SARS-CoV-2 Mpro inhibitor ensitrelvir: asymmetrical cross-resistance with nirmatrelvir and emerging resistance hotspots
et al., Emerging Microbes & Infections, doi:10.1080/22221751.2025.2552716, Oct 2025
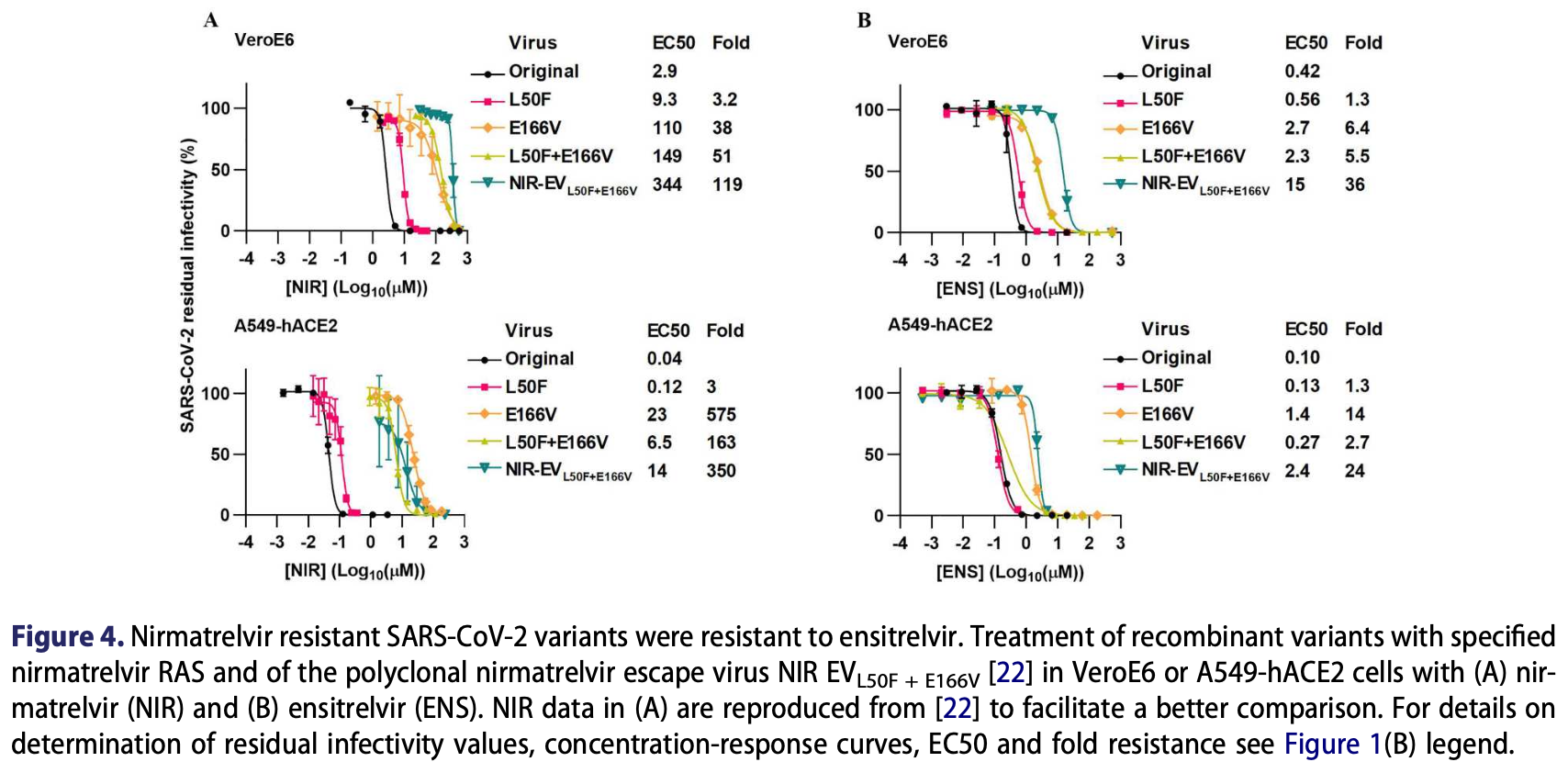
In vitro study showing that SARS-CoV-2 develops high-fitness resistance to ensitrelvir through Mpro mutations. Authors found asymmetrical cross-resistance, with ensitrelvir-resistant variants showing minimal cross-resistance to nirmatrelvir, while nirmatrelvir-resistant variants showed substantial cross-resistance to ensitrelvir.
Study covers ensitrelvir and paxlovid.
Zhou et al., 14 Oct 2025, peer-reviewed, 13 authors.
Contact: jgottwein@sund.ku.dk.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
SARS-CoV-2 Mpro inhibitor ensitrelvir: asymmetrical cross-resistance with nirmatrelvir and emerging resistance hotspots
Emerging Microbes & Infections, doi:10.1080/22221751.2025.2552716
SARS-CoV-2 main protease (Mpro) inhibitors are the first-line COVID-19 treatment. Nirmatrelvir is used worldwide, while ensitrelvir, licensed in Japan and Singapore, has received FDA fast-track designation. To facilitate population monitoring for viral resistance and guide next-generation inhibitor design, we investigated SARS-CoV-2 resistance and crossresistance to ensitrelvir and nirmatrelvir. SARS-CoV-2 escape variants with high fitness and high ensitrelvir resistance were selected under clinically relevant concentrations in infectious cell culture assays. Using infectious cell culture, replicon, and Mpro assays, reverse genetics revealed synergistic combinations of resistance-associated substitutions (RAS), specifically M49L + S144A and M49L + S144A + T169I, that conferred high resistance with a low fitness cost. Molecular dynamics simulations confirmed that M49L + S144A or M49L + S144A + T169I weakened ensitrelvir-Mpro binding. M49L + S144A and M49L + S144A + T169I exhibited a lower fitness cost and conferred higher resistance than the previously identified ensitrelvir RAS M49L + E166A. Cross-resistance between these ensitrelvir RAS and previously described nirmatrelvir RAS L50F + E166V was asymmetrical, with nirmatrelvir RAS showing greater resistance to ensitrelvir than vice versa. Amino acid changes at Mpro-position 166, an emerging resistance hotspot with natural variation, had differential impacts on viral fitness and Mpro inhibitor resistance in infectious cell culture assays. The most frequently naturally occurring substitution, E166Q, did not confer significant resistance to either ensitrelvir or nirmatrelvir. However, the second most frequent substitution, E166H, conferred high resistance to nirmatrelvir, but not to ensitrelvir. This comparative resistance analysis can inform COVID-19 treatment strategies and contribute to pandemic preparedness.
Author contributions YZ, JMG contributed to the conception and design of the study. YZ, KAG, HDT, LAR, AO, AC, ZD, LVP, UF, GHJP, SR, JB, JMG generated assays and experimental data. Data was verified by GHJP, SR, JB, JMG. YZ, KAG, HDT, LAR, JMG drafted the original manuscript. All authors read and approved the final version of the manuscript.
References
Abdelnabi, Jochmans, Donckers, Nirmatrelvir-resistant SARS-CoV-2 is efficiently transmitted in female Syrian hamsters and retains partial susceptibility to treatment, Nat Commun, doi:10.1038/s41467-023-37773-6
Abraham, Murtola, Schulz, GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers, SoftwareX, doi:10.1016/J.SOFTX.2015.06.001
Alam, Rana, Hosen, Modified thymidine derivatives as potential inhibitors of SARS-CoV: PASS, In vitro antimicrobial physicochemical and molecular docking studies, Phys Chem Res, doi:10.22036/PCR.2022.317494.1996
Amin, Dey, Methyl β-D-galactopyranoside esters as potential inhibitors for SARS-CoV-2 protease enzyme: synthesis, antimicrobial, PASS, molecular docking, molecular dynamics simulations and quantum computations, Glycoconj J, doi:10.1007/S10719-021-10039-3
Bager, Svalgaard, Lomholt, The hospital and mortality burden of COVID-19 compared with influenza in Denmark: a national observational cohort study, 2022-24, Lancet Infect Dis, doi:10.1016/S1473-3099(24)00806-5
Barkan, Garland, Zhang, Identification of potent, broad-spectrum coronavirus main protease inhibitors for pandemic preparedness, J Med Chem, doi:10.1021/ACS.JMEDCHEM.4C01404
Beigel, Tomashek, Dodd, Remdesivir for the treatment of COVID-19 -final report, N Engl J Med, doi:10.1056/NEJMOA2007764
Bernal, Da Silva, Musungaie, Molnupiravir for oral treatment of COVID-19 in nonhospitalized patients, N Engl J Med, doi:10.1056/nejmoa2116044
Best, Zhu, Shim, Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and sidechain χ(1) and χ(2) dihedral angles, J Chem Theory Comput, doi:10.1021/CT300400X
Bouzidi, Driouich, Klitting, Generation and evaluation of protease inhibitor-resistant SARS-CoV-2 strains, Antiviral Res, doi:10.1016/j.antiviral.2024.105814
Cao, Wang, Jian, Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies, Nature, doi:10.1038/s41586-021-04385-3
Cao, Wang, Lu, Oral simnotrelvir for adult patients with mild-to-moderate COVID-19, N Engl J Med, doi:10.1056/nejmoa2301425
Cortese, Voglino, Hackenbrock, The ionic strength of the intermembrane space of intact mitochondria is not affected by the pH or volume of the intermembrane space, Biochim Biophys Acta, doi:10.1016/0005-2728(92)90081-C
Detomasi, Degotte, Huang, Structurebased discovery of highly bioavailable, covalent, broad-spectrum coronavirus MPro inhibitors with potent in vivo efficacy, Sci Adv, doi:10.1126/sciadv.adt7836
Duan, Zhou, Liu, Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir, Nature, doi:10.1038/S41586-023-06609-0
Eltayb, Abdalla, Rabie, Novel investigational anti-SARS-CoV-2 agent ensitrelvir 'S-217622': A very promising potential universal broad-spectrum antiviral at the therapeutic frontline of coronavirus species, ACS Omega, doi:10.1021/ACSOMEGA.2C03881
Esler, Shi, Rollie, Structural basis for varying drug resistance of SARS-CoV-2 mpro E166 variants, mBio, doi:10.1128/MBIO.02624-24
Fahnøe, Pham, Fernandez-Antunez, Versatile SARS-CoV-2 reverse-genetics systems for the study of antiviral resistance and replication, Viruses, doi:10.3390/v14020172
Gammeltoft, Zhou, Hernandez, Hepatitis C virus protease inhibitors show differential efficacy and interactions with remdesivir for treatment of SARS-CoV-2 in vitro, Antimicrob Agents Chemother, doi:10.1128/AAC.02680-20
Gammeltoft, Zhou, Ryberg, Substitutions in SARS-CoV-2 mpro selected by protease inhibitor boceprevir confer resistance to nirmatrelvir, Viruses, doi:10.3390/v15091970
Godwin, Polsonetti, Caron, Remdesivir for the treatment of COVID-19: A narrative review, Infect Dis Ther, doi:10.1007/S40121-023-00900-3/TABLES/4
Hammond, Leister-Tebbe, Gardner, Oral nirmatrelvir for high-risk, nonhospitalized adults with COVID-19, N Engl J Med, doi:10.1056/NEJMOA2118542
Hirotsu, Kobayashi, Kakizaki, Multidrugresistant mutations to antiviral and antibody therapy in an immunocompromised patient infected with SARS-CoV-2, Med, doi:10.1016/J.MEDJ.2023.08.001
Hosen, Munia, Al-Ghorbani, Synthesis, antimicrobial, molecular docking and molecular dynamics studies of lauroyl thymidine analogs against SARS-CoV-2: POM study and identification of the pharmacophore sites, Bioorg Chem, doi:10.1016/J.BIOORG.2022.105850
Hu, Lewandowski, Tan, Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir, ACS Cent Sci, doi:10.1021/ACSCENTSCI.3C00538
Huang, Rauscher, Nawrocki, CHARMM36m: an improved force field for folded and intrinsically disordered proteins, Nat Methods, doi:10.1038/NMETH.4067
Humphrey, Dalke, Schulten, VMD: visual molecular dynamics, J Mol Graph, doi:10.1016/0263-7855(96)00018-5
Iketani, Mohri, Culbertson, Multiple pathways for SARS-CoV-2 resistance to nirmatrelvir, Nature, doi:10.1038/s41586-022-05514-2
Ip, Chu, Chan, Global prevalence of SARS-CoV-2 3CL protease mutations associated with nirmatrelvir or ensitrelvir resistance, EBioMedicine, doi:10.1016/J.EBIOM.2023.104559
Jiang, Su, Shang, Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir, Nat Commun, doi:10.1038/s41467-023-42102-y
Jo, Kim, Iyer, CHARMM-GUI: A web-based graphical user interface for CHARMM, J Comput Chem, doi:10.1002/JCC.20945
Jochmans, Liu, Donckers, The substitutions L50F, E166A, and L167F in SARS-CoV-2 3CLpro Are selected by a protease inhibitor In vitro and confer resistance To nirmatrelvir, mBio, doi:10.1128/mbio.02815-22
Kawsar, Hossain, Saha, Nucleoside-Based drug target with general antimicrobial screening and specific computational studies against SARS-CoV-2 main protease, ChemistrySelect, doi:10.1002/SLCT.202304774
Kiso, Yamayoshi, Iida, In vitro and in vivo characterization of SARS-CoV-2 resistance to ensitrelvir, Nat Commun, doi:10.1038/s41467-023-40018-1
Lee, Cheng, Swails, CHARMM-GUI Input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field, J Chem Theory Comput, doi:10.1021/ACS.JCTC.5B00935
Lee, Worrall, Vuckovic, Crystallographic structure of wild-type SARS-CoV-2 main protease acyl-enzyme intermediate with physiological C-terminal autoprocessing site, Nat Commun, doi:10.1038/s41467-020-19662-4
Lewandowski, Zhang, Tan, Distal protein-protein interactions contribute to nirmatrelvir resistance, Nat Commun, doi:10.1038/s41467-025-56651-x
Lin, Zeng, Duan, Molecular mechanism of ensitrelvir inhibiting SARS-CoV-2 main protease and its variants, Communications Biology, doi:10.1038/s42003-023-05071-y
Lo, Kariv, Hao, Replication capacity and susceptibility of nirmatrelvir-resistant mutants to next-generation mpro inhibitors in a SARS-CoV-2 replicon system, Antiviral Res, doi:10.1016/j.antiviral.2024.106022
Macdonald, Frey, Namchuk, Recognition of divergent viral substrates by the SARS-CoV-2 main protease, ACS Infect Dis, doi:10.1021/ACSINFECDIS.1C00237
Messore, Malune, Patacchini, New thiazolidine-4-One derivatives as SARS-CoV-2 main protease inhibitors, Pharmaceuticals, doi:10.3390/PH17050650/S1
Mukae, Yotsuyanagi, Ohmagari, Efficacy and safety of ensitrelvir in patients With mild-to-moderate coronavirus disease 2019: The phase 2b part of a randomized, placebo-controlled, phase 2/3 study, Clin Infect Dis, doi:10.1093/CID/CIAC933
Noske, De Souza Silva, De Godoy, Mo, Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease, J Biol Chem, doi:10.1016/j.jbc.2023.103004
Offersgaard, Hernandez, Feng, An inactivated SARS-CoV-2 vaccine induced cross-neutralizing persisting antibodies and protected against challenge in small animals, iScience, doi:10.1016/J.ISCI.2023.105949
Offersgaard, Hernandez, Zhou, An inactivated SARS-CoV-2 vaccine based on a Vero cell culture-adapted high-titer virus confers crossprotection in small animals, Sci Rep, doi:10.1038/S41598-024-67570-0
Owen, Allerton, Anderson, An oral SARS-CoV-2 mpro inhibitor clinical candidate for the treatment of COVID-19, Science, doi:10.1126/science.abl4784
Pham, Underwood, Binderup, Neutralisation resistance of SARS-CoV-2 spike-variants is primarily mediated by synergistic receptor binding domain substitutions, Emerg Microbes Infect, doi:10.1080/22221751.2024.2412643
Ramirez, Fernandez-Antunez, Galli, Overcoming culture restriction for SARS-CoV-2 in human cells facilitates the screening of compounds inhibiting viral replication, Antimicrob Agents Chemother, doi:10.1128/AAC.00097-21
Sacco, Hu, Gongora, The P132H mutation in the main protease of omicron SARS-CoV-2 decreases thermal stability without compromising catalysis or small-molecule drug inhibition, Cell Res, doi:10.1038/S41422-022-00640-Y
Shimizu, Sonoyama, Fukuhara, Safety, tolerability, and pharmacokinetics of the novel antiviral agent ensitrelvir fumaric acid, a SARS-CoV-2 3CL protease inhibitor, in healthy adults, Antimicrob Agents Chemother, doi:10.1128/aac.00632-22
Shionogi, FDA fast track designation for ensitrelvir fumaric acid, an investigational oral antiviral for COVID-19
Sridhara, Gungor, Erol, Lack of effectiveness of bebtelovimab monoclonal antibody among high-risk patients with SARS-Cov-2 omicron during BA.2, BA.2.12.1 and BA.5 subvariants dominated era, PLoS One, doi:10.1371/journal.pone.0279326
Sun, Sun, Yang, A novel, covalent broadspectrum inhibitor targeting human coronavirus mpro, Nat Commun, doi:10.1038/s41467-025-59870-4
Tamura, Choudhary, Deo, Emerging SARS-CoV-2 resistance after antiviral treatment, JAMA Netw Open, doi:10.1001/JAMANETWORKOPEN.2024.35431
Uehara, Yotsuyanagi, Ohmagari, Ensitrelvir treatment-emergent amino acid substitutions in SARS-CoV-2 3CLpro detected in the SCORPIO-SR phase 3 trial, Antiviral Res, doi:10.1016/J.ANTIVIRAL.2025.106097
Van Der Spoel, Lindahl, Hess, GROMACS: fast, flexible, and free, J Comput Chem, doi:10.1002/JCC.20291
Yaghi, Wylie, Andrews, An investigation of nirmatrelvir (paxlovid) resistance in SARS-CoV-2 mpro, ACS Bio and Med Chem Au, doi:10.1021/acsbiomedchemau.4c00045
Zhou, Gammeltoft, Galli, Efficacy of ionchannel inhibitors amantadine, memantine and rimantadine for the treatment of SARS-CoV-2 in vitro, Viruses, doi:10.3390/v13102082
Zhou, Gammeltoft, Ryberg, Nirmatrelvir-resistant SARS-CoV-2 variants with high fitness in an infectious cell culture system, Sci Adv, doi:10.1126/sciadv.add7197
Zhou, Gilmore, Ramirez, In vitro efficacy of artemisinin-based treatments against SARS-CoV-2, Sci Rep, doi:10.1038/s41598-021-93361-y
Zhu, Yurgelonis, Noell, In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance, Sci Adv, doi:10.1126/sciadv.adl4013
Zuckerman, Bucris, Keidar-Friedman, Nirmatrelvir resistance -de novo E166V/L50V mutations in an immunocompromised patient treated with prolonged nirmatrelvir/ritonavir monotherapy leading to clinical and virological treatment failurea case report, Clin Infect Dis, doi:10.1093/cid/ciad494
DOI record:
{
"DOI": "10.1080/22221751.2025.2552716",
"ISSN": [
"2222-1751"
],
"URL": "http://dx.doi.org/10.1080/22221751.2025.2552716",
"alternative-id": [
"10.1080/22221751.2025.2552716"
],
"article-number": "2552716",
"assertion": [
{
"label": "Peer Review Statement",
"name": "peerreview_statement",
"order": 1,
"value": "The publishing and review policy for this title is described in its Aims & Scope."
},
{
"URL": "http://www.tandfonline.com/action/journalInformation?show=aimsScope&journalCode=temi20",
"label": "Aim & Scope",
"name": "aims_and_scope_url",
"order": 2,
"value": "http://www.tandfonline.com/action/journalInformation?show=aimsScope&journalCode=temi20"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Received",
"name": "received",
"order": 0,
"value": "2025-04-01"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Revised",
"name": "revised",
"order": 1,
"value": "2025-07-22"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "2025-08-22"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Published",
"name": "published",
"order": 3,
"value": "2025-10-14"
}
],
"author": [
{
"affiliation": [
{
"name": "Copenhagen University Hospital–Hvidovre",
"place": [
"Hvidovre, Denmark"
]
},
{
"name": "University of Copenhagen",
"place": [
"Copenhagen, Denmark"
]
}
],
"family": "Zhou",
"given": "Yuyong",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Copenhagen University Hospital–Hvidovre",
"place": [
"Hvidovre, Denmark"
]
},
{
"name": "University of Copenhagen",
"place": [
"Copenhagen, Denmark"
]
}
],
"family": "Gammeltoft",
"given": "Karen A.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Copenhagen University Hospital–Hvidovre",
"place": [
"Hvidovre, Denmark"
]
},
{
"name": "University of Copenhagen",
"place": [
"Copenhagen, Denmark"
]
}
],
"family": "Tjørnelund-Sjursen",
"given": "Helena D.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Copenhagen University Hospital–Hvidovre",
"place": [
"Hvidovre, Denmark"
]
},
{
"name": "University of Copenhagen",
"place": [
"Copenhagen, Denmark"
]
}
],
"family": "Ryberg",
"given": "Line A.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Copenhagen University Hospital–Hvidovre",
"place": [
"Hvidovre, Denmark"
]
},
{
"name": "University of Copenhagen",
"place": [
"Copenhagen, Denmark"
]
}
],
"family": "Offersgaard",
"given": "Anna",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Copenhagen University Hospital–Hvidovre",
"place": [
"Hvidovre, Denmark"
]
},
{
"name": "University of Copenhagen",
"place": [
"Copenhagen, Denmark"
]
}
],
"family": "Czarnota",
"given": "Anna",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Copenhagen University Hospital–Hvidovre",
"place": [
"Hvidovre, Denmark"
]
},
{
"name": "University of Copenhagen",
"place": [
"Copenhagen, Denmark"
]
}
],
"family": "Duan",
"given": "Zhe",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Copenhagen University Hospital–Hvidovre",
"place": [
"Hvidovre, Denmark"
]
},
{
"name": "University of Copenhagen",
"place": [
"Copenhagen, Denmark"
]
}
],
"family": "Pham",
"given": "Long V.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Copenhagen University Hospital–Hvidovre",
"place": [
"Hvidovre, Denmark"
]
},
{
"name": "University of Copenhagen",
"place": [
"Copenhagen, Denmark"
]
}
],
"family": "Fahnøe",
"given": "Ulrik",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Technical University of Denmark",
"place": [
"Kongens Lyngby, Denmark"
]
}
],
"family": "Peters",
"given": "Günther H.J.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Copenhagen University Hospital–Hvidovre",
"place": [
"Hvidovre, Denmark"
]
},
{
"name": "University of Copenhagen",
"place": [
"Copenhagen, Denmark"
]
}
],
"family": "Ramirez",
"given": "Santseharay",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Copenhagen University Hospital–Hvidovre",
"place": [
"Hvidovre, Denmark"
]
},
{
"name": "University of Copenhagen",
"place": [
"Copenhagen, Denmark"
]
}
],
"family": "Bukh",
"given": "Jens",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Copenhagen University Hospital–Hvidovre",
"place": [
"Hvidovre, Denmark"
]
},
{
"name": "University of Copenhagen",
"place": [
"Copenhagen, Denmark"
]
}
],
"family": "Gottwein",
"given": "Judith M.",
"sequence": "additional"
}
],
"container-title": "Emerging Microbes & Infections",
"container-title-short": "Emerging Microbes & Infections",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"www.tandfonline.com"
]
},
"created": {
"date-parts": [
[
2025,
10,
15
]
],
"date-time": "2025-10-15T04:22:55Z",
"timestamp": 1760502175000
},
"deposited": {
"date-parts": [
[
2025,
10,
15
]
],
"date-time": "2025-10-15T04:22:57Z",
"timestamp": 1760502177000
},
"funder": [
{
"DOI": "10.13039/501100004543",
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100004543",
"id-type": "DOI"
}
],
"name": "China Scholarship Council"
},
{
"DOI": "10.13039/501100011958",
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100011958",
"id-type": "DOI"
}
],
"name": "Independent Research Fund Denmark"
},
{
"DOI": "10.13039/100012774",
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/100012774",
"id-type": "DOI"
}
],
"name": "Innovation Fund Denmark"
},
{
"DOI": "10.13039/501100009708",
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100009708",
"id-type": "DOI"
}
],
"name": "Novo Nordisk Foundation"
},
{
"name": "Sygeforsikring “danmark”, Mauritzen La Fontaine Fonden"
},
{
"DOI": "10.13039/100009584",
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/100009584",
"id-type": "DOI"
}
],
"name": "Toyota Foundation"
},
{
"DOI": "10.13039/501100013826",
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100013826",
"id-type": "DOI"
}
],
"name": "Candys Foundation"
}
],
"indexed": {
"date-parts": [
[
2025,
10,
16
]
],
"date-time": "2025-10-16T00:13:43Z",
"timestamp": 1760573623393,
"version": "build-2065373602"
},
"is-referenced-by-count": 0,
"issue": "1",
"issued": {
"date-parts": [
[
2025,
10,
14
]
]
},
"journal-issue": {
"issue": "1",
"published-print": {
"date-parts": [
[
2025,
12,
31
]
]
}
},
"language": "en",
"license": [
{
"URL": "http://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
10,
14
]
],
"date-time": "2025-10-14T00:00:00Z",
"timestamp": 1760400000000
}
}
],
"link": [
{
"URL": "https://www.tandfonline.com/doi/pdf/10.1080/22221751.2025.2552716",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "301",
"original-title": [],
"prefix": "10.1080",
"published": {
"date-parts": [
[
2025,
10,
14
]
]
},
"published-online": {
"date-parts": [
[
2025,
10,
14
]
]
},
"published-print": {
"date-parts": [
[
2025,
12,
31
]
]
},
"publisher": "Informa UK Limited",
"reference": [
{
"DOI": "10.1016/S1473-3099(24)00806-5",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_2_1"
},
{
"DOI": "10.1056/NEJMOA2118542",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_3_1"
},
{
"DOI": "10.1126/science.abl4784",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_4_1"
},
{
"DOI": "10.1093/CID/CIAC933",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_5_1"
},
{
"key": "e_1_3_4_6_1",
"unstructured": "Shionogi receives U.S. FDA fast track designation for ensitrelvir fumaric acid an investigational oral antiviral for COVID-19. https://www.shionogi.com/global/en/news/2023/04/20230404.htm."
},
{
"DOI": "10.1021/ACSOMEGA.2C03881",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_7_1"
},
{
"DOI": "10.1038/s41467-023-42102-y",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_8_1"
},
{
"DOI": "10.1056/nejmoa2301425",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_9_1"
},
{
"DOI": "10.3390/PH17050650/S1",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_10_1"
},
{
"DOI": "10.1016/J.BIOORG.2022.105850",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_11_1"
},
{
"DOI": "10.1007/S10719-021-10039-3",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_12_1"
},
{
"DOI": "10.1002/SLCT.202304774",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_13_1"
},
{
"DOI": "10.22036/PCR.2022.317494.1996",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_14_1"
},
{
"DOI": "10.1038/s41467-025-59870-4",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_15_1"
},
{
"DOI": "10.1021/ACS.JMEDCHEM.4C01404",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_16_1"
},
{
"DOI": "10.1126/sciadv.adt7836",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_17_1"
},
{
"DOI": "10.1056/NEJMOA2007764",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_18_1"
},
{
"DOI": "10.1007/S40121-023-00900-3/TABLES/4",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_19_1"
},
{
"DOI": "10.1038/s41586-021-04385-3",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_20_1"
},
{
"DOI": "10.1371/journal.pone.0279326",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_21_1"
},
{
"DOI": "10.1056/nejmoa2116044",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_22_1"
},
{
"DOI": "10.1126/sciadv.add7197",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_23_1"
},
{
"DOI": "10.1038/s41586-022-05514-2",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_24_1"
},
{
"DOI": "10.1128/mbio.02815-22",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_25_1"
},
{
"DOI": "10.1126/sciadv.adl4013",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_26_1"
},
{
"DOI": "10.1093/cid/ciad494",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_27_1"
},
{
"DOI": "10.1038/S41422-022-00640-Y",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_28_1"
},
{
"DOI": "10.1016/J.EBIOM.2023.104559",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_29_1"
},
{
"DOI": "10.1038/s41467-025-56651-x",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_30_1"
},
{
"DOI": "10.1021/acsbiomedchemau.4c00045",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_31_1"
},
{
"DOI": "10.3390/v15091970",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_32_1"
},
{
"DOI": "10.3390/v14020172",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_33_1"
},
{
"DOI": "10.1080/22221751.2024.2412643",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_34_1"
},
{
"DOI": "10.3390/v13102082",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_35_1"
},
{
"DOI": "10.1128/AAC.02680-20",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_36_1"
},
{
"DOI": "10.1128/AAC.00097-21",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_37_1"
},
{
"DOI": "10.1038/S41598-024-67570-0",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_38_1"
},
{
"DOI": "10.1016/J.ISCI.2023.105949",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_39_1"
},
{
"DOI": "10.1038/s41598-021-93361-y",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_40_1"
},
{
"DOI": "10.1016/j.antiviral.2024.106022",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_41_1"
},
{
"DOI": "10.1021/ACSINFECDIS.1C00237",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_42_1"
},
{
"DOI": "10.1038/S41586-023-06609-0",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_43_1"
},
{
"DOI": "10.1038/s41467-020-19662-4",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_44_1"
},
{
"DOI": "10.1016/0005-2728(92)90081-C",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_45_1"
},
{
"DOI": "10.1038/NMETH.4067",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_46_1"
},
{
"DOI": "10.1021/CT300400X",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_47_1"
},
{
"DOI": "10.1002/JCC.20945",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_48_1"
},
{
"DOI": "10.1021/ACS.JCTC.5B00935",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_49_1"
},
{
"DOI": "10.1002/JCC.20291",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_50_1"
},
{
"DOI": "10.1016/J.SOFTX.2015.06.001",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_51_1"
},
{
"DOI": "10.1016/0263-7855(96)00018-5",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_52_1"
},
{
"DOI": "10.1038/s41467-023-40018-1",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_53_1"
},
{
"DOI": "10.1038/s42003-023-05071-y",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_54_1"
},
{
"DOI": "10.1016/j.jbc.2023.103004",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_55_1"
},
{
"DOI": "10.1016/j.antiviral.2024.105814",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_56_1"
},
{
"DOI": "10.1128/aac.00632-22",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_57_1"
},
{
"DOI": "10.1001/JAMANETWORKOPEN.2024.35431",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_58_1"
},
{
"DOI": "10.1016/J.ANTIVIRAL.2025.106097",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_59_1"
},
{
"DOI": "10.1021/ACSCENTSCI.3C00538",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_60_1"
},
{
"DOI": "10.1128/MBIO.02624-24",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_61_1"
},
{
"DOI": "10.1016/J.MEDJ.2023.08.001",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_62_1"
},
{
"DOI": "10.1038/s41467-023-37773-6",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_63_1"
}
],
"reference-count": 62,
"references-count": 62,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.tandfonline.com/doi/full/10.1080/22221751.2025.2552716"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "SARS-CoV-2 Mpro inhibitor ensitrelvir: asymmetrical cross-resistance with nirmatrelvir and emerging resistance hotspots",
"type": "journal-article",
"update-policy": "https://doi.org/10.1080/tandf_crossmark_01",
"volume": "14"
}
zhou26