Generation and evaluation of protease inhibitor-resistant SARS-CoV-2 strains
et al., Antiviral Research, doi:10.1016/j.antiviral.2024.105814, Feb 2024
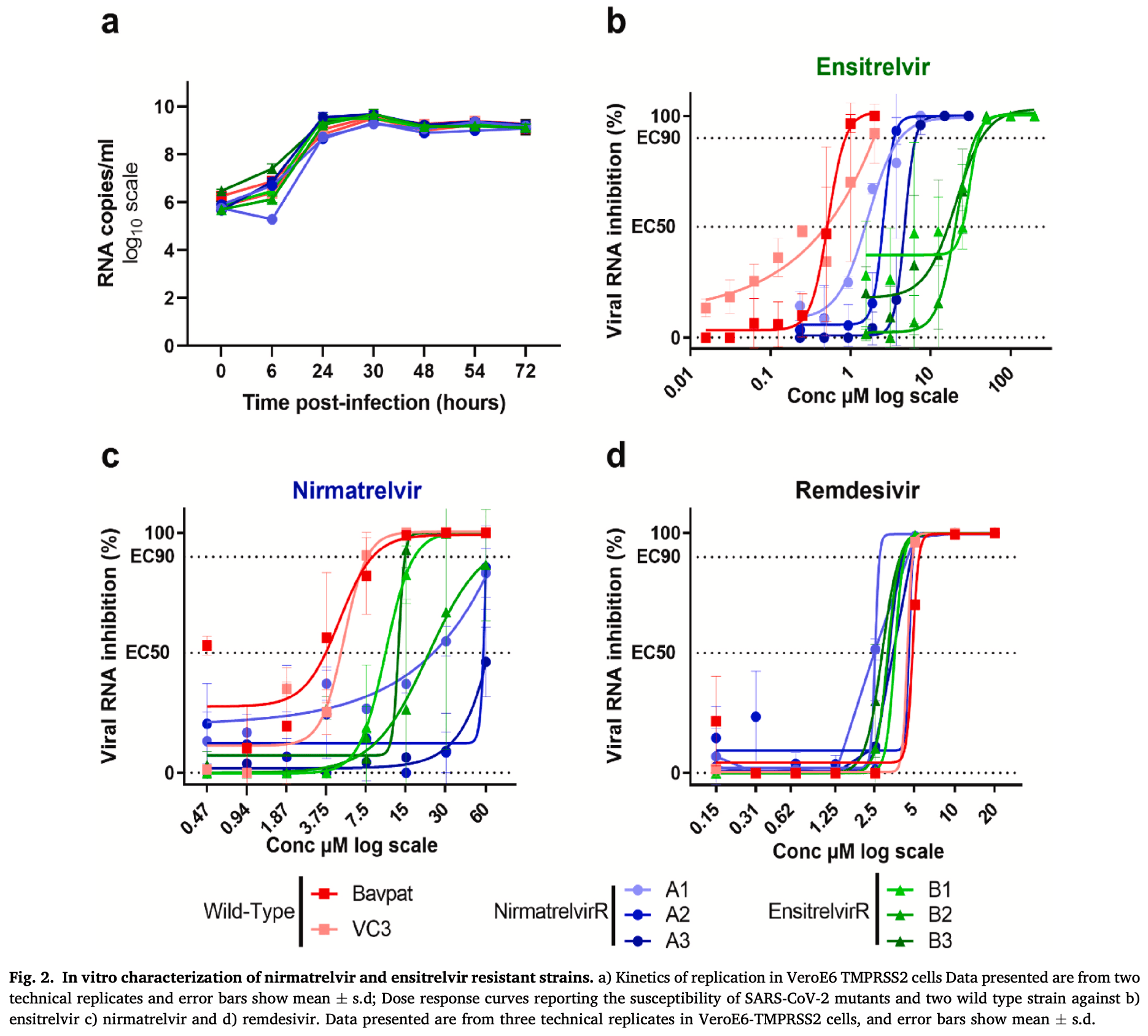
In vitro and animal study showing that SARS-CoV-2 can develop resistance to nirmatrelvir and ensitrelvir. Authors generated resistant viral strains through repeated passaging with both drugs. For nirmatrelvir, they identified three different mutation sets conferring 6-17 fold resistance, while for ensitrelvir, all strains developed the same mutation causing approximately 40-fold resistance.
Study covers ensitrelvir and paxlovid.
Bouzidi et al., 29 Feb 2024, peer-reviewed, 14 authors, study period July 2023 - October 2023.
Contact: franck.touret@univ-amu.fr.
Generation and evaluation of protease inhibitor-resistant SARS-CoV-2 strains
Antiviral Research, doi:10.1016/j.antiviral.2024.105814
Since the start of the SARS-CoV-2 pandemic, the search for antiviral therapies has been at the forefront of medical research. To date, the 3CLpro inhibitor nirmatrelvir (Paxlovid®) has shown the best results in clinical trials and the greatest robustness against variants. A second SARS-CoV-2 protease inhibitor, ensitrelvir (Xocova®), has been developed. Ensitrelvir, currently in Phase 3, was approved in Japan under the emergency regulatory approval procedure in November 2022, and is available since March 31, 2023. One of the limitations for the use of antiviral monotherapies is the emergence of resistance mutations. Here, we experimentally generated mutants resistant to nirmatrelvir and ensitrelvir in vitro following repeating passages of SARS-CoV-2 in the presence of both antivirals. For both molecules, we demonstrated a loss of sensitivity for resistance mutants in vitro. Using a Syrian golden hamster infection model, we showed that the ensitrelvir M49L mutation, in the multipassage strain, confers a high level of in vivo resistance. Finally, we identified a recent increase in the prevalence of M49L-carrying sequences, which appears to be associated with multiple repeated emergence events in Japan and may be related to the use of Xocova® in the country since November 2022. These results highlight the strategic importance of genetic monitoring of circulating SARS-CoV-2 strains to ensure that treatments administered retain their full effectiveness.
continuous monitoring in countries where ensitrelvir is, or will soon be, approved for use. These findings argues in favor of genetic monitoring of circulating strains to ensure that the treatments administered retain their full effectiveness. In addition, the range of antiviral treatments available against SARS-CoV-2 has become worryingly limited, with most commercial monoclonal drugs losing their activity against the most recent variants. Obviously, there is still a need to develop other antivirals targeting other viral proteins to broaden the therapeutic armory, and possibly to develop dual therapies as has been done for other viral diseases.
CRediT authorship contribution statement
Hawa
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data Supplementary data to this article can be found online at https://doi. org/10.1016/j.antiviral.2024.105814 .
References
Beigel, Tomashek, Dodd, Mehta, Zingman et al., Remdesivir for the treatment of Covid-19 -final report, N. Engl. J. Med, doi:10.1056/NEJMoa2007764
Bloom, Gong, Baltimore, Permissive secondary mutations enable the Evolution of Influenza Oseltamivir resistance, Science, doi:10.1126/science.1187816
Boras, Jones, Anson, Arenson, Aschenbrenner et al., Preclinical characterization of an intravenous coronavirus 3CL protease inhibitor for the potential treatment of COVID19, Nat. Commun, doi:10.1038/s41467-021-26239-2
Driouich, Cochin, Lingas, Moureau, Touret et al., Favipiravir antiviral efficacy against SARS-CoV-2 in a hamster model, Nat. Commun, doi:10.1038/s41467-021-21992-w
Driouich, Cochin, Touret, Petit, Gilles et al., Pre-clinical evaluation of antiviral activity of nitazoxanide against SARS-CoV-2, EBioMedicine, doi:10.1016/j.ebiom.2022.104148
Hammond, Leister-Tebbe, Gardner, Abreu, Bao et al., Oral nirmatrelvir for high-risk, nonhospitalized adults with covid-19
Heilmann, Costacurta, Moghadasi, Ye, Pavan et al., SARS-CoV-2 3CLpro mutations selected in a VSVbased system confer resistance to nirmatrelvir, ensitrelvir, and GC376, Sci. Transl. Med, doi:10.1126/scitranslmed.abq7360
Hoertel, Boulware, Sánchez-Rico, Burgun, Limosin, Prevalence of contraindications to nirmatrelvir-ritonavir among hospitalized patients with COVID-19 at risk for progression to severe disease, JAMA Netw. Open, doi:10.1001/jamanetworkopen.2022.42140
Hu, Lewandowski, Tan, Zhang, Xiaoming et al., Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir, ACS Cent. Sci, doi:10.1021/acscentsci.3c00538
Hunter, Matplotlib: a 2D graphics environment, Comput. Sci. Eng, doi:10.1109/MCSE.2007.55
Iketani, Mohri, Culbertson, Hong, Duan et al., Multiple pathways for SARS-CoV-2 resistance to nirmatrelvir, Nature, doi:10.1038/s41586-022-05514-2
Imai, Ito, Kiso, Yamayoshi, Uraki et al., Efficacy of antiviral agents against omicron subvariants BQ.1.1 and XBB, New England J. Med, doi:10.1056/NEJMc2214302
Ip, Wing-Ho, Chu, Chan, Cheuk-Ying et al., Global prevalence of SARS-CoV-2 3CL protease mutations associated with nirmatrelvir or ensitrelvir resistance, doi:10.1016/j.ebiom.2023.104559
Jin, Du, Xu, Deng, Liu et al., Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors, Nature, doi:10.1038/s41586-020-2223-y
Katoh, Standley, MAFFT multiple sequence alignment software version 7: improvements in performance and usability, Mol. Biol. Evol, doi:10.1093/molbev/mst010
Kawashima, Matsui, Adachi, Morikawa, Inoue et al., Ensitrelvir is effective against SARS-CoV-2 3CL protease mutants circulating globally, Biochem. Biophys. Res. Commun, doi:10.1016/j.bbrc.2023.01.040
Kiso, Furusawa, Uraki, Imai, Yamayoshi et al., In vitro and in vivo characterization of SARS-CoV-2 strains resistant to nirmatrelvir, Nat. Commun, doi:10.1038/s41467-023-39704-x
Kiso, Yamayoshi, Iida, Furusawa, Hirata et al., In vitro and in vivo characterization of SARS-CoV-2 resistance to ensitrelvir, Nat. Commun, doi:10.1038/s41467-023-40018-1
Korber, Fischer, Gnanakaran, Yoon, Theiler et al., Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus, Cell, doi:10.1016/j.cell.2020.06.043
Minh, Nguyen, Von Haeseler, Ultrafast approximation for phylogenetic bootstrap, Mol. Biol. Evol, doi:10.1093/molbev/mst024
Minh, Schmidt, Chernomor, Schrempf, Woodhams et al., IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era, Mol. Biol. Evol, doi:10.1093/molbev/msaa015
Moghadasi, Biswas, Harki, Harris, Rapid resistance profiling of SARS-CoV-2 protease inhibitors, npj Antimicrob Resist, doi:10.1038/s44259-023-00009-0
Moghadasi, Heilmann, Khalil, Nnabuife, Kearns et al., Transmissible SARS-CoV-2 variants with resistance to clinical protease inhibitors, Sci. Adv, doi:10.1126/sciadv.ade8778
Noske, Silva, De, Godoy, De et al., Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease, J. Biol. Chem, doi:10.1016/j.jbc.2023.103004
O'toole, Scher, Underwood, Jackson, Hill et al., Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool, Virus Evol, doi:10.1093/ve/veab064
Owen, Allerton, Anderson, Aschenbrenner, Avery et al., An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19, Science, doi:10.1126/science.abl4784
Pinto, Park, Beltramello, Walls, Tortorici et al., Crossneutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody, Nature, doi:10.1038/s41586-020-2349-y
Reed, Muench, A simple method of estimating fifty per cent ENDPOINTS12, Am. J. Epidemiol, doi:10.1093/oxfordjournals.aje.a118408
Rockett, Basile, Maddocks, Fong, Agius et al., Resistance mutations in SARS-CoV-2 Delta variant after sotrovimab use, N. Engl. J. Med, doi:10.1056/NEJMc2120219
Sasaki, Tabata, Kishimoto, Itakura, Kobayashi et al., S-217622, a SARS-CoV-2 main protease inhibitor, decreases viral load and ameliorates COVID-19 severity in hamsters, Sci. Transl. Med, doi:10.1126/scitranslmed.abq4064
Shionogi, A Phase 3, Multicenter, Randomized, Double-Blind, 24-Week Study of the Clinical and Antiviral Effect of S-217622 Compared with Placebo in Nonhospitalized Participants with COVID-19
Starr, Czudnochowski, Liu, Zatta, Park et al., SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape, Nature, doi:10.1038/s41586-021-03807-6
Takashita, Kinoshita, Yamayoshi, Sakai-Tagawa, Fujisaki et al., Efficacy of antiviral agents against the SARS-CoV-2 omicron subvariant BA.2, N. Engl. J. Med, doi:10.1056/NEJMc2201933
Touret, Baronti, Bouzidi, De Lamballerie, In vitro evaluation of therapeutic antibodies against a SARS-CoV-2 Omicron B.1.1.529 isolate, Sci. Rep, doi:10.1038/s41598-022-08559-5
Touret, Baronti, Goethals, Van Loock, De Lamballerie et al., Phylogenetically based establishment of a dengue virus panel, representing all available genotypes, as a tool in dengue drug discovery, Antivir. Res, doi:10.1016/j.antiviral.2019.05.005
Touret, Driouich, Cochin, Petit, Gilles et al., Preclinical evaluation of Imatinib does not support its use as an antiviral drug against SARS-CoV-2, Antivir. Res, doi:10.1016/j.antiviral.2021.105137
Touret, Driouich, Cochin, Petit, Gilles et al., Preclinical evaluation of Imatinib does not support its use as an antiviral drug against SARS-CoV-2, Antivir. Res, doi:10.1016/j.antiviral.2021.105137
Touret, Gilles, Barral, Nougairède, Van Helden et al., In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication, Sci. Rep, doi:10.1038/s41598-020-70143-6
Touret, Giraud, Bourret, Donati, Tran-Rajau et al., Enhanced neutralization escape to therapeutic monoclonal antibodies by SARS-CoV-2 omicron sub-lineages, iScience, doi:10.1016/j.isci.2023.106413
Unoh, Uehara, Nakahara, Nobori, Yamatsu et al., Discovery of S-217622, a noncovalent oral SARS-CoV-2 3CL protease inhibitor clinical candidate for treating COVID-19, J. Med. Chem, doi:10.1021/acs.jmedchem.2c00117
Vangeel, Chiu, De Jonghe, Maes, Slechten et al., Remdesivir, Molnupiravir and Nirmatrelvir remain active against SARS-CoV-2 Omicron and other variants of concern, Antivir. Res, doi:10.1016/j.antiviral.2022.105252
Wang, Cao, Zhang, Yang, Liu et al., Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro, Cell Res, doi:10.1038/s41422-020-0282-0
Williamson, Feldmann, Schwarz, Meade-White, Porter et al., Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2, Nature, doi:10.1038/s41586-020-2423-5
DOI record:
{
"DOI": "10.1016/j.antiviral.2024.105814",
"ISSN": [
"0166-3542"
],
"URL": "http://dx.doi.org/10.1016/j.antiviral.2024.105814",
"alternative-id": [
"S0166354224000226"
],
"article-number": "105814",
"assertion": [
{
"label": "This article is maintained by",
"name": "publisher",
"value": "Elsevier"
},
{
"label": "Article Title",
"name": "articletitle",
"value": "Generation and evaluation of protease inhibitor-resistant SARS-CoV-2 strains"
},
{
"label": "Journal Title",
"name": "journaltitle",
"value": "Antiviral Research"
},
{
"label": "CrossRef DOI link to publisher maintained version",
"name": "articlelink",
"value": "https://doi.org/10.1016/j.antiviral.2024.105814"
},
{
"label": "Content Type",
"name": "content_type",
"value": "article"
},
{
"label": "Copyright",
"name": "copyright",
"value": "© 2024 The Authors. Published by Elsevier B.V."
}
],
"author": [
{
"affiliation": [],
"family": "Bouzidi",
"given": "Hawa Sophia",
"sequence": "first"
},
{
"ORCID": "https://orcid.org/0000-0003-2326-2938",
"affiliation": [],
"authenticated-orcid": false,
"family": "Driouich",
"given": "Jean-Sélim",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-7777-0308",
"affiliation": [],
"authenticated-orcid": false,
"family": "Klitting",
"given": "Raphaëlle",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-2102-8198",
"affiliation": [],
"authenticated-orcid": false,
"family": "Bernadin",
"given": "Ornéllie",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Piorkowski",
"given": "Géraldine",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Amaral",
"given": "Rayane",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Fraisse",
"given": "Laurent",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Mowbray",
"given": "Charles E.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Scandale",
"given": "Ivan",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-9177-9755",
"affiliation": [],
"authenticated-orcid": false,
"family": "Escudié",
"given": "Fanny",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-3158-7743",
"affiliation": [],
"authenticated-orcid": false,
"family": "Chatelain",
"given": "Eric",
"sequence": "additional"
},
{
"affiliation": [],
"family": "de Lamballerie",
"given": "Xavier",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Nougairède",
"given": "Antoine",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-4734-2249",
"affiliation": [],
"authenticated-orcid": false,
"family": "Touret",
"given": "Franck",
"sequence": "additional"
}
],
"container-title": "Antiviral Research",
"container-title-short": "Antiviral Research",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"elsevier.com",
"sciencedirect.com"
]
},
"created": {
"date-parts": [
[
2024,
1,
24
]
],
"date-time": "2024-01-24T02:42:50Z",
"timestamp": 1706064170000
},
"deposited": {
"date-parts": [
[
2024,
7,
5
]
],
"date-time": "2024-07-05T17:56:42Z",
"timestamp": 1720202202000
},
"indexed": {
"date-parts": [
[
2025,
7,
30
]
],
"date-time": "2025-07-30T14:15:50Z",
"timestamp": 1753884950897,
"version": "3.40.5"
},
"is-referenced-by-count": 17,
"issued": {
"date-parts": [
[
2024,
2
]
]
},
"language": "en",
"license": [
{
"URL": "https://www.elsevier.com/tdm/userlicense/1.0/",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
2,
1
]
],
"date-time": "2024-02-01T00:00:00Z",
"timestamp": 1706745600000
}
},
{
"URL": "https://www.elsevier.com/legal/tdmrep-license",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
2,
1
]
],
"date-time": "2024-02-01T00:00:00Z",
"timestamp": 1706745600000
}
},
{
"URL": "http://creativecommons.org/licenses/by-nc-nd/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
1,
23
]
],
"date-time": "2024-01-23T00:00:00Z",
"timestamp": 1705968000000
}
}
],
"link": [
{
"URL": "https://api.elsevier.com/content/article/PII:S0166354224000226?httpAccept=text/xml",
"content-type": "text/xml",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://api.elsevier.com/content/article/PII:S0166354224000226?httpAccept=text/plain",
"content-type": "text/plain",
"content-version": "vor",
"intended-application": "text-mining"
}
],
"member": "78",
"original-title": [],
"page": "105814",
"prefix": "10.1016",
"published": {
"date-parts": [
[
2024,
2
]
]
},
"published-print": {
"date-parts": [
[
2024,
2
]
]
},
"publisher": "Elsevier BV",
"reference": [
{
"DOI": "10.1056/NEJMoa2007764",
"article-title": "Remdesivir for the treatment of Covid-19 — final report",
"author": "Beigel",
"doi-asserted-by": "crossref",
"first-page": "1813",
"journal-title": "N. Engl. J. Med.",
"key": "10.1016/j.antiviral.2024.105814_bib1",
"volume": "383",
"year": "2020"
},
{
"DOI": "10.1126/science.1187816",
"article-title": "Permissive secondary mutations enable the Evolution of Influenza Oseltamivir resistance",
"author": "Bloom",
"doi-asserted-by": "crossref",
"first-page": "1272",
"journal-title": "Science",
"key": "10.1016/j.antiviral.2024.105814_bib2",
"volume": "328",
"year": "2010"
},
{
"article-title": "Preclinical characterization of an intravenous coronavirus 3CL protease inhibitor for the potential treatment of COVID19",
"author": "Boras",
"issue": "6055",
"journal-title": "Nat. Commun.",
"key": "10.1016/j.antiviral.2024.105814_bib3",
"volume": "12",
"year": "2021"
},
{
"article-title": "Favipiravir antiviral efficacy against SARS-CoV-2 in a hamster model",
"author": "Driouich",
"issue": "1735",
"journal-title": "Nat. Commun.",
"key": "10.1016/j.antiviral.2024.105814_bib4",
"volume": "12",
"year": "2021"
},
{
"article-title": "Pre-clinical evaluation of antiviral activity of nitazoxanide against SARS-CoV-2",
"author": "Driouich",
"issue": "104148",
"journal-title": "EBioMedicine",
"key": "10.1016/j.antiviral.2024.105814_bib5",
"volume": "82",
"year": "2022"
},
{
"DOI": "10.1056/NEJMoa2118542",
"article-title": "Oral nirmatrelvir for high-risk, nonhospitalized adults with covid-19",
"author": "Hammond",
"doi-asserted-by": "crossref",
"first-page": "1397",
"journal-title": "N. Engl. J. Med.",
"key": "10.1016/j.antiviral.2024.105814_bib7",
"volume": "386",
"year": "2022"
},
{
"article-title": "SARS-CoV-2 3CLpro mutations selected in a VSV-based system confer resistance to nirmatrelvir, ensitrelvir, and GC376",
"author": "Heilmann",
"journal-title": "Sci. Transl. Med. 15, eabq7360",
"key": "10.1016/j.antiviral.2024.105814_bib8",
"year": "2022"
},
{
"DOI": "10.1001/jamanetworkopen.2022.42140",
"article-title": "Prevalence of contraindications to nirmatrelvir-ritonavir among hospitalized patients with COVID-19 at risk for progression to severe disease",
"author": "Hoertel",
"doi-asserted-by": "crossref",
"journal-title": "JAMA Netw. Open",
"key": "10.1016/j.antiviral.2024.105814_bib9",
"volume": "5",
"year": "2022"
},
{
"DOI": "10.1021/acscentsci.3c00538",
"article-title": "Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir",
"author": "Hu",
"doi-asserted-by": "crossref",
"first-page": "1658",
"journal-title": "ACS Cent. Sci.",
"key": "10.1016/j.antiviral.2024.105814_bib10",
"volume": "9",
"year": "2023"
},
{
"DOI": "10.1109/MCSE.2007.55",
"article-title": "Matplotlib: a 2D graphics environment",
"author": "Hunter",
"doi-asserted-by": "crossref",
"first-page": "90",
"journal-title": "Comput. Sci. Eng.",
"key": "10.1016/j.antiviral.2024.105814_bib11",
"volume": "9",
"year": "2007"
},
{
"DOI": "10.1038/s41586-022-05514-2",
"article-title": "Multiple pathways for SARS-CoV-2 resistance to nirmatrelvir",
"author": "Iketani",
"doi-asserted-by": "crossref",
"first-page": "558",
"journal-title": "Nature",
"key": "10.1016/j.antiviral.2024.105814_bib12",
"volume": "613",
"year": "2023"
},
{
"article-title": "Efficacy of antiviral agents against omicron subvariants BQ.1.1 and XBB",
"author": "Imai",
"journal-title": "New England J. Med. 0, null",
"key": "10.1016/j.antiviral.2024.105814_bib13",
"year": "2022"
},
{
"article-title": "Global prevalence of SARS-CoV-2 3CL protease mutations associated with nirmatrelvir or ensitrelvir resistance",
"author": "Ip",
"issue": "104559",
"journal-title": "EBioMedicine",
"key": "10.1016/j.antiviral.2024.105814_bib14",
"volume": "91",
"year": "2023"
},
{
"DOI": "10.1038/s41586-020-2223-y",
"article-title": "Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors",
"author": "Jin",
"doi-asserted-by": "crossref",
"first-page": "289",
"journal-title": "Nature",
"key": "10.1016/j.antiviral.2024.105814_bib16",
"volume": "582",
"year": "2020"
},
{
"DOI": "10.1093/molbev/mst010",
"article-title": "MAFFT multiple sequence alignment software version 7: improvements in performance and usability",
"author": "Katoh",
"doi-asserted-by": "crossref",
"first-page": "772",
"journal-title": "Mol. Biol. Evol.",
"key": "10.1016/j.antiviral.2024.105814_bib17",
"volume": "30",
"year": "2013"
},
{
"DOI": "10.1016/j.bbrc.2023.01.040",
"article-title": "Ensitrelvir is effective against SARS-CoV-2 3CL protease mutants circulating globally",
"author": "Kawashima",
"doi-asserted-by": "crossref",
"first-page": "132",
"journal-title": "Biochem. Biophys. Res. Commun.",
"key": "10.1016/j.antiviral.2024.105814_bib18",
"volume": "645",
"year": "2023"
},
{
"article-title": "In vitro and in vivo characterization of SARS-CoV-2 strains resistant to nirmatrelvir",
"author": "Kiso",
"issue": "3952",
"journal-title": "Nat. Commun.",
"key": "10.1016/j.antiviral.2024.105814_bib19",
"volume": "14",
"year": "2023"
},
{
"article-title": "In vitro and in vivo characterization of SARS-CoV-2 resistance to ensitrelvir",
"author": "Kiso",
"issue": "4231",
"journal-title": "Nat. Commun.",
"key": "10.1016/j.antiviral.2024.105814_bib20",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.1016/j.cell.2020.06.043",
"article-title": "Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus",
"author": "Korber",
"doi-asserted-by": "crossref",
"first-page": "812",
"journal-title": "Cell",
"key": "10.1016/j.antiviral.2024.105814_bib21",
"volume": "182",
"year": "2020"
},
{
"DOI": "10.1093/molbev/mst024",
"article-title": "Ultrafast approximation for phylogenetic bootstrap",
"author": "Minh",
"doi-asserted-by": "crossref",
"first-page": "1188",
"journal-title": "Mol. Biol. Evol.",
"key": "10.1016/j.antiviral.2024.105814_bib22",
"volume": "30",
"year": "2013"
},
{
"DOI": "10.1093/molbev/msaa015",
"article-title": "IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era",
"author": "Minh",
"doi-asserted-by": "crossref",
"first-page": "1530",
"journal-title": "Mol. Biol. Evol.",
"key": "10.1016/j.antiviral.2024.105814_bib23",
"volume": "37",
"year": "2020"
},
{
"DOI": "10.1038/s44259-023-00009-0",
"article-title": "Rapid resistance profiling of SARS-CoV-2 protease inhibitors",
"author": "Moghadasi",
"doi-asserted-by": "crossref",
"first-page": "1",
"journal-title": "npj Antimicrob Resist",
"key": "10.1016/j.antiviral.2024.105814_bib24",
"volume": "1",
"year": "2023"
},
{
"DOI": "10.1126/sciadv.ade8778",
"article-title": "Transmissible SARS-CoV-2 variants with resistance to clinical protease inhibitors",
"author": "Moghadasi",
"doi-asserted-by": "crossref",
"journal-title": "Sci. Adv.",
"key": "10.1016/j.antiviral.2024.105814_bib25",
"volume": "9",
"year": "2023"
},
{
"DOI": "10.1016/j.jbc.2023.103004",
"article-title": "Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease",
"author": "Noske",
"doi-asserted-by": "crossref",
"journal-title": "J. Biol. Chem.",
"key": "10.1016/j.antiviral.2024.105814_bib26",
"volume": "299",
"year": "2023"
},
{
"DOI": "10.1126/science.abl4784",
"article-title": "An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19",
"author": "Owen",
"doi-asserted-by": "crossref",
"first-page": "1586",
"journal-title": "Science",
"key": "10.1016/j.antiviral.2024.105814_bib27",
"volume": "374",
"year": "2021"
},
{
"author": "O'Toole",
"key": "10.1016/j.antiviral.2024.105814_bib28"
},
{
"DOI": "10.1038/s41586-020-2349-y",
"article-title": "Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody",
"author": "Pinto",
"doi-asserted-by": "crossref",
"first-page": "290",
"journal-title": "Nature",
"key": "10.1016/j.antiviral.2024.105814_bib29",
"volume": "583",
"year": "2020"
},
{
"DOI": "10.1093/oxfordjournals.aje.a118408",
"article-title": "A simple method of estimating fifty per cent ENDPOINTS12",
"author": "REED",
"doi-asserted-by": "crossref",
"first-page": "493",
"journal-title": "Am. J. Epidemiol.",
"key": "10.1016/j.antiviral.2024.105814_bib30",
"volume": "27",
"year": "1938"
},
{
"DOI": "10.1056/NEJMc2120219",
"article-title": "Resistance mutations in SARS-CoV-2 Delta variant after sotrovimab use",
"author": "Rockett",
"doi-asserted-by": "crossref",
"first-page": "1477",
"journal-title": "N. Engl. J. Med.",
"key": "10.1016/j.antiviral.2024.105814_bib31",
"volume": "386",
"year": "2022"
},
{
"article-title": "S-217622, a SARS-CoV-2 main protease inhibitor, decreases viral load and ameliorates COVID-19 severity in hamsters",
"author": "Sasaki",
"journal-title": "Sci. Transl. Med. 15, eabq4064",
"key": "10.1016/j.antiviral.2024.105814_bib33",
"year": "2022"
},
{
"author": "Shionogi",
"key": "10.1016/j.antiviral.2024.105814_bib34",
"series-title": "A Phase 3, Multicenter, Randomized, Double-Blind, 24-Week Study of the Clinical and Antiviral Effect of S-217622 Compared with Placebo in Non-hospitalized Participants with COVID-19 (Clinical Trial Registration No. NCT05305547). clinicaltrials.Gov",
"year": "2023"
},
{
"DOI": "10.1038/s41586-021-03807-6",
"article-title": "SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape",
"author": "Starr",
"doi-asserted-by": "crossref",
"first-page": "97",
"journal-title": "Nature",
"key": "10.1016/j.antiviral.2024.105814_bib35",
"volume": "597",
"year": "2021"
},
{
"DOI": "10.1056/NEJMc2201933",
"article-title": "Efficacy of antiviral agents against the SARS-CoV-2 omicron subvariant BA.2",
"author": "Takashita",
"doi-asserted-by": "crossref",
"first-page": "1475",
"journal-title": "N. Engl. J. Med.",
"key": "10.1016/j.antiviral.2024.105814_bib36",
"volume": "386",
"year": "2022"
},
{
"DOI": "10.1016/j.antiviral.2019.05.005",
"article-title": "Phylogenetically based establishment of a dengue virus panel, representing all available genotypes, as a tool in dengue drug discovery",
"author": "Touret",
"doi-asserted-by": "crossref",
"first-page": "109",
"journal-title": "Antivir. Res.",
"key": "10.1016/j.antiviral.2024.105814_bib37",
"volume": "168",
"year": "2019"
},
{
"article-title": "In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication",
"author": "Touret",
"issue": "13093",
"journal-title": "Sci. Rep.",
"key": "10.1016/j.antiviral.2024.105814_bib38",
"volume": "10",
"year": "2020"
},
{
"article-title": "Preclinical evaluation of Imatinib does not support its use as an antiviral drug against SARS-CoV-2",
"author": "Touret",
"issue": "105137",
"journal-title": "Antivir. Res.",
"key": "10.1016/j.antiviral.2024.105814_bib40",
"volume": "193",
"year": "2021"
},
{
"article-title": "Preclinical evaluation of Imatinib does not support its use as an antiviral drug against SARS-CoV-2",
"author": "Touret",
"issue": "105137",
"journal-title": "Antivir. Res.",
"key": "10.1016/j.antiviral.2024.105814_bib41",
"volume": "193",
"year": "2021"
},
{
"article-title": "In vitro evaluation of therapeutic antibodies against a SARS-CoV-2 Omicron B.1.1.529 isolate",
"author": "Touret",
"issue": "4683",
"journal-title": "Sci. Rep.",
"key": "10.1016/j.antiviral.2024.105814_bib42",
"volume": "12",
"year": "2022"
},
{
"article-title": "Enhanced neutralization escape to therapeutic monoclonal antibodies by SARS-CoV-2 omicron sub-lineages",
"author": "Touret",
"issue": "106413",
"journal-title": "iScience",
"key": "10.1016/j.antiviral.2024.105814_bib43",
"volume": "26",
"year": "2023"
},
{
"key": "10.1016/j.antiviral.2024.105814_bib44",
"series-title": "Strategies and Treatments for Respiratory Infections & Viral Emergencies (STRIVE): Shionogi Protease Inhibitor (Clinical Trial Registration No. NCT05605093). clinicaltrials.Gov",
"year": "2023"
},
{
"DOI": "10.1021/acs.jmedchem.2c00117",
"article-title": "Discovery of S-217622, a noncovalent oral SARS-CoV-2 3CL protease inhibitor clinical candidate for treating COVID-19",
"author": "Unoh",
"doi-asserted-by": "crossref",
"first-page": "6499",
"journal-title": "J. Med. Chem.",
"key": "10.1016/j.antiviral.2024.105814_bib45",
"volume": "65",
"year": "2022"
},
{
"article-title": "Remdesivir, Molnupiravir and Nirmatrelvir remain active against SARS-CoV-2 Omicron and other variants of concern",
"author": "Vangeel",
"issue": "105252",
"journal-title": "Antivir. Res.",
"key": "10.1016/j.antiviral.2024.105814_bib46",
"volume": "198",
"year": "2022"
},
{
"article-title": "Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro",
"author": "Wang",
"first-page": "1",
"journal-title": "Cell Res.",
"key": "10.1016/j.antiviral.2024.105814_bib47",
"year": "2020"
},
{
"DOI": "10.1038/s41586-020-2423-5",
"article-title": "Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2",
"author": "Williamson",
"doi-asserted-by": "crossref",
"first-page": "273",
"journal-title": "Nature",
"key": "10.1016/j.antiviral.2024.105814_bib48",
"volume": "585",
"year": "2020"
}
],
"reference-count": 44,
"references-count": 44,
"relation": {
"has-preprint": [
{
"asserted-by": "object",
"id": "10.1101/2023.11.22.568013",
"id-type": "doi"
}
]
},
"resource": {
"primary": {
"URL": "https://linkinghub.elsevier.com/retrieve/pii/S0166354224000226"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"special_numbering": "C",
"subject": [],
"subtitle": [],
"title": "Generation and evaluation of protease inhibitor-resistant SARS-CoV-2 strains",
"type": "journal-article",
"update-policy": "https://doi.org/10.1016/elsevier_cm_policy",
"volume": "222"
}
bouzidi