Messina, Italy"
}
],
"family": "Chines",
"given": "Valeria",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Clinical and Experimental Medicine, University Hospital of Messina, 98124 Messina, Italy"
}
],
"family": "Caminiti",
"given": "Giuseppe",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0009-0006-5434-6738",
"affiliation": [
{
"name": "Department of Clinical and Experimental Medicine, University Hospital of Messina, 98124 Messina, Italy"
}
],
"authenticated-orcid": false,
"family": "Palermo",
"given": "Claudia",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-7721-6799",
"affiliation": [
{
"name": "Department of Clinical and Experimental Medicine, University Hospital of Messina, 98124 Messina, Italy"
}
],
"authenticated-orcid": false,
"family": "Cacciola",
"given": "Irene",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Clinical and Experimental Medicine, University Hospital of Messina, 98124 Messina, Italy"
}
],
"family": "Raffa",
"given": "Giuseppina",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Clinical and Experimental Medicine, University Hospital of Messina, 98124 Messina, Italy"
}
],
"family": "Pollicino",
"given": "Teresa",
"sequence": "additional"
}
],
"container-title": "Microorganisms",
"container-title-short": "Microorganisms",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2023,
12,
19
]
],
"date-time": "2023-12-19T09:18:37Z",
"timestamp": 1702977517000
},
"deposited": {
"date-parts": [
[
2023,
12,
19
]
],
"date-time": "2023-12-19T10:45:34Z",
"timestamp": 1702982734000
},
"indexed": {
"date-parts": [
[
2025,
5,
9
]
],
"date-time": "2025-05-09T05:54:11Z",
"timestamp": 1746770051090,
"version": "3.37.3"
},
"is-referenced-by-count": 4,
"issue": "1",
"issued": {
"date-parts": [
[
2023,
12,
19
]
]
},
"journal-issue": {
"issue": "1",
"published-online": {
"date-parts": [
[
2024,
1
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2023,
12,
19
]
],
"date-time": "2023-12-19T00:00:00Z",
"timestamp": 1702944000000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/2076-2607/12/1/2/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "2",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2023,
12,
19
]
]
},
"published-online": {
"date-parts": [
[
2023,
12,
19
]
]
},
"publisher": "MDPI AG",
"reference": [
{
"DOI": "10.1038/s41580-021-00432-z",
"article-title": "Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design",
"author": "Malone",
"doi-asserted-by": "crossref",
"first-page": "21",
"journal-title": "Nat. Rev. Mol. Cell Biol.",
"key": "ref_1",
"volume": "23",
"year": "2022"
},
{
"DOI": "10.1038/s41467-021-26884-7",
"article-title": "Organ-specific genome diversity of replication-competent SARS-CoV-2",
"author": "Lambrechts",
"doi-asserted-by": "crossref",
"first-page": "6612",
"journal-title": "Nat. Commun.",
"key": "ref_2",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1371/journal.ppat.1009037",
"doi-asserted-by": "crossref",
"key": "ref_3",
"unstructured": "Trypsteen, W., Van Cleemput, J., Snippenberg, W., and Gerlo SVandekerckhove, L. (2020). On the whereabouts of SARS-CoV-2 in the human body: A systematic review. PLoS Pathog., 16."
},
{
"DOI": "10.1146/annurev-biochem-052521-115653",
"article-title": "The Life of SARS-CoV-2 Inside Cells: Replication-Transcription Complex Assembly and Function",
"author": "Lou",
"doi-asserted-by": "crossref",
"first-page": "381",
"journal-title": "Annu. Rev. Biochem.",
"key": "ref_4",
"volume": "91",
"year": "2022"
},
{
"DOI": "10.1128/CMR.00109-21",
"doi-asserted-by": "crossref",
"key": "ref_5",
"unstructured": "Tao, K., Tzou, P.L., Nouhin, J., Bonilla, H., Jagannathan, P., and Shafer, R.W. (2021). SARS-CoV-2 Antiviral Therapy. Clin. Microbiol. Rev., 34."
},
{
"DOI": "10.1038/s41594-021-00657-8",
"article-title": "Molnupiravir: Coding for catastrophe",
"author": "Malone",
"doi-asserted-by": "crossref",
"first-page": "706",
"journal-title": "Nat. Struct. Mol. Biol.",
"key": "ref_6",
"volume": "28",
"year": "2021"
},
{
"DOI": "10.1056/NEJMp2032369",
"article-title": "FDA Approval of Remdesivir—A Step in the Right Direction",
"author": "Rubin",
"doi-asserted-by": "crossref",
"first-page": "2598",
"journal-title": "N. Engl. J. Med.",
"key": "ref_7",
"volume": "383",
"year": "2020"
},
{
"DOI": "10.1136/bmj.m2610",
"article-title": "COVID-19: Remdesivir is recommended for authorisation by European Medicines Agency",
"author": "Wise",
"doi-asserted-by": "crossref",
"first-page": "m2610",
"journal-title": "BMJ",
"key": "ref_8",
"volume": "369",
"year": "2020"
},
{
"DOI": "10.1080/13543776.2021.1880568",
"article-title": "SARS-CoV-2 RNA-dependent RNA polymerase as a therapeutic target for COVID-19",
"author": "Vicenti",
"doi-asserted-by": "crossref",
"first-page": "325",
"journal-title": "Expert Opin. Ther. Pat.",
"key": "ref_9",
"volume": "31",
"year": "2021"
},
{
"DOI": "10.1038/s41586-020-2368-8",
"article-title": "Structure of replicating SARS-CoV-2 polymerase",
"author": "Hillen",
"doi-asserted-by": "crossref",
"first-page": "154",
"journal-title": "Nature",
"key": "ref_10",
"volume": "584",
"year": "2020"
},
{
"DOI": "10.1126/science.abb7498",
"article-title": "Structure of the RNA-dependent RNA polymerase from COVID-19 virus",
"author": "Gao",
"doi-asserted-by": "crossref",
"first-page": "779",
"journal-title": "Science",
"key": "ref_11",
"volume": "368",
"year": "2020"
},
{
"DOI": "10.1016/j.molcel.2021.01.035",
"article-title": "Remdesivir is a delayed translocation inhibitor of SARS-CoV-2 replication",
"author": "Bravo",
"doi-asserted-by": "crossref",
"first-page": "1548",
"journal-title": "Mol. Cell",
"key": "ref_12",
"volume": "81",
"year": "2021"
},
{
"DOI": "10.1038/s41467-020-20542-0",
"article-title": "Mechanism of SARS-CoV-2 polymerase stalling by remdesivir",
"author": "Kokic",
"doi-asserted-by": "crossref",
"first-page": "279",
"journal-title": "Nat. Commun.",
"key": "ref_13",
"volume": "12",
"year": "2021"
},
{
"key": "ref_14",
"unstructured": "Medicines and Healthcare Products Regulatory Agency (2021, November 04). Regulatory Approval of Lagevrio (Molnupiravir), Available online: https://www.gov.uk/government/publications/regulatory-approval-of-lagevrio-molnupiravir."
},
{
"key": "ref_15",
"unstructured": "Food and Drug Administration (2021, December 23). Coronavirus (COVID-19) Update: FDA Authorizes Additional Oral Antiviral for Treatment of COVID-19 in Certain Adults, Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-additional-oral-antiviral-treatment-covid-19-certain."
},
{
"DOI": "10.1038/s41594-021-00651-0",
"article-title": "Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis",
"author": "Kabinger",
"doi-asserted-by": "crossref",
"first-page": "740",
"journal-title": "Nat. Struct. Mol. Biol.",
"key": "ref_16",
"volume": "28",
"year": "2021"
},
{
"DOI": "10.1136/bmj.n2422",
"article-title": "COVID-19: Molnupiravir reduces risk of hospital admission or death by 50% in patients at risk, MSD reports",
"author": "Mahase",
"doi-asserted-by": "crossref",
"first-page": "n2422",
"journal-title": "BMJ",
"key": "ref_17",
"volume": "375",
"year": "2021"
},
{
"key": "ref_18",
"unstructured": "European Medicines Agency (2023, January 21). Withdrawal of Marketing Authorizarion Application for LAGEVRIO, Molnupiravir, 200 mg Hard Capsules, EMEA/H/C/005789. Available online: https://www.ema.europa.eu/en/documents/withdrawal-letter/withdrawal-letter-lagevrio_en.pdf."
},
{
"key": "ref_19",
"unstructured": "Food and Drug Administration (2021, December 22). Coronavirus (COVID-19) Update: FDA Authorizes First Oral Antiviral for Treatment of COVID-19, Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-first-oral-antiviral-treatment-covid-19."
},
{
"key": "ref_20",
"unstructured": "European Medicines Agency (2022, January 27). COVID-19: EMA Recommends Conditional Marketing Authorisation for Paxlovid. Available online: https://www.ema.europa.eu/en/news/covid-19-ema-recommends-conditional-marketing-authorisation-paxlovid#:~:text=COVID%2D19%3A%20EMA%20recommends%20conditional%20marketing%20authorisation%20for%20Paxlovid,-Share&text=Update%3A%20Paxlovid%20is%20now%20authorised,Commission%20on%2028%20January%202022."
},
{
"DOI": "10.1038/s41467-020-16954-7",
"article-title": "Structural plasticity of SARS-CoV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography",
"author": "Kneller",
"doi-asserted-by": "crossref",
"first-page": "3202",
"journal-title": "Nat. Commun.",
"key": "ref_21",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1038/s41467-020-19662-4",
"article-title": "Crystallographic structure of wild-type SARS-CoV-2 main protease acyl-enzyme intermediate with physiological C-terminal autoprocessing site",
"author": "Lee",
"doi-asserted-by": "crossref",
"first-page": "5877",
"journal-title": "Nat. Commun.",
"key": "ref_22",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1136/bmj.n2713",
"article-title": "COVID-19: Pfizer’s paxlovid is 89% effective in patients at risk of serious illness, company reports",
"author": "Mahase",
"doi-asserted-by": "crossref",
"first-page": "n2713",
"journal-title": "BMJ",
"key": "ref_23",
"volume": "375",
"year": "2021"
},
{
"DOI": "10.1038/s41586-020-2223-y",
"article-title": "Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors",
"author": "Jin",
"doi-asserted-by": "crossref",
"first-page": "289",
"journal-title": "Nature",
"key": "ref_24",
"volume": "582",
"year": "2020"
},
{
"key": "ref_25",
"unstructured": "Vere Hodge, A., and Field, H.J. (2011). Genetics and Evolution of Infectious Disease, Elsevier."
},
{
"DOI": "10.2471/BLT.20.253591",
"article-title": "Variant analysis of SARS-CoV-2 genomes",
"author": "Koyama",
"doi-asserted-by": "crossref",
"first-page": "495",
"journal-title": "Bull. World Health Organ.",
"key": "ref_26",
"volume": "98",
"year": "2020"
},
{
"DOI": "10.1016/j.antiviral.2018.07.020",
"article-title": "Comparison of antiviral resistance across acute and chronic viral infections",
"author": "Mason",
"doi-asserted-by": "crossref",
"first-page": "103",
"journal-title": "Antivir. Res.",
"key": "ref_27",
"volume": "158",
"year": "2018"
},
{
"DOI": "10.3390/molecules27175413",
"doi-asserted-by": "crossref",
"key": "ref_28",
"unstructured": "Patel, D., Ono, S.K., Bassit, L., Verma, K., Amblard, F., and Schinazi, R.F. (2022). Assessment of a Computational Approach to Predict Drug Resistance Mutations for HIV, HBV and SARS-CoV-2. Molecules, 27."
},
{
"DOI": "10.1186/s12967-020-02344-6",
"article-title": "Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant",
"author": "Pachetti",
"doi-asserted-by": "crossref",
"first-page": "179",
"journal-title": "J. Transl. Med.",
"key": "ref_29",
"volume": "18",
"year": "2020"
},
{
"DOI": "10.7717/peerj.9703",
"article-title": "Mutation density changes in SARS-CoV-2 are related to the pandemic stage but to a lesser extent in the dominant strain with mutations in spike and RdRp",
"author": "Eskier",
"doi-asserted-by": "crossref",
"first-page": "e9703",
"journal-title": "PeerJ",
"key": "ref_30",
"volume": "8",
"year": "2020"
},
{
"DOI": "10.1093/cid/ciaa1474",
"article-title": "Emerging RNA-Dependent RNA Polymerase Mutation in a Remdesivir-Treated B-cell Immunodeficient Patient with Protracted Coronavirus Disease 2019",
"author": "Martinot",
"doi-asserted-by": "crossref",
"first-page": "e1762",
"journal-title": "Clin. Infect. Dis.",
"key": "ref_31",
"volume": "73",
"year": "2021"
},
{
"DOI": "10.1371/journal.ppat.1009929",
"doi-asserted-by": "crossref",
"key": "ref_32",
"unstructured": "Szemiel, A.M., Merits, A., Orton, R.J., MacLean, O.A., Pinto, R.M., Wickenhagen, A., Lieber, G., Turnbull, M.L., Wang, S., and Furnon, W. (2021). In vitro selection of Remdesivir resistance suggests evolutionary predictability of SARS-CoV-2. PLoS Pathog., 17."
},
{
"DOI": "10.1016/j.ebiom.2023.104559",
"doi-asserted-by": "crossref",
"key": "ref_33",
"unstructured": "Ip, J.D., Chu, A.W.-H., Chan, W.-M., Leung, R.C.-Y., Abdullah, S.M.U., Sun, Y., and To, K.K.-W. (2023). Global prevalence of SARS-CoV-2 3CL protease mutations associated with nirmatrelvir or ensitrelvir resistance. EBioMedicine, 91."
},
{
"DOI": "10.1038/s41431-020-0596-x",
"article-title": "Disruptive and avoidable: GDPR challenges to secondary research uses of data",
"author": "Peloquin",
"doi-asserted-by": "crossref",
"first-page": "697",
"journal-title": "Eur. J. Hum. Genet.",
"key": "ref_34",
"volume": "28",
"year": "2020"
},
{
"key": "ref_35",
"unstructured": "Italian Data Protection Authority (2016, December 15). Authorization n. 9/2016—General Authorization to Process Personal Data for Scientific Research Purposes. Available online: https://www.garanteprivacy.it/home/docweb/-/docweb-display/docweb/5805552."
},
{
"DOI": "10.1101/2021.01.16.425365",
"doi-asserted-by": "crossref",
"key": "ref_36",
"unstructured": "De Sabato, L., Vaccari, G., Knijn, A., Ianiro, G., Di Bartolo, I., and Morabito, S. (2021). SARS-CoV-2 RECoVERY: A multi-platform open-source bioinformatic pipeline for the automatic construction and analysis of SARS-CoV-2 genomes from NGS sequencing data. bioRxiv."
},
{
"DOI": "10.1093/molbev/msy096",
"article-title": "MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms",
"author": "Kumar",
"doi-asserted-by": "crossref",
"first-page": "1547",
"journal-title": "Mol. Biol. Evol.",
"key": "ref_37",
"volume": "35",
"year": "2018"
},
{
"article-title": "Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees",
"author": "Tamura",
"first-page": "512",
"journal-title": "Mol. Biol. Evol.",
"key": "ref_38",
"volume": "10",
"year": "1993"
},
{
"DOI": "10.2307/2408678",
"article-title": "Confidence limits on phylogenies: An approach using the bootstrap",
"author": "Felsenstein",
"doi-asserted-by": "crossref",
"first-page": "783",
"journal-title": "Evolution",
"key": "ref_39",
"volume": "39",
"year": "1985"
},
{
"DOI": "10.1093/nar/gkab301",
"article-title": "Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation",
"author": "Letunic",
"doi-asserted-by": "crossref",
"first-page": "W293",
"journal-title": "Nucleic Acids Res.",
"key": "ref_40",
"volume": "49",
"year": "2021"
},
{
"DOI": "10.1002/cbic.202100346",
"article-title": "Hidden in Plain Sight: Natural Products of Commensal Microbiota as an Environmental Selection Pressure for the Rise of New Variants of SARS-CoV-2",
"author": "Dragelj",
"doi-asserted-by": "crossref",
"first-page": "2946",
"journal-title": "Chembiochem",
"key": "ref_41",
"volume": "22",
"year": "2021"
},
{
"DOI": "10.3389/fcimb.2022.736397",
"doi-asserted-by": "crossref",
"key": "ref_42",
"unstructured": "De, R., and Dutta, S. (2022). Role of the Microbiome in the Pathogenesis of COVID-19. Front. Cell. Infect. Microbiol., 12."
},
{
"DOI": "10.1101/2023.11.16.567485",
"doi-asserted-by": "crossref",
"key": "ref_43",
"unstructured": "Cao, B., Wang, X., Yin, W., Gao, Z., and Xia, B. (2023). Human microbiota is a reservoir of SARS-CoV-2 advantageous mutations. bioRxiv."
},
{
"DOI": "10.7717/peerj.9492",
"article-title": "Identification of novel mutations in RNA-dependent RNA polymerases of SARS-CoV-2 and their implications on its protein structure",
"author": "Chand",
"doi-asserted-by": "crossref",
"first-page": "e9492",
"journal-title": "PeerJ",
"key": "ref_44",
"volume": "8",
"year": "2020"
},
{
"DOI": "10.1002/jmv.26791",
"article-title": "Mutations in SARS-CoV-2 Nsp7 and Nsp8 proteins and their predicted impact on replication/transcription complex structure",
"author": "Reshamwala",
"doi-asserted-by": "crossref",
"first-page": "4616",
"journal-title": "J. Med. Virol.",
"key": "ref_45",
"volume": "93",
"year": "2021"
},
{
"DOI": "10.1038/s41598-021-91662-w",
"article-title": "Concurrent mutations in RNA-dependent RNA polymerase and spike protein emerged as the epidemiologically most successful SARS-CoV-2 variant",
"author": "Abdul",
"doi-asserted-by": "crossref",
"first-page": "13705",
"journal-title": "Sci. Rep.",
"key": "ref_46",
"volume": "11",
"year": "2021"
},
{
"DOI": "10.3390/biom11070919",
"doi-asserted-by": "crossref",
"key": "ref_47",
"unstructured": "Mohammad, A., Al-Mulla, F., Wei, D.Q., Abubaker, J., and Remdesivir, M.D. (2021). Simulations Suggest a More Favourable Binding to SARS-CoV-2 RNA Dependent RNA Polymerase Mutant P323L Than Wild-Type. Biomolecules, 11."
},
{
"DOI": "10.1016/j.celrep.2023.113077",
"article-title": "SARS-CoV-2 variants with NSP12 P323L/G671S mutations display enhanced virus replication in ferret upper airways and higher transmissibility",
"author": "Kim",
"doi-asserted-by": "crossref",
"first-page": "113077",
"journal-title": "Cell Rep.",
"key": "ref_48",
"volume": "42",
"year": "2023"
},
{
"DOI": "10.1016/j.celrep.2020.107774",
"article-title": "Structural and Biochemical Characterization of the Nsp12-Nsp7-Nsp8 Core Polymerase Complex from SARS-CoV-2",
"author": "Peng",
"doi-asserted-by": "crossref",
"first-page": "107774",
"journal-title": "Cell Rep.",
"key": "ref_49",
"volume": "31",
"year": "2020"
}
],
"reference-count": 49,
"references-count": 49,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.mdpi.com/2076-2607/12/1/2"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Assessing Genomic Mutations in SARS-CoV-2: Potential Resistance to Antiviral Drugs in Viral Populations from Untreated COVID-19 Patients",
"type": "journal-article",
"volume": "12"
}