Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir
et al., Nature Communications, doi:10.1038/s41467-023-42102-y, Oct 2023
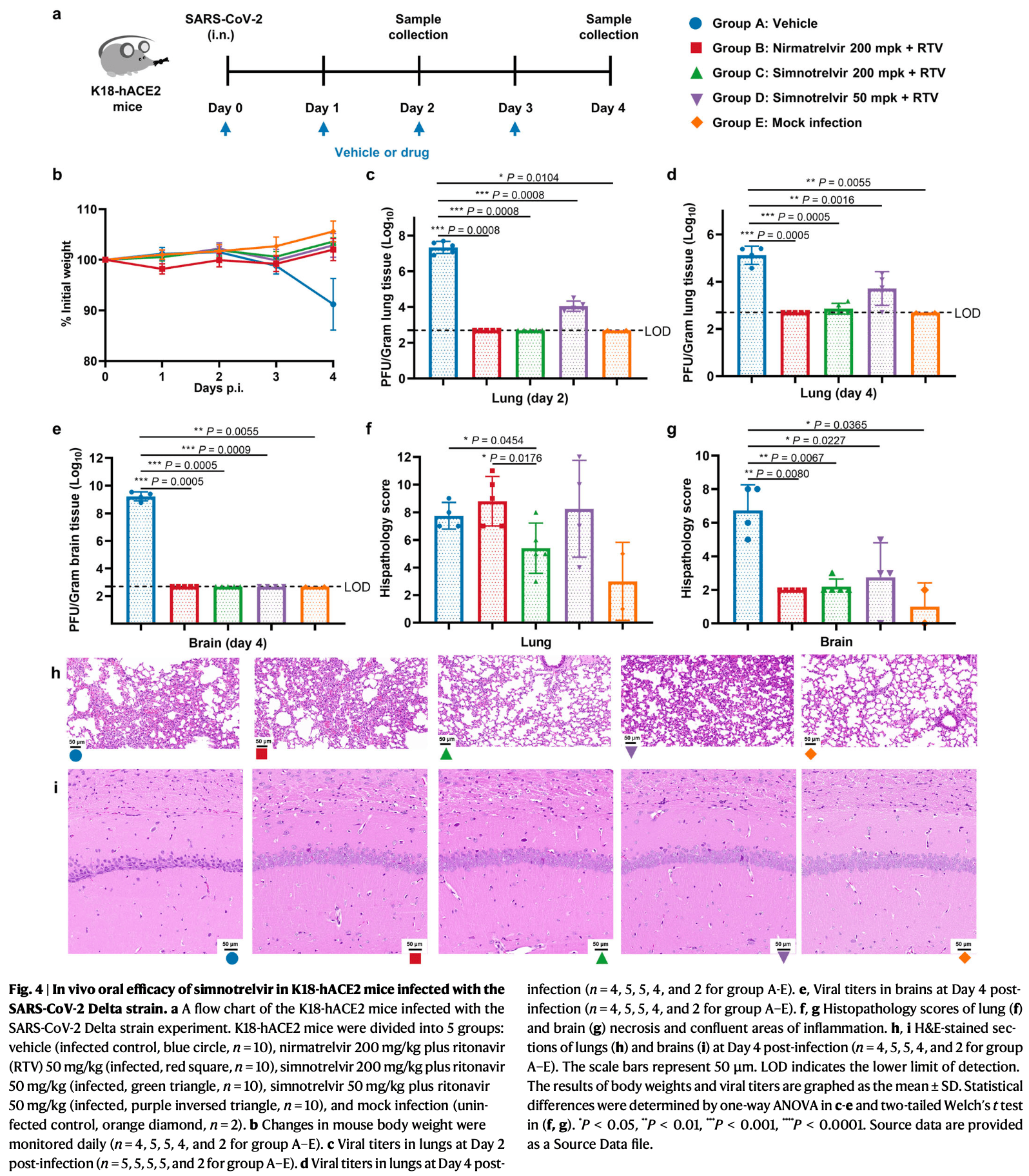
In vitro and mouse study showing the development and preclinical evaluation of the SARS-CoV-2 3C-like protease (3CLpro) inhibitor simnotrelvir (SSD8432, SIM0417, part of xiannuoxin) as an orally bioavailable COVID-19 therapeutic agent. Structure-based optimization of the HCV protease inhibitor boceprevir led to identification of simnotrelvir, which covalently inhibits 3CLpro from SARS-CoV-2 and other coronaviruses. Simnotrelvir demonstrated potent antiviral activity against SARS-CoV-2 variants in enzymatic and cell-based assays and showed favorable pharmacokinetics, safety profiles, and robust oral efficacy in a mouse model of SARS-CoV-2 delta infection, significantly reducing lung viral loads and eliminating virus from brains.
3 preclinical studies support the efficacy of xiannuoxin for COVID-19:
Jiang et al., 13 Oct 2023, China, peer-reviewed, 22 authors.
Contact: ycxu@simm.ac.cn, renhong.tang@simceregroup.com, zhangleike@wh.iov.cn, shenjingshan@simm.ac.cn.
Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir
Nature Communications, doi:10.1038/s41467-023-42102-y
The persistent pandemic of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its variants accentuates the great demand for developing effective therapeutic agents. Here, we report the development of an orally bioavailable SARS-CoV-2 3C-like protease (3CL pro ) inhibitor, namely simnotrelvir, and its preclinical evaluation, which lay the foundation for clinical trials studies as well as the conditional approval of simnotrelvir in combination with ritonavir for the treatment of COVID-19. The structure-based optimization of boceprevir, an approved HCV protease inhibitor, leads to identification of simnotrelvir that covalently inhibits SARS-CoV-2 3CL pro with an enthalpy-driven thermodynamic binding signature. Multiple enzymatic assays reveal that simnotrelvir is a potent pan-CoV 3CL pro inhibitor but has high selectivity. It effectively blocks replications of SARS-CoV-2 variants in cell-based assays and exhibits good pharmacokinetic and safety profiles in male and female rats and monkeys, leading to robust oral efficacy in a male mouse model of SARS-CoV-2 Delta infection in which it not only significantly reduces lung viral loads but also eliminates the virus from brains. The discovery of simnotrelvir thereby highlights the utility of structure-based development of marked protease inhibitors for providing a small molecule therapeutic effectively combatting human coronaviruses. The catastrophic coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its variants has resulted in an unbearable number of infections and deaths worldwide, and posed an unprecedented threat to global public health and economies. Thus, great efforts have been devoted to developing vaccines as well as antiviral agents for the treatment and prevention of COVID-19, but the clinically significant therapeutic options for COVID-19 are limited. According to therapeutic strategies against other viruses, such as HCV and HIV, orally effective small molecule drugs targeting viral proteins that are essential for virus replication have been particularly pursued to combat the ongoing pandemic and future SARS-like zoonotic coronavirus outbreaks.
Reporting summary Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability The atomic coordinates and structure factors have been deposited into the Protein Data Bank with accession codes 8IFP (SARS-CoV-2 3 CL pro in complex with compound 1), 8IFQ (SARS-CoV-2 3CL pro in complex with compound 2), 8IFR (SARS-CoV-2 3CL pro in complex with compound 3), 8IFS (SARS-CoV-2 3CL pro in complex with compound 7), 8IFT (SARS-CoV-2 3CL pro in complex with compound 10), 8IGX (SARS-CoV-2 3CL pro in complex with simnotrelvir), and 8IGY (SARS-CoV-2 3CL pro in complex with nirmatrelvir). The structure of SARS-CoV-2 3CL pro in complex with boceprevir (PDB code: 6XQU) was obtained from Protein Data Bank. The cDNA of 3CL pro s of SARS-CoV-2 (Gen-Bank: MN908947.3), SARS-CoV (Gen-Bank: AAP13442.1), MERS-CoV (Gen-Bank: MT387202.1), H229E-CoV (Gen-Bank: AF304460.1), HKU1-CoV (Gen-Bank: AY597011.2), NL63-CoV (Gen-Bank: AY567487.2), and OC43-CoV (Gen-Bank: AY903459.1) were obtained from Genbank [https://https.ncbi.nlm.nih.gov/genbank/]. Source data are provided with this paper. All the raw data generated in this study are provided in the Source Data file. Source data are provided with this paper.
Author contributions Y.X., X.J., and J.S. conceived and design the project. Y.X., X.J., J.S., H.J., L.Z., H.S., F.Z., and R.T. designed the experiments; Y.X., X.J., J.S., H.S., and M.X. performed the drug design; X.J., J.S.,..
References
Adams, PHENIX: building new software for automated crystallographic structure determination, Acta Crystallogr. D
Baum, Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies, Science
Boike, Henning, Nomura, Advances in covalent drug discovery, Nat. Rev. Drug Discov
Bowes, Reducing safety-related drug attrition: the use of in vitro pharmacological profiling, Nat. Rev. Drug Discov
Cannalire, Targeting SARS-CoV-2 proteases and polymerase for COVID-19 treatment: state of the art and future opportunities, J. Med. Chem
Collaborative, The CCP4 suite: programs for protein crystallography, Acta Crystallogr. D
Dai, Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease, Science
De Vries, A comparative analysis of SARS-CoV-2 antivirals characterizes 3CLpro inhibitor PF-00835231 as a potential new treatment for COVID-19, J. Virol
Emsley, Cowtan, Coot: model-building tools for molecular graphics, Acta Crystallogr. D
Freire, Do enthalpy and entropy distinguish first in class from best in class?, Drug Discov. Today
Fu, Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease, Nat. Commun
Hilgenfeld, From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design, FEBS J
Hoffman, Discovery of ketone-based covalent inhibitors of coronavirus 3CL proteases for the potential therapeutic treatment of COVID-19, J. Med. Chem
Jiang, Su, Shang, Zhou, Zhang et al., 12 Deceased: Hualiang Jiang. e-mail: renhong.tang@simceregroup
Jin, Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors, Nature
Kneller, Covalent narlaprevir-and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease, Nat. Commun
Kneller, Malleability of the SARS-CoV-2 3CL Mpro activesite cavity facilitates binding of clinical antivirals, Structure
Minor, HKL-3000: the integration of data reduction and structure solution-from diffraction images to an initial model in minutes, Acta Crystallogr. D
Njoroge, Challenges in modern drug discovery: a case study of boceprevir, an HCV protease inhibitor for the treatment of hepatitis C virus infection, Acc. Chem. Res
Owen, An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19, Science
Pillaiyar, An overview of severe acute respiratory syndromecoronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy, J. Med. Chem
Qiao, SARS-CoV-2 M(pro) inhibitors with antiviral activity in a transgenic mouse model, Science
Schon, Freire, Enthalpy screen of drug candidates, Anal. Biochem
Su, Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients, Acta Pharmacol. Sin
Su, Xu, Jiang, Drug discovery and development targeting the life cycle of SARS-CoV-2, Fund. Res
Tarcsay, Keseru, Is there a link between selectivity and binding thermodynamics profiles?, Drug Discov. Today
Tyndall, S-217622, a 3CL protease inhibitor and clinical candidate for SARS-CoV-2, J. Med. Chem
Van Den Ent, Lowe, RF cloning: a restriction-free method for inserting target genes into plasmids, J. Biochem. Biophys. Methods
Wang, Covalent inhibition mechanism of antidiabetic drugs-vildagliptin vs saxagliptin, Acs Catal
Xiong, What coronavirus 3C-like protease tells us: from structure, substrate selectivity, to inhibitor design, Med. Res. Rev
Yu, Aquarium: an automatic data-processing and experiment information management system for biological macromolecular crystallography beamlines, J. Appl. Crystallogr
Zhang, The protein complex crystallography beamline (BL19U1) at the shanghai synchrotron radiation facility, Nucl. Sci. Tech
DOI record:
{
"DOI": "10.1038/s41467-023-42102-y",
"ISSN": [
"2041-1723"
],
"URL": "http://dx.doi.org/10.1038/s41467-023-42102-y",
"abstract": "<jats:title>Abstract</jats:title><jats:p>The persistent pandemic of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its variants accentuates the great demand for developing effective therapeutic agents. Here, we report the development of an orally bioavailable SARS-CoV-2 3C-like protease (3CL<jats:sup>pro</jats:sup>) inhibitor, namely simnotrelvir, and its preclinical evaluation, which lay the foundation for clinical trials studies as well as the conditional approval of simnotrelvir in combination with ritonavir for the treatment of COVID-19. The structure-based optimization of boceprevir, an approved HCV protease inhibitor, leads to identification of simnotrelvir that covalently inhibits SARS-CoV-2 3CL<jats:sup>pro</jats:sup> with an enthalpy-driven thermodynamic binding signature. Multiple enzymatic assays reveal that simnotrelvir is a potent pan-CoV 3CL<jats:sup>pro</jats:sup> inhibitor but has high selectivity. It effectively blocks replications of SARS-CoV-2 variants in cell-based assays and exhibits good pharmacokinetic and safety profiles in male and female rats and monkeys, leading to robust oral efficacy in a male mouse model of SARS-CoV-2 Delta infection in which it not only significantly reduces lung viral loads but also eliminates the virus from brains. The discovery of simnotrelvir thereby highlights the utility of structure-based development of marked protease inhibitors for providing a small molecule therapeutic effectively combatting human coronaviruses.</jats:p>",
"alternative-id": [
"42102"
],
"article-number": "6463",
"assertion": [
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Received",
"name": "received",
"order": 1,
"value": "18 April 2023"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "25 September 2023"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "First Online",
"name": "first_online",
"order": 3,
"value": "13 October 2023"
},
{
"group": {
"label": "Competing interests",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 1,
"value": "X.J., Y.X., L.Z., H.S., Q.Z., W.Z., W.S., J.S., G.X., and H.J. are co-inventors of the patent (CN202111168232.4, China) that cover 3CL<sup>pro</sup> inhibitors included in this study. F. Z., L.J., X.H., P.C., S.P., and R.T. are employers of Simcere Co. Ltd. F. Z., L.J., X.H., P.C., S.P., and R.T. are shareholders in Simcere Co. Ltd. All other authors declare no competing interests."
}
],
"author": [
{
"affiliation": [],
"family": "Jiang",
"given": "Xiangrui",
"sequence": "first"
},
{
"affiliation": [],
"family": "Su",
"given": "Haixia",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Shang",
"given": "Weijuan",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0009-0007-3050-6239",
"affiliation": [],
"authenticated-orcid": false,
"family": "Zhou",
"given": "Feng",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Zhang",
"given": "Yan",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Zhao",
"given": "Wenfeng",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Zhang",
"given": "Qiumeng",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Xie",
"given": "Hang",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Jiang",
"given": "Lei",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Nie",
"given": "Tianqing",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Yang",
"given": "Feipu",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Xiong",
"given": "Muya",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Huang",
"given": "Xiaoxing",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Li",
"given": "Minjun",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Chen",
"given": "Ping",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Peng",
"given": "Shaoping",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-9401-235X",
"affiliation": [],
"authenticated-orcid": false,
"family": "Xiao",
"given": "Gengfu",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-0656-6315",
"affiliation": [],
"authenticated-orcid": false,
"family": "Jiang",
"given": "Hualiang",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Tang",
"given": "Renhong",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-2593-2571",
"affiliation": [],
"authenticated-orcid": false,
"family": "Zhang",
"given": "Leike",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-9679-9934",
"affiliation": [],
"authenticated-orcid": false,
"family": "Shen",
"given": "Jingshan",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-1581-6155",
"affiliation": [],
"authenticated-orcid": false,
"family": "Xu",
"given": "Yechun",
"sequence": "additional"
}
],
"container-title": "Nature Communications",
"container-title-short": "Nat Commun",
"content-domain": {
"crossmark-restriction": false,
"domain": [
"link.springer.com"
]
},
"created": {
"date-parts": [
[
2023,
10,
13
]
],
"date-time": "2023-10-13T18:03:06Z",
"timestamp": 1697220186000
},
"deposited": {
"date-parts": [
[
2023,
11,
17
]
],
"date-time": "2023-11-17T06:15:50Z",
"timestamp": 1700201750000
},
"funder": [
{
"DOI": "10.13039/100012543",
"award": [
"No. 22YF1457300"
],
"doi-asserted-by": "publisher",
"name": "Shanghai Science and Technology Development Foundation"
}
],
"indexed": {
"date-parts": [
[
2024,
2,
22
]
],
"date-time": "2024-02-22T10:04:41Z",
"timestamp": 1708596281697
},
"is-referenced-by-count": 8,
"issue": "1",
"issued": {
"date-parts": [
[
2023,
10,
13
]
]
},
"journal-issue": {
"issue": "1",
"published-online": {
"date-parts": [
[
2023,
12
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2023,
10,
13
]
],
"date-time": "2023-10-13T00:00:00Z",
"timestamp": 1697155200000
}
},
{
"URL": "https://creativecommons.org/licenses/by/4.0",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2023,
10,
13
]
],
"date-time": "2023-10-13T00:00:00Z",
"timestamp": 1697155200000
}
}
],
"link": [
{
"URL": "https://www.nature.com/articles/s41467-023-42102-y.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.nature.com/articles/s41467-023-42102-y",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.nature.com/articles/s41467-023-42102-y.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "297",
"original-title": [],
"prefix": "10.1038",
"published": {
"date-parts": [
[
2023,
10,
13
]
]
},
"published-online": {
"date-parts": [
[
2023,
10,
13
]
]
},
"publisher": "Springer Science and Business Media LLC",
"reference": [
{
"author": "H Su",
"first-page": "151",
"journal-title": "Fund. Res.",
"key": "42102_CR1",
"unstructured": "Su, H., Xu, Y. & Jiang, H. Drug discovery and development targeting the life cycle of SARS-CoV-2. Fund. Res. 1, 151–165 (2021).",
"volume": "1",
"year": "2021"
},
{
"DOI": "10.1002/med.21783",
"author": "M Xiong",
"doi-asserted-by": "publisher",
"first-page": "1965",
"journal-title": "Med. Res. Rev.",
"key": "42102_CR2",
"unstructured": "Xiong, M. et al. What coronavirus 3C-like protease tells us: from structure, substrate selectivity, to inhibitor design. Med. Res. Rev. 41, 1965–1998 (2021).",
"volume": "41",
"year": "2021"
},
{
"DOI": "10.1126/science.abd0831",
"author": "A Baum",
"doi-asserted-by": "publisher",
"first-page": "1014",
"journal-title": "Science",
"key": "42102_CR3",
"unstructured": "Baum, A. et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 369, 1014–1018 (2020).",
"volume": "369",
"year": "2020"
},
{
"DOI": "10.1126/science.abl4784",
"author": "DR Owen",
"doi-asserted-by": "publisher",
"first-page": "1586",
"journal-title": "Science",
"key": "42102_CR4",
"unstructured": "Owen, D. R. et al. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science 374, 1586–1593 (2021).",
"volume": "374",
"year": "2021"
},
{
"DOI": "10.1021/acs.jmedchem.2c00624",
"author": "JDA Tyndall",
"doi-asserted-by": "publisher",
"first-page": "6496",
"journal-title": "J. Med. Chem.",
"key": "42102_CR5",
"unstructured": "Tyndall, J. D. A. S-217622, a 3CL protease inhibitor and clinical candidate for SARS-CoV-2. J. Med. Chem. 65, 6496–6498 (2022).",
"volume": "65",
"year": "2022"
},
{
"DOI": "10.1126/science.abb4489",
"author": "W Dai",
"doi-asserted-by": "publisher",
"first-page": "1331",
"journal-title": "Science",
"key": "42102_CR6",
"unstructured": "Dai, W. et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science 368, 1331–1335 (2020).",
"volume": "368",
"year": "2020"
},
{
"DOI": "10.1021/ar700109k",
"author": "FG Njoroge",
"doi-asserted-by": "publisher",
"first-page": "50",
"journal-title": "Acc. Chem. Res.",
"key": "42102_CR7",
"unstructured": "Njoroge, F. G. et al. Challenges in modern drug discovery: a case study of boceprevir, an HCV protease inhibitor for the treatment of hepatitis C virus infection. Acc. Chem. Res. 41, 50–59 (2008).",
"volume": "41",
"year": "2008"
},
{
"DOI": "10.1016/j.str.2020.10.007",
"author": "DW Kneller",
"doi-asserted-by": "publisher",
"first-page": "1313",
"journal-title": "Structure",
"key": "42102_CR8",
"unstructured": "Kneller, D. W. et al. Malleability of the SARS-CoV-2 3CL Mpro active-site cavity facilitates binding of clinical antivirals. Structure 28, 1313–1320 (2020).",
"volume": "28",
"year": "2020"
},
{
"DOI": "10.1038/s41467-020-18233-x",
"author": "L Fu",
"doi-asserted-by": "publisher",
"journal-title": "Nat. Commun.",
"key": "42102_CR9",
"unstructured": "Fu, L. et al. Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 11, 4417 (2020).",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1021/acs.jmedchem.0c01063",
"author": "RL Hoffman",
"doi-asserted-by": "publisher",
"first-page": "12725",
"journal-title": "J. Med. Chem.",
"key": "42102_CR10",
"unstructured": "Hoffman, R. L. et al. Discovery of ketone-based covalent inhibitors of coronavirus 3CL proteases for the potential therapeutic treatment of COVID-19. J. Med. Chem. 63, 12725–12747 (2020).",
"volume": "63",
"year": "2020"
},
{
"DOI": "10.1111/febs.12936",
"author": "R Hilgenfeld",
"doi-asserted-by": "publisher",
"first-page": "4085",
"journal-title": "FEBS J.",
"key": "42102_CR11",
"unstructured": "Hilgenfeld, R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 281, 4085–4096 (2014).",
"volume": "281",
"year": "2014"
},
{
"DOI": "10.1021/acs.jmedchem.5b01461",
"author": "T Pillaiyar",
"doi-asserted-by": "publisher",
"first-page": "6595",
"journal-title": "J. Med. Chem.",
"key": "42102_CR12",
"unstructured": "Pillaiyar, T. et al. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 59, 6595–6628 (2016).",
"volume": "59",
"year": "2016"
},
{
"DOI": "10.1021/acs.jmedchem.0c01140",
"author": "R Cannalire",
"doi-asserted-by": "publisher",
"first-page": "2716",
"journal-title": "J. Med. Chem.",
"key": "42102_CR13",
"unstructured": "Cannalire, R. et al. Targeting SARS-CoV-2 proteases and polymerase for COVID-19 treatment: state of the art and future opportunities. J. Med. Chem. 65, 2716–2746 (2022).",
"volume": "65",
"year": "2022"
},
{
"DOI": "10.1128/JVI.01819-20",
"author": "M de Vries",
"doi-asserted-by": "publisher",
"first-page": "e01819",
"journal-title": "J. Virol.",
"key": "42102_CR14",
"unstructured": "de Vries, M. et al. A comparative analysis of SARS-CoV-2 antivirals characterizes 3CLpro inhibitor PF-00835231 as a potential new treatment for COVID-19. J. Virol. 95, e01819 (2021).",
"volume": "95",
"year": "2021"
},
{
"DOI": "10.1016/j.drudis.2014.09.014",
"author": "A Tarcsay",
"doi-asserted-by": "publisher",
"first-page": "86",
"journal-title": "Drug Discov. Today",
"key": "42102_CR15",
"unstructured": "Tarcsay, A. & Keseru, G. M. Is there a link between selectivity and binding thermodynamics profiles? Drug Discov. Today 20, 86–94 (2015).",
"volume": "20",
"year": "2015"
},
{
"DOI": "10.1016/j.ab.2016.08.023",
"author": "A Schon",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Anal. Biochem.",
"key": "42102_CR16",
"unstructured": "Schon, A. & Freire, E. Enthalpy screen of drug candidates. Anal. Biochem. 513, 1–6 (2016).",
"volume": "513",
"year": "2016"
},
{
"DOI": "10.1016/j.drudis.2008.07.005",
"author": "E Freire",
"doi-asserted-by": "publisher",
"first-page": "869",
"journal-title": "Drug Discov. Today",
"key": "42102_CR17",
"unstructured": "Freire, E. Do enthalpy and entropy distinguish first in class from best in class? Drug Discov. Today 13, 869–874 (2008).",
"volume": "13",
"year": "2008"
},
{
"DOI": "10.1038/s41586-020-2223-y",
"author": "Z Jin",
"doi-asserted-by": "publisher",
"first-page": "289",
"journal-title": "Nature",
"key": "42102_CR18",
"unstructured": "Jin, Z. et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 582, 289–293 (2020).",
"volume": "582",
"year": "2020"
},
{
"DOI": "10.1038/nrd3845",
"author": "J Bowes",
"doi-asserted-by": "publisher",
"first-page": "909",
"journal-title": "Nat. Rev. Drug Discov.",
"key": "42102_CR19",
"unstructured": "Bowes, J. et al. Reducing safety-related drug attrition: the use of in vitro pharmacological profiling. Nat. Rev. Drug Discov. 11, 909–922 (2012).",
"volume": "11",
"year": "2012"
},
{
"DOI": "10.1126/science.abf1611",
"author": "J Qiao",
"doi-asserted-by": "publisher",
"first-page": "1374",
"journal-title": "Science",
"key": "42102_CR20",
"unstructured": "Qiao, J. et al. SARS-CoV-2 M(pro) inhibitors with antiviral activity in a transgenic mouse model. Science 371, 1374–1378 (2021).",
"volume": "371",
"year": "2021"
},
{
"DOI": "10.1038/s41467-022-29915-z",
"author": "DW Kneller",
"doi-asserted-by": "publisher",
"journal-title": "Nat. Commun.",
"key": "42102_CR21",
"unstructured": "Kneller, D. W. et al. Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease. Nat. Commun. 13, 2268 (2022).",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.1021/acscatal.8b05051",
"author": "YH Wang",
"doi-asserted-by": "publisher",
"first-page": "2292",
"journal-title": "Acs Catal.",
"key": "42102_CR22",
"unstructured": "Wang, Y. H. et al. Covalent inhibition mechanism of antidiabetic drugs-vildagliptin vs saxagliptin. Acs Catal. 9, 2292–2302 (2019).",
"volume": "9",
"year": "2019"
},
{
"DOI": "10.1038/s41573-022-00542-z",
"author": "L Boike",
"doi-asserted-by": "publisher",
"first-page": "881",
"journal-title": "Nat. Rev. Drug Discov.",
"key": "42102_CR23",
"unstructured": "Boike, L., Henning, N. J. & Nomura, D. K. Advances in covalent drug discovery. Nat. Rev. Drug Discov. 21, 881–898 (2022).",
"volume": "21",
"year": "2022"
},
{
"DOI": "10.1016/j.jbbm.2005.12.008",
"author": "F van den Ent",
"doi-asserted-by": "publisher",
"first-page": "67",
"journal-title": "J. Biochem. Biophys. Methods",
"key": "42102_CR24",
"unstructured": "van den Ent, F. & Lowe, J. RF cloning: a restriction-free method for inserting target genes into plasmids. J. Biochem. Biophys. Methods 67, 67–74 (2006).",
"volume": "67",
"year": "2006"
},
{
"DOI": "10.1007/s41365-019-0683-2",
"author": "W Zhang",
"doi-asserted-by": "publisher",
"first-page": "170",
"journal-title": "Nucl. Sci. Tech.",
"key": "42102_CR25",
"unstructured": "Zhang, W. et al. The protein complex crystallography beamline (BL19U1) at the shanghai synchrotron radiation facility. Nucl. Sci. Tech. 30, 170 (2019).",
"volume": "30",
"year": "2019"
},
{
"DOI": "10.1107/S1600576719001183",
"author": "F Yu",
"doi-asserted-by": "publisher",
"first-page": "472",
"journal-title": "J. Appl. Crystallogr.",
"key": "42102_CR26",
"unstructured": "Yu, F. et al. Aquarium: an automatic data-processing and experiment information management system for biological macromolecular crystallography beamlines. J. Appl. Crystallogr. 52, 472–477 (2019).",
"volume": "52",
"year": "2019"
},
{
"DOI": "10.1107/S0907444906019949",
"author": "W Minor",
"doi-asserted-by": "publisher",
"first-page": "859",
"journal-title": "Acta Crystallogr. D",
"key": "42102_CR27",
"unstructured": "Minor, W. et al. HKL-3000: the integration of data reduction and structure solution-from diffraction images to an initial model in minutes. Acta Crystallogr. D 62, 859–866 (2006).",
"volume": "62",
"year": "2006"
},
{
"DOI": "10.1107/S0907444994003112",
"author": "Collaborative C. P.",
"doi-asserted-by": "publisher",
"first-page": "760",
"journal-title": "Acta Crystallogr. D",
"key": "42102_CR28",
"unstructured": "Collaborative C. P. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).",
"volume": "50",
"year": "1994"
},
{
"DOI": "10.1038/s41401-020-0483-6",
"author": "H Su",
"doi-asserted-by": "publisher",
"first-page": "1167",
"journal-title": "Acta Pharmacol. Sin.",
"key": "42102_CR29",
"unstructured": "Su, H. et al. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol. Sin. 41, 1167–1177 (2020).",
"volume": "41",
"year": "2020"
},
{
"DOI": "10.1107/S0907444904019158",
"author": "P Emsley",
"doi-asserted-by": "publisher",
"first-page": "2126",
"journal-title": "Acta Crystallogr. D",
"key": "42102_CR30",
"unstructured": "Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).",
"volume": "60",
"year": "2004"
},
{
"DOI": "10.1107/S0907444902016657",
"author": "PD Adams",
"doi-asserted-by": "publisher",
"first-page": "1948",
"journal-title": "Acta Crystallogr. D",
"key": "42102_CR31",
"unstructured": "Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002).",
"volume": "58",
"year": "2002"
}
],
"reference-count": 31,
"references-count": 31,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.nature.com/articles/s41467-023-42102-y"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"General Physics and Astronomy",
"General Biochemistry, Genetics and Molecular Biology",
"General Chemistry",
"Multidisciplinary"
],
"subtitle": [],
"title": "Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1007/springer_crossmark_policy",
"volume": "14"
}