SARS-CoV-2 3CLpro mutations selected in a VSV-based system confer resistance to nirmatrelvir, ensitrelvir, and GC376
et al., Science Translational Medicine, doi:10.1126/scitranslmed.abq7360, Jan 2023
In vitro and in silico study showing selection of resistant mutations with nirmatrelvir use.
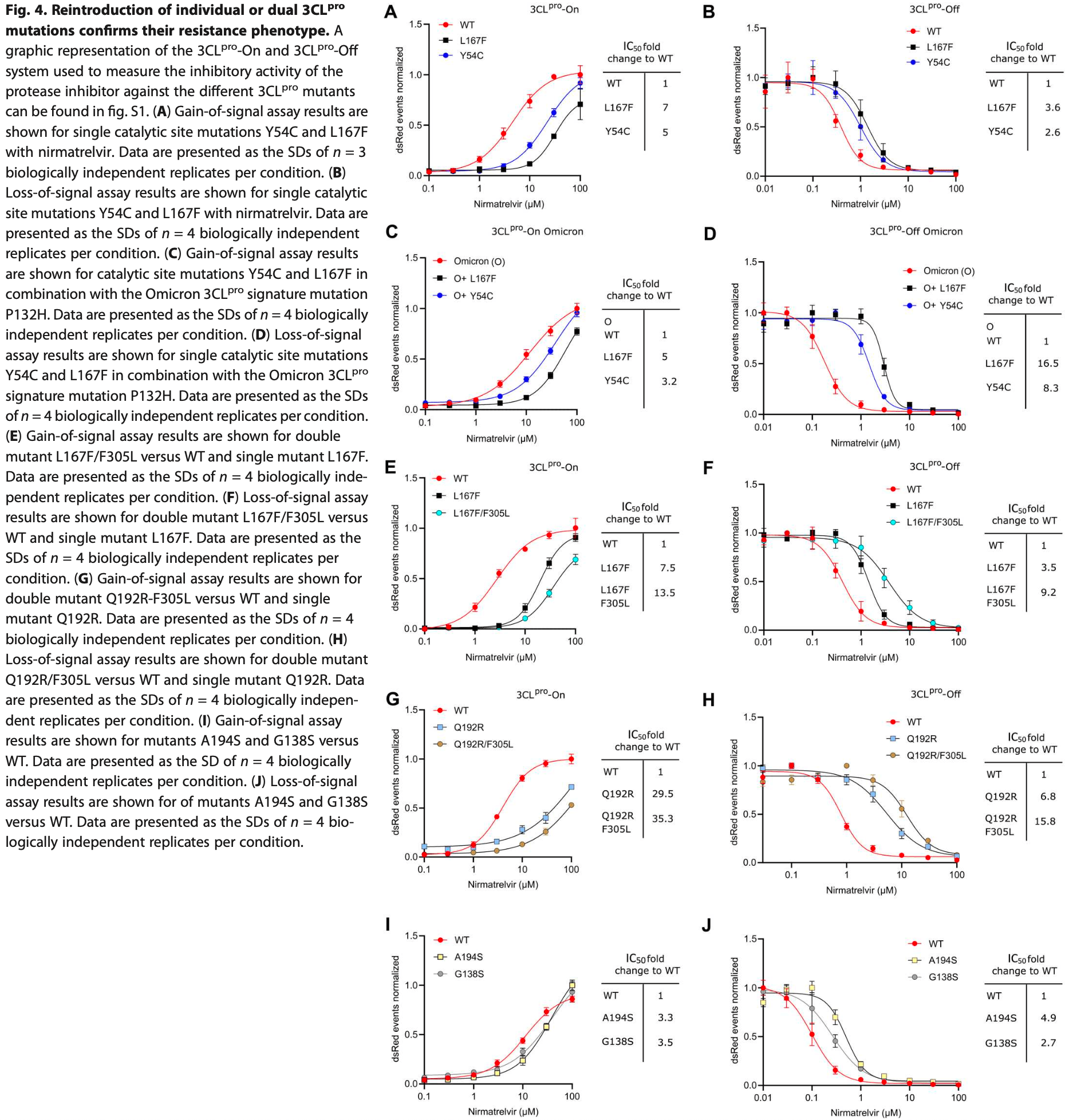
Several mutations were identified that confer resistance to 3CLpro inhibitors nirmatrelvir, ensitrelvir, and GC376. Authors note that most of these have already been found in existing SARS-CoV-2 sequences.
Authors argue for highly selective use because extensive, unselective use is expected to rapidly lead to emergence of drug resistance.
Heilmann et al., 11 Jan 2023, peer-reviewed, 15 authors.
Contact: emmanuel.heilmann@i-med.ac.at, von-laer@i-med.ac.at.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
SARS-CoV-2 3CL pro mutations selected in a VSV-based system confer resistance to nirmatrelvir, ensitrelvir, and GC376
Protease inhibitors are among the most powerful antiviral drugs. Nirmatrelvir is the first protease inhibitor specifically developed against the SARS-CoV-2 protease 3CL pro that has been licensed for clinical use. To identify mutations that confer resistance to this protease inhibitor, we engineered a chimeric vesicular stomatitis virus (VSV) that expressed a polyprotein composed of the VSV glycoprotein (G), the SARS-CoV-2 3CL pro , and the VSV polymerase (L). Viral replication was thus dependent on the autocatalytic processing of this precursor protein by 3CL pro and release of the functional viral proteins G and L, and replication of this chimeric VSV was effectively inhibited by nirmatrelvir. Using this system, we applied nirmatrelvir to select for resistance mutations. Resistance was confirmed by retesting nirmatrelvir against the selected mutations in additional VSV-based systems, in an independently developed cellular system, in a biochemical assay, and in a recombinant SARS-CoV-2 system. We demonstrate that some mutants are cross-resistant to ensitrelvir and GC376, whereas others are less resistant to these compounds. Furthermore, we found that most of these resistance mutations already existed in SARS-CoV-2 sequences that have been deposited in the NCBI and GISAID databases, indicating that these mutations were present in circulating SARS-CoV-2 strains.
KTSAVLQSGFRKME-EDANS. IC 50 and EC 50 calculations and statistical analysis for all assays were performed with GraphPad Prism 9 (see the "Statistical analysis" section).
Nanopore sequencing of recombinant SARS-CoV-2 (rWA1) expressing mCherry To validate the sequence of the recombinant SARS-CoV-2 (rWA1) expressing mCherry, we used the Nanopore sequencing "Midnight protocol," version 6 (57) . Primer pools generating 1200-base pair (bp) overlapping amplicons were purchased from Integrated DNA Technologies, as referenced in the abovementioned protocol. The sequencing reactions were prepared using the Rapid Barcoding Kit SQK-RBK110.96 (Oxford Nanopore Technologies) and were performed in a sequencer (MinION Mk1B) using a proprietary flow cell (R9.4.1, Oxford Nanopore Technologies). Electrical signals were translated into nucleotide sequences (basecalling). Sequenced reads were sorted into separate files for each sample (demultiplexing). Demultiplexing was done using the super highaccuracy model in Guppy 6.1.5. Output sequences generated socalled fastq files, and sequences below 200 and above 1200 bp were removed. Sequences between 200 and 1200 bp were assembled with the algorithm epi2me-labs/wf-artic v0.3.18 in Nextflow 22.04.4. The SARS-CoV-2 lineage pangolin 4.1.1 was used to map the sequences. A visualization application (Nextclade 2.4.0) was used to check mutations.
Protein structure preparation for molecular modeling The three-dimensional structure of..
References
Anand, Nagarajan, Mukherjee, Chandra, ABS-Scan: In silico alanine scanning mutagenesis for binding site residues in protein-ligand complex, F1000Res
Baek, Dimaio, Anishchenko, Dauparas, Ovchinnikov et al., Accurate prediction of protein structures and interactions using a three-track neural network, Science
Brogi, Ibba, Rossi, Butini, Calderone et al., Covalent reversible inhibitors of cysteine proteases containing the nitrile warhead: Recent advancement in the field of viral and parasitic diseases, Molecules
Case, Darden, Cheatham, Simmerling, Wang et al., Amber 10: User's Manual
Dold, Rodriguez Urbiola, Wollmann, Egerer, Muik et al., Application of interferon modulators to overcome partial resistance of human ovarian cancers to VSV-GP oncolytic viral therapy, Mol. Ther. Oncolytics
Drake, Holland, Mutation rates among RNA viruses, Proc. Natl. Acad. Sci. U.S.A
Elbe, Buckland-Merrett, Data, disease and diplomacy: GISAID's innovative contribution to global health, Glob. Chall
Fan, Wei, Feng, Chen, Huang et al., Biosynthesis, purification, and substrate specificity of severe acute respiratory syndrome coronavirus 3C-like proteinase, J. Biol. Chem
Freed, Vlková, Faisal, Silander, Rapid and inexpensive whole-genome sequencing of SARS-CoV-2 using 1200 bp tiled amplicons and Oxford Nanopore Rapid Barcoding, Biol. Methods Protoc
Gibson, Young, Chuang, Venter, Hutchison et al., Enzymatic assembly of DNA molecules up to several hundred kilobases, Nat. Methods
Greasley, Noell, Plotnikova, Ferre, Liu et al., Structural basis for the in vitro efficacy of nirmatrelvir against SARS-CoV-2 variants, J. Biol. Chem
Harcourt, Jukneliene, Kanjanahaluethai, Bechill, Severson et al., Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity, J. Virol
Harrington, Carpenter, Hit HIV-1 hard, but only when necessary, Lancet
Hatcher, Zhdanov, Bao, Blinkova, Nawrocki et al., Virus Variation Resource-improved response to emergent viral outbreaks, Nucleic Acids Res
Heilmann, Costacurta, Geley, Mogadashi, Volland et al., A VSV-based assay quantifies coronavirus Mpro/3CLpro/Nsp5 main protease activity and chemical inhibition, Commun. Biol
Heilmann, Costacurta, Seyed, Moghadasi, Ye et al., None, Sci. Transl. Med, doi:10.1126/scitranslmed.abq7360
Heilmann, None, Sci. Transl. Med
Hsu, Kuo, Chang, Chang, Chou et al., Mechanism of the maturation process of SARS-CoV 3CL protease, J. Biol. Chem
Hu, Lewandowski, Tan, Morgan, Zhang et al., Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir, bioRxiv, doi:10.1101/2022.06.28.497978
Iketani, Mohri, Culbertson, Hong, Duan et al., Multiple pathways for SARS-CoV-2 resistance to nirmatrelvir, bioRxiv, doi:10.1101/2022.08.07.499047
Jaskolski, Dauter, Shabalin, Gilski, Brzezinski et al., Crystallographic models of SARS-CoV-2 3CLpro: In-depth assessment of structure quality and validation, IUCrJ
Jochmans, Liu, Donckers, Stoycheva, Boland et al., The substitutions L50F, E166A and L167F in SARS-CoV-2 3CLpro are selected by a protease inhibitor in vitro and confer resistance to nirmatrelvir, bioRxiv, doi:10.1101/2022.06.07.495116
Kaufer, Theis, Lau, Gray, Rawlinson, Laboratory biosafety measures involving SARS-CoV-2 and the classification as a Risk Group 3 biological agent, Pathology
Khare, Gurry, Freitas, Schultz, Bach et al., GISAID's role in pandemic response, China CDC Wkly
Kim, Chivian, Baker, Protein structure prediction and analysis using the Robetta server, Nucleic Acids Res
Kärber, Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Naunyn Schmiedebergs, Arch. Exp. Pathol. Pharmakol
Labute, LowModeMD-Implicit low-mode velocity filtering applied to conformational search of macrocycles and protein loops, J. Chem. Inf. Model
Labute, The generalized born/volume integral implicit solvent model: Estimation of the free energy of hydration using London dispersion instead of atomic surface area, J. Comput. Chem
Lamb, Nirmatrelvir plus ritonavir: First approval, Drugs
Liu, Peng, Zhou, Zhang, Zhang, Computational alanine scanning with interaction entropy for protein-ligand binding free energies, J. Chem. Theory Comput
Liu, Vanblargan, Bloyet, Rothlauf, Chen et al., Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization, Cell Host Microbe
Manandhar, Blass, Colussi, Almi, Abou-Gharbia et al., Targeting SARS-CoV-2 M3CLpro by HCV NS3/4a inhibitors: In silico modeling and in vitro screening, J. Chem. Inf. Model
Manandhar, Srinivasulu, Hamad, Tarazi, Omar et al., Discovery of novel smallmolecule inhibitors of SARS-CoV-2 main protease as potential leads for COVID-19 treatment, J. Chem. Inf. Model
Moghadasi, Esler, Otsuka, Becker, Moraes et al., Gain-of-signal assays for probing inhibition of SARS-CoV-2 M pro /3CL pro in living cells, MBio
Moghadasi, Heilmann, Moraes, Kearns, Laer et al., Transmissible SARS-CoV-2 variants with resistance to clinical protease inhibitors, bioRxiv, doi:10.1101/2022.08.07.503099
Morrison, Weiss, Combinatorial alanine-scanning, Curr. Opin. Chem. Biol
Mukae, Yotsuyanagi, Ohmagari, Doi, Imamura et al., A randomized phase 2/3
Muramatsu, Takemoto, Kim, Wang, Nishii et al., SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity, Proc. Natl. Acad. Sci
Onufriev, Case, Bashford, Effective Born radii in the generalized Born approximation: The importance of being perfect, J. Comput. Chem
Owen, Allerton, Anderson, Aschenbrenner, Avery et al., An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19, Science
Owen, Allerton, Anderson, Aschenbrenner, Avery et al., An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19, Science
Padhi, Tripathi, Hotspot residues and resistance mutations in the nirmatrelvirbinding site of SARS-CoV-2 main protease: Design, identification, and correlation with globally circulating viral genomes, Biochem. Biophys. Res. Commun
Panda, Dinh, Beura, Pattnaik, Induction of interferon and interferon signaling pathways by replication of defective interfering particle RNA in cells constitutively expressing vesicular stomatitis virus replication proteins, J. Virol
Pavan, Bolcato, Bassani, Sturlese, Moro, Supervised molecular dynamics (SuMD) insights into the mechanism of action of SARS-CoV-2 main protease inhibitor PF-07321332, J. Enzyme Inhib. Med. Chem
Pettersen, Goddard, Huang, Couch, Greenblatt et al., UCSF Chimera-A visualization system for exploratory research and analysis, J. Comput. Chem
Rut, Groborz, Zhang, Sun, Zmudzinski et al., SARS-CoV-2 M pro inhibitors and activitybased probes for patient-sample imaging, Nat. Chem. Biol
S C I E N C E T R A N S L At I O N A L M E D I C I N E | R E S E A R C H A R T I C L E, None
Schmidt, Weisblum, Rutkowska, Poston, Dasilva et al., High genetic barrier to SARS-CoV-2 polyclonal neutralizing antibody escape, Nature
Schnell, Buonocore, Whitt, Rose, The minimal conserved transcription stop-start signal promotes stable expression of a foreign gene in vesicular stomatitis virus, J. Virol
Shu, Mccauley, GISAID: Global initiative on sharing all influenza data -from vision to reality, Euro Surveill
Silvestrini, Belhaj, Comez, Gerelli, Lauria et al., The dimer-monomer equilibrium of SARS-CoV-2 main protease is affected by small molecule inhibitors, Sci. Rep
St, West, Suite #910
Steinhauer, Domingo, Holland, Lack of evidence for proofreading mechanisms associated with an RNA virus polymerase, Gene
Ullrich, Ekanayake, Otting, Nitsche, Main protease mutants of SARS-CoV-2 variants remain susceptible to nirmatrelvir, Bioorg. Med. Chem. Lett
Ullrich, Nitsche, The SARS-CoV-2 main protease as drug target, Bioorg. Med. Chem. Lett
Voelker, Combination drug for HCV infection, JAMA
Whittaker, Groth, Mynarcik, Pluzek, Gadsbøll et al., Alanine scanning mutagenesis of a Type 1 insulin-like growth factor receptor ligand binding site, J. Biol. Chem
Witko, Kotash, Nowak, Johnson, Boutilier et al., An efficient helper-virus-free method for rescue of recombinant paramyxoviruses and rhadoviruses from a cell line suitable for vaccine development, J. Virol. Methods
Yahalom-Ronen, Erez, Fisher, Tamir, Politi et al., Neutralization of SARS-CoV-2 variants by rVSV-∆G-spike-elicited human sera, Vaccines
Ye, Chiem, Park, Oladunni, Platt et al., Rescue of SARS-CoV-2 from a single bacterial artificial chromosome, mBio
Ye, Chiem, Park, Silvas, Vasquez et al., Analysis of SARS-CoV-2 infection dynamic in vivo using reporter-expressing viruses, Proc. Natl. Acad. Sci. U.S.A
Zhang, Lin, Sun, Curth, Drosten et al., Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved aketoamide inhibitors, Science
Zhao, Fang, Zhang, Zhang, Zhao et al., Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332, Protein Cell
Zhou, Yang, Wang, Hu, Zhang et al., A pneumonia outbreak associated with a new coronavirus of probable bat origin, Nature
DOI record:
{
"DOI": "10.1126/scitranslmed.abq7360",
"ISSN": [
"1946-6234",
"1946-6242"
],
"URL": "http://dx.doi.org/10.1126/scitranslmed.abq7360",
"abstract": "<jats:p>\n Protease inhibitors are among the most powerful antiviral drugs. Nirmatrelvir is the first protease inhibitor specifically developed against the SARS-CoV-2 protease 3CL\n <jats:sup>pro</jats:sup>\n that has been licensed for clinical use. To identify mutations that confer resistance to this protease inhibitor, we engineered a chimeric vesicular stomatitis virus (VSV) that expressed a polyprotein composed of the VSV glycoprotein (G), the SARS-CoV-2 3CL\n <jats:sup>pro</jats:sup>\n , and the VSV polymerase (L). Viral replication was thus dependent on the autocatalytic processing of this precursor protein by 3CL\n <jats:sup>pro</jats:sup>\n and release of the functional viral proteins G and L, and replication of this chimeric VSV was effectively inhibited by nirmatrelvir. Using this system, we applied nirmatrelvir to select for resistance mutations. Resistance was confirmed by retesting nirmatrelvir against the selected mutations in additional VSV-based systems, in an independently developed cellular system, in a biochemical assay, and in a recombinant SARS-CoV-2 system. We demonstrate that some mutants are cross-resistant to ensitrelvir and GC376, whereas others are less resistant to these compounds. Furthermore, we found that most of these resistance mutations already existed in SARS-CoV-2 sequences that have been deposited in the NCBI and GISAID databases, indicating that these mutations were present in circulating SARS-CoV-2 strains.\n </jats:p>",
"alternative-id": [
"10.1126/scitranslmed.abq7360"
],
"author": [
{
"ORCID": "http://orcid.org/0000-0001-8148-9490",
"affiliation": [
{
"name": "Institute of Virology, Medical University of Innsbruck, Innsbruck, 6020, Austria."
}
],
"authenticated-orcid": true,
"family": "Heilmann",
"given": "Emmanuel",
"sequence": "first"
},
{
"ORCID": "http://orcid.org/0000-0002-4556-5967",
"affiliation": [
{
"name": "Institute of Virology, Medical University of Innsbruck, Innsbruck, 6020, Austria."
}
],
"authenticated-orcid": true,
"family": "Costacurta",
"given": "Francesco",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-6598-8994",
"affiliation": [
{
"name": "Department of Biochemistry, Molecular Biology and Biophysics, Institute for Molecular Virology, University of Minnesota, Minneapolis, MN 55455, USA."
}
],
"authenticated-orcid": true,
"family": "Moghadasi",
"given": "Seyed Arad",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-1934-9494",
"affiliation": [
{
"name": "Texas Biomedical Research Institute, San Antonio, TX 78229, USA."
}
],
"authenticated-orcid": true,
"family": "Ye",
"given": "Chengjin",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-6234-5934",
"affiliation": [
{
"name": "Molecular Modeling Section (MMS), Department of Pharmaceutical and Pharmacological Sciences, University of Padua, Via F. Marzolo 5, 35131, Padova, Italy."
}
],
"authenticated-orcid": true,
"family": "Pavan",
"given": "Matteo",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-9057-6297",
"affiliation": [
{
"name": "Molecular Modeling Section (MMS), Department of Pharmaceutical and Pharmacological Sciences, University of Padua, Via F. Marzolo 5, 35131, Padova, Italy."
}
],
"authenticated-orcid": true,
"family": "Bassani",
"given": "Davide",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-2805-8215",
"affiliation": [
{
"name": "Institute of Virology, Medical University of Innsbruck, Innsbruck, 6020, Austria."
}
],
"authenticated-orcid": true,
"family": "Volland",
"given": "Andre",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-0183-917X",
"affiliation": [
{
"name": "Institute for Biomedical Aging Research, University of Innsbruck, Innsbruck, 6020, Austria."
}
],
"authenticated-orcid": true,
"family": "Ascher",
"given": "Claudia",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-0166-8630",
"affiliation": [
{
"name": "Institute for Biomedical Aging Research, University of Innsbruck, Innsbruck, 6020, Austria."
}
],
"authenticated-orcid": true,
"family": "Weiss",
"given": "Alexander Kurt Hermann",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-5617-8022",
"affiliation": [
{
"name": "Institute of Virology, Medical University of Innsbruck, Innsbruck, 6020, Austria."
}
],
"authenticated-orcid": true,
"family": "Bante",
"given": "David",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-9034-9112",
"affiliation": [
{
"name": "Department of Biochemistry, Molecular Biology and Biophysics, Institute for Molecular Virology, University of Minnesota, Minneapolis, MN 55455, USA."
},
{
"name": "Department of Biochemistry and Structural Biology, University of Texas Health San Antonio, San Antonio, TX 78229, USA."
},
{
"name": "Howard Hughes Medical Institute, University of Texas Health San Antonio, San Antonio, TX 78229, USA."
}
],
"authenticated-orcid": true,
"family": "Harris",
"given": "Reuben S.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-7514-3802",
"affiliation": [
{
"name": "Molecular Modeling Section (MMS), Department of Pharmaceutical and Pharmacological Sciences, University of Padua, Via F. Marzolo 5, 35131, Padova, Italy."
}
],
"authenticated-orcid": true,
"family": "Moro",
"given": "Stefano",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-3300-6965",
"affiliation": [
{
"name": "Division of Genetic Epidemiology, Medical University of Innsbruck, Innsbruck, 6020, Austria."
},
{
"name": "k.-k. Hofkristallamt, San Diego, CA 92084, USA."
}
],
"authenticated-orcid": true,
"family": "Rupp",
"given": "Bernhard",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-7084-0804",
"affiliation": [
{
"name": "Texas Biomedical Research Institute, San Antonio, TX 78229, USA."
}
],
"authenticated-orcid": true,
"family": "Martinez-Sobrido",
"given": "Luis",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-5825-7237",
"affiliation": [
{
"name": "Institute of Virology, Medical University of Innsbruck, Innsbruck, 6020, Austria."
}
],
"authenticated-orcid": true,
"family": "von Laer",
"given": "Dorothee",
"sequence": "additional"
}
],
"container-title": "Science Translational Medicine",
"container-title-short": "Sci. Transl. Med.",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2022,
10,
4
]
],
"date-time": "2022-10-04T13:58:07Z",
"timestamp": 1664891887000
},
"deposited": {
"date-parts": [
[
2023,
1,
11
]
],
"date-time": "2023-01-11T18:58:34Z",
"timestamp": 1673463514000
},
"indexed": {
"date-parts": [
[
2023,
1,
12
]
],
"date-time": "2023-01-12T06:59:18Z",
"timestamp": 1673506758279
},
"is-referenced-by-count": 7,
"issue": "678",
"issued": {
"date-parts": [
[
2023,
1,
11
]
]
},
"journal-issue": {
"issue": "678",
"published-print": {
"date-parts": [
[
2023,
1,
11
]
]
}
},
"language": "en",
"member": "221",
"original-title": [],
"prefix": "10.1126",
"published": {
"date-parts": [
[
2023,
1,
11
]
]
},
"published-print": {
"date-parts": [
[
2023,
1,
11
]
]
},
"publisher": "American Association for the Advancement of Science (AAAS)",
"reference": [
{
"DOI": "10.1038/s41586-020-2012-7",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_2_2"
},
{
"DOI": "10.1001/jama.2017.10930",
"article-title": "Combination drug for HCV infection",
"author": "Voelker R.",
"doi-asserted-by": "crossref",
"first-page": "790",
"journal-title": "JAMA",
"key": "e_1_3_3_3_2",
"unstructured": "R. Voelker, Combination drug for HCV infection. JAMA 318, 790 (2017).",
"volume": "318",
"year": "2017"
},
{
"DOI": "10.1016/S0140-6736(00)02388-6",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_4_2"
},
{
"DOI": "10.1074/jbc.M310875200",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_5_2"
},
{
"DOI": "10.1128/JVI.78.24.13600-13612.2004",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_6_2"
},
{
"DOI": "10.1007/s40265-022-01692-5",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_7_2"
},
{
"DOI": "10.3390/molecules27082561",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_8_2"
},
{
"DOI": "10.1016/j.jbc.2022.101972",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_9_2"
},
{
"DOI": "10.1101/2022.08.07.499047",
"doi-asserted-by": "crossref",
"key": "e_1_3_3_10_2",
"unstructured": "S. Iketani H. Mohri B. Culbertson S. J. Hong Y. Duan M. I. Luck M. K. Annavajhala Y. Guo Z. Sheng A.-C. Uhlemann S. P. Goff Y. Sabo H. Yang A. Chavez D. D. Ho Multiple pathways for SARS-CoV-2 resistance to nirmatrelvir. bioRxiv 2022.08.07.499047 [ Preprint ] (2022)."
},
{
"DOI": "10.1101/2022.06.07.495116",
"doi-asserted-by": "crossref",
"key": "e_1_3_3_11_2",
"unstructured": "D. Jochmans C. Liu K. Donckers A. Stoycheva S. Boland S. K. Stevens C. De Vita B. Vanmechelen P. Maes B. Trüeb N. Ebert V. Thiel S. De Jonghe L. Vangeel D. Bardiot A. Jekle L. M. Blatt L. Beigelman J. A. Symons P. Raboisson P. Chaltin A. Marchand J. Neyts J. Deval K. Vandyck The substitutions L50F E166A and L167F in SARS-CoV-2 3CLpro are selected by a protease inhibitor in vitro and confer resistance to nirmatrelvir. bioRxiv 2022.06.07.495116 [ Preprint ] (2022)."
},
{
"DOI": "10.1101/2022.08.07.503099",
"doi-asserted-by": "crossref",
"key": "e_1_3_3_12_2",
"unstructured": "S. A. Moghadasi E. Heilmann S. N. Moraes F. L. Kearns D. von Laer R. E. Amaro R. S. Harris Transmissible SARS-CoV-2 variants with resistance to clinical protease inhibitors. bioRxiv 2022.08.07.503099 [ Preprint ] (2022)."
},
{
"DOI": "10.1101/2022.06.28.497978",
"doi-asserted-by": "crossref",
"key": "e_1_3_3_13_2",
"unstructured": "Y. Hu E. M. Lewandowski H. Tan R. T. Morgan X. Zhang L. M. C. Jacobs S. G. Butler M. V. Mongora J. Choy Y. Chen J. Wang Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir. bioRxiv 2022.06.28.497978 [ Preprint ] (2022)."
},
{
"DOI": "10.1016/j.bbrc.2022.09.010",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_14_2"
},
{
"DOI": "10.1016/j.pathol.2020.09.006",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_15_2"
},
{
"DOI": "10.1093/nar/gkh468",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_16_2"
},
{
"key": "e_1_3_3_17_2",
"unstructured": "Chemical Computing Group ULC Molecular Operating Environment (MOE) 2022.02 1010 Sherbooke St. West Suite #910 Montreal QC Canada H3A 2R7 (2022)."
},
{
"DOI": "10.1038/s42003-022-03277-0",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_18_2"
},
{
"DOI": "10.1107/S2052252521001159",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_19_2"
},
{
"DOI": "10.1038/s41598-021-88630-9",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_20_2"
},
{
"DOI": "10.1073/pnas.96.24.13910",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_21_2"
},
{
"DOI": "10.1016/0378-1119(92)90216-C",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_22_2"
},
{
"DOI": "10.2807/1560-7917.ES.2017.22.13.30494",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_23_2"
},
{
"DOI": "10.1002/gch2.1018",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_24_2"
},
{
"DOI": "10.46234/ccdcw2021.255",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_25_2"
},
{
"DOI": "10.1093/nar/gkw1065",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_26_2"
},
{
"key": "e_1_3_3_27_2",
"unstructured": "U.S. Food and Drug Administration Fact sheet for healthcare providers Emergency Use Authorization (EUA) for Paxlovid (2021); www.fda.gov/media/143823/download."
},
{
"DOI": "10.1128/mbio.00784-22",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_28_2"
},
{
"DOI": "10.1128/aac.00697-22",
"article-title": "A randomized phase 2/3 Study of Ensitrelvir, a novel oral SARS-CoV-2 3C-like PROTEASE INHIBITOR, in Japanese patients with mild-to-moderate COVID-19 or asymptomatic SARS-CoV-2 infection: Results of the phase 2a part",
"author": "Mukae H.",
"doi-asserted-by": "crossref",
"first-page": "e0069722",
"journal-title": "Antimicrob. Agents Chemother.",
"key": "e_1_3_3_29_2",
"unstructured": "H. Mukae, H. Yotsuyanagi, N. Ohmagari, Y. Doi, T. Imamura, T. Sonoyama, T. Fukuhara, G. Ichihashi, T. Sanaki, K. Baba, Y. Takeda, Y. Tsuge, T. Uehara, A randomized phase 2/3 Study of Ensitrelvir, a novel oral SARS-CoV-2 3C-like PROTEASE INHIBITOR, in Japanese patients with mild-to-moderate COVID-19 or asymptomatic SARS-CoV-2 infection: Results of the phase 2a part. Antimicrob. Agents Chemother. ,e0069722 (2022).",
"year": "2022"
},
{
"DOI": "10.1073/pnas.2111593118",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_30_2"
},
{
"DOI": "10.1128/mBio.02168-20",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_31_2"
},
{
"DOI": "10.1126/science.abj8754",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_32_2"
},
{
"DOI": "10.1126/science.abl4784",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_33_2"
},
{
"DOI": "10.1007/s13238-021-00883-2",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_34_2"
},
{
"DOI": "10.1016/S1367-5931(00)00206-4",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_35_2"
},
{
"DOI": "10.1074/jbc.M102863200",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_36_2"
},
{
"DOI": "10.1021/acs.jctc.7b01295",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_37_2"
},
{
"DOI": "10.12688/f1000research.5165.1",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_38_2"
},
{
"DOI": "10.1080/14756366.2021.1954919",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_39_2"
},
{
"DOI": "10.1016/j.chom.2021.01.014",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_40_2"
},
{
"DOI": "10.1038/s41586-021-04005-0",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_41_2"
},
{
"DOI": "10.3390/vaccines10020291",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_42_2"
},
{
"DOI": "10.1016/j.bmcl.2020.127377",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_43_2"
},
{
"DOI": "10.1016/j.bmcl.2022.128629",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_44_2"
},
{
"DOI": "10.1038/s41589-020-00689-z",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_45_2"
},
{
"DOI": "10.1021/acs.jcim.1c00684",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_46_2"
},
{
"DOI": "10.1126/science.abl4784",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_47_2"
},
{
"DOI": "10.1021/acs.jcim.0c01457",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_48_2"
},
{
"DOI": "10.1074/jbc.M502577200",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_49_2"
},
{
"DOI": "10.1073/pnas.1601327113",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_50_2"
},
{
"DOI": "10.1038/nmeth.1318",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_51_2"
},
{
"DOI": "10.1128/jvi.70.4.2318-2323.1996",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_52_2"
},
{
"DOI": "10.1128/JVI.02701-09",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_53_2"
},
{
"DOI": "10.1016/j.jviromet.2006.02.006",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_54_2"
},
{
"DOI": "10.1038/mto.2016.21",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_55_2"
},
{
"DOI": "10.1007/BF01863914",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_56_2"
},
{
"DOI": "10.1126/science.abb3405",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_57_2"
},
{
"DOI": "10.1093/biomethods/bpaa014",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_58_2"
},
{
"key": "e_1_3_3_59_2",
"unstructured": "D. A. Case T. Darden T. E. Cheatham III C. Simmerling J. Wang R. E. Duke R. Luo M. Crowley R. Walker W. Zhang K. M. Merz B. Wang S. Hayik A. Roitberg G. Seabra I. Kolossváry K. F. Wong F. Paesani J. Vanicek X. Wu S. R. Brozell T. Steinbrecher H. Gohlke L. Yang C. Tan J. Mongan V. Hornak G. Cui D. H. Mathews M. G. Seetin C. Sagui V. Babin P. A. Kollman Amber 10: User’s Manual (University of California San Francisco 2008)."
},
{
"DOI": "10.1002/jcc.20084",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_60_2"
},
{
"DOI": "10.1021/ci900508k",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_61_2"
},
{
"DOI": "10.1002/jcc.10126",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_62_2"
},
{
"DOI": "10.1002/jcc.20933",
"doi-asserted-by": "publisher",
"key": "e_1_3_3_63_2"
}
],
"reference-count": 62,
"references-count": 62,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.science.org/doi/10.1126/scitranslmed.abq7360"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"General Medicine"
],
"subtitle": [],
"title": "SARS-CoV-2 3CL\n <sup>pro</sup>\n mutations selected in a VSV-based system confer resistance to nirmatrelvir, ensitrelvir, and GC376",
"type": "journal-article",
"volume": "15"
}