Clinical advancement of patients infected with SARS-CoV-2 Omicron variant in Shanghai, China
et al., medRxiv, doi:10.1101/2022.07.03.22277169, Jul 2022
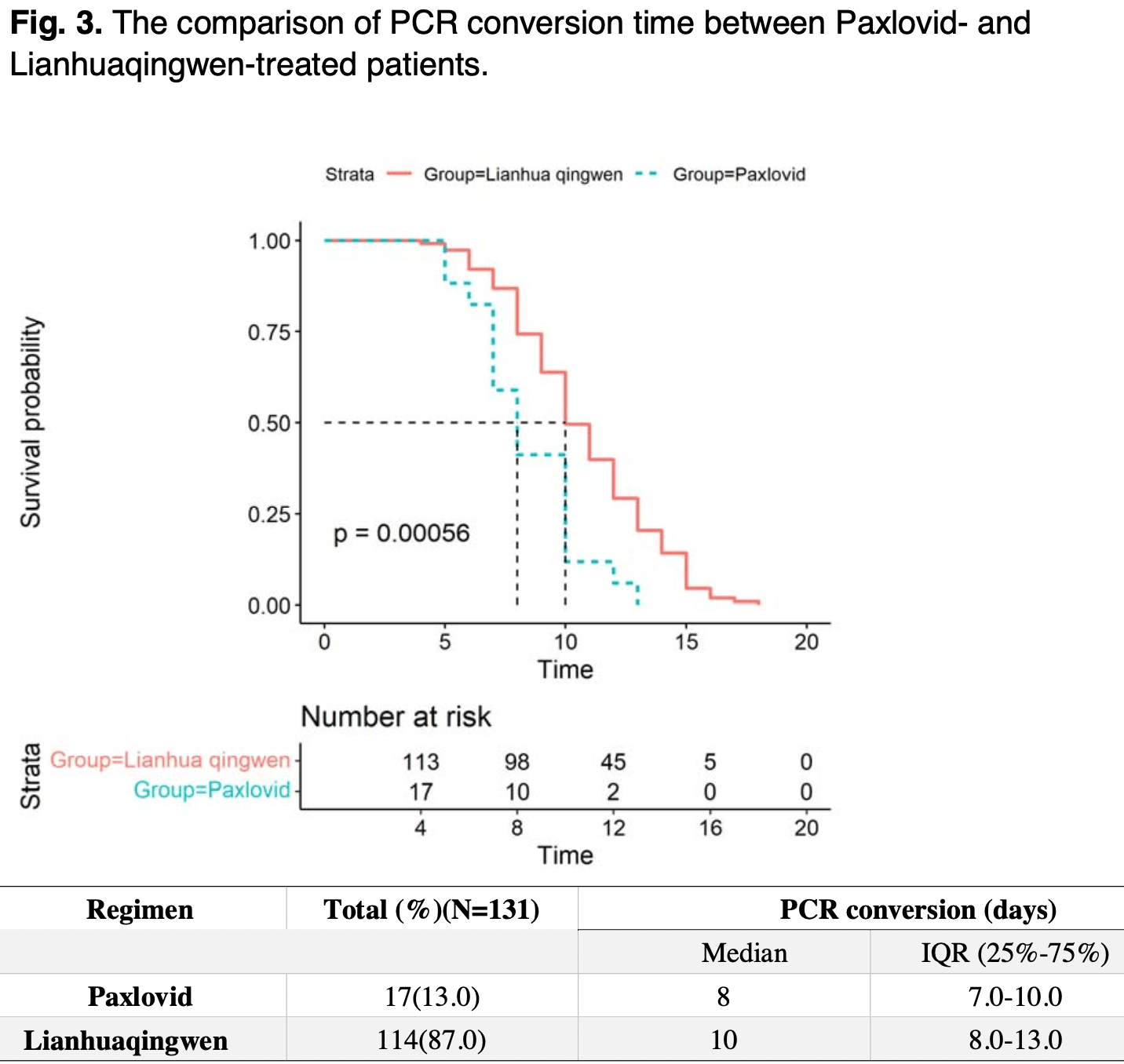
Retrospective 17 paxlovid and 114 lianhuaqingwen patients in China, showing faster viral clearance with paxlovid.
Resistance. Variants may be resistant to paxlovid1-8. Use may promote the emergence of variants that weaken host immunity and potentially contribute to long COVID9. Confounding by contraindication. Hoertel et al. find that over 50% of patients that died had a contraindication for the use of Paxlovid10. Retrospective studies that do not exclude contraindicated patients may significantly overestimate efficacy. Black box warning. The FDA notes that severe, life-threatening, and/or fatal adverse reactions due to drug interactions have been reported in patients treated with paxlovid11. Kidney and liver injury. Studies show significantly increased risk of acute kidney injury12 and liver injury13,14. Viral rebound. Studies show significantly increased risk of replication-competent viral rebound15-17.
Standard of Care (SOC) for COVID-19 in the study country,
China, is average with moderate efficacy for approved treatments18.
This study is excluded in the after exclusion results of meta-analysis:
unadjusted results with no group details.
|
time to viral-, 20.0% lower, relative time 0.80, p < 0.001, treatment 17, control 114.
|
| Effect extraction follows pre-specified rules prioritizing more serious outcomes. Submit updates |
1.
Zhou et al., Nirmatrelvir-resistant SARS-CoV-2 variants with high fitness in an infectious cell culture system, Science Advances, doi:10.1126/sciadv.add7197.
2.
Moghadasi et al., Rapid resistance profiling of SARS-CoV-2 protease inhibitors, npj Antimicrobials and Resistance, doi:10.1038/s44259-023-00009-0.
3.
Jochmans et al., The Substitutions L50F, E166A, and L167F in SARS-CoV-2 3CLpro Are Selected by a Protease Inhibitor In Vitro and Confer Resistance To Nirmatrelvir, mBio, doi:10.1128/mbio.02815-22.
4.
Lopez et al., SARS-CoV-2 Resistance to Small Molecule Inhibitors, Current Clinical Microbiology Reports, doi:10.1007/s40588-024-00229-6.
5.
Zvornicanin et al., Molecular Mechanisms of Drug Resistance and Compensation in SARS-CoV-2 Main Protease: The Interplay Between E166 and L50, bioRxiv, doi:10.1101/2025.01.24.634813.
6.
Vukovikj et al., Impact of SARS-CoV-2 variant mutations on susceptibility to monoclonal antibodies and antiviral drugs: a non-systematic review, April 2022 to October 2024, Eurosurveillance, doi:10.2807/1560-7917.ES.2025.30.10.2400252.
7.
Deschenes et al., Functional and structural characterization of treatment-emergent nirmatrelvir resistance mutations at low frequencies in the main protease (Mpro) reveals a unique evolutionary route for SARS-CoV-2 to gain resistance, The Journal of Infectious Diseases, doi:10.1093/infdis/jiaf294.
8.
Zhou (B) et al., SARS-CoV-2 Mpro inhibitor ensitrelvir: asymmetrical cross-resistance with nirmatrelvir and emerging resistance hotspots, Emerging Microbes & Infections, doi:10.1080/22221751.2025.2552716.
9.
Thomas et al., Nirmatrelvir-Resistant Mutations in SARS-CoV-2 Mpro Enhance Host Immune Evasion via Cleavage of NF-κB Essential Modulator, bioRxiv, doi:10.1101/2024.10.18.619137.
10.
Hoertel et al., Prevalence of Contraindications to Nirmatrelvir-Ritonavir Among Hospitalized Patients With COVID-19 at Risk for Progression to Severe Disease, JAMA Network Open, doi:10.1001/jamanetworkopen.2022.42140.
11.
FDA, Fact sheet for healthcare providers: emergency use authorization for paxlovid, www.fda.gov/media/155050/download.
12.
Kamo et al., Association of Antiviral Drugs for the Treatment of COVID-19 With Acute Renal Failure, In Vivo, doi:10.21873/invivo.13637.
13.
Wang et al., Development and validation of a nomogram to assess the occurrence of liver dysfunction in patients with COVID-19 pneumonia in the ICU, BMC Infectious Diseases, doi:10.1186/s12879-025-10684-1.
14.
Siby et al., Temporal Trends in Serious Adverse Events Associated with Oral Antivirals During the COVID-19 Pandemic: Insights from the FAERS Database (2020–2023), Open Forum Infectious Diseases, doi:10.1093/ofid/ofaf695.1825.
15.
Edelstein et al., SARS-CoV-2 virologic rebound with nirmatrelvir-ritonavir therapy, medRxiv, doi:10.1101/2023.06.23.23288598.
16.
Shah et al., SARS-CoV-2 infectious shedding and rebound among adults with and without oral antiviral use: two case-ascertained prospective household studies, The Lancet Microbe, doi:10.1016/j.lanmic.2025.101227.
Shao et al., 6 Jul 2022, retrospective, China, preprint, median age 52.0, 12 authors, study period 6 April, 2022 - 11 May, 2022, this trial compares with another treatment - results may be better when compared to placebo.
Contact: huanggang@sumhs.edu.cn, hujianrong@jdhospital.com.
Clinical advancement of patients infected with SARS-CoV-2 Omicron variant in Shanghai, China
doi:10.1101/2022.07.03.22277169
All rights reserved. No reuse allowed without permission. (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity.
References
Abas, Marfuah, Idroes, Kusumawaty, Fatimawali et al., Can the SARS-CoV-2 Omicron Variant Confer Natural Immunity against COVID-19?, Molecules, doi:10.3390/molecules2707222
Anjos, Fiaccadori, Servian, Da Fonseca, Guilarde et al., SARS-CoV-2 loads in urine, sera and stool specimens in association with clinical features of COVID-19 patients, J Clin Virol Plus, doi:10.1016/j.jcvp.2021.100059
Bennasrallah, Zemni, Dhouib, Sriha, Mezhoud et al., Factors associated with a prolonged negative conversion of viral RNA in patients with COVID-19, Int J Infect Dis, doi:10.1016/j.ijid.2021.02.089
Butt, Dargham, Loka, Shaik, Chemaitelly et al., COVID-19 Disease Severity in Children Infected with the Omicron Variant, Clin Infect Dis, doi:10.1093/cid/ciac275
Cann, Fever: Could A Cardinal Sign of COVID-19 Infection Reduce Mortality?, Am J Med Sci, doi:10.1016/j.amjms.2021.01.004
Chen, Qi, Liu, Ling, Qian et al., Clinical progression of patients with COVID-19 in Shanghai, China, J Infect, doi:10.1016/j.jinf.2020.03.004
Choi, Kang, Kim, Choi, Joh et al., Clinical Presentation and Outcomes of Middle East Respiratory Syndrome in the Republic of Korea, Infect Chemother
Esr, None
Graham, Daily briefing: Omicron struggles to infect the lungs, Nature, doi:10.3947/ic.2016.48.2.118
Gupta, SARS-CoV-2 Omicron spike mediated immune escape and tropism shift, Res Sq
He, Lau, Wu, Deng, Wang et al., Temporal dynamics in viral shedding and transmissibility of COVID-19, Nat Med, doi:10.1038/s41591-020-0869-5
Huang, Zeng, Letter to the editor: Epidemiology of the SARS-CoV-2 variant Omicron BA.2 -vigilance needed, Euro Surveill, doi:10.2807/1560-7917.ES.2022.27.13.2200254
Hui, Ho, Cheung, Ng, Ching et al., SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo, Nature
Kim, Lee, Choi, Um, Lee et al., Clinical Characteristics of 40 Patients Infected With the SARS-CoV-2 Omicron Variant in Korea, J Korean Med Sci, doi:10.3346/jkms.2022.37.e31
Lee, Choe, Jeong, Kim, Kim et al., Importation and Transmission of SARS-CoV-2 B.1.1.529 (Omicron) Variant of Concern in Korea, November 2021, J Korean Med Sci, doi:10.3346/jkms.2021.36.e346
Liang, Hui, Liu, Qiao, Li et al., Insights into forsythia honeysuckle (Lianhuaqingwen) capsules: A Chinese herbal medicine repurposed for COVID-19 pandemic, Phytomed Plus, doi:10.1016/j.phyplu.2021.100027
Liu, Gao, Yuan, Yang, Shi et al., Efficacy and safety of herbal medicine (Lianhuaqingwen) for treating COVID-19: A systematic review and meta-analysis
Martin, Dewitt, Russell, Sanchez-Pinto, Haendel et al., Acute Upper Airway Disease in Children With the Omicron (B.1.1.529) Variant of SARS-CoV-2-A Report From the US National COVID Cohort Collaborative, JAMA Pediatr, doi:10.1001/jamapediatrics.2022.1110
Maslo, Friedland, Toubkin, Laubscher, Akaloo et al., Characteristics and Outcomes of Hospitalized Patients in South Africa During the COVID-19 Omicron Wave Compared With Previous Waves, JAMA, doi:10.1001/jama.2021.24868
Mefsin, Bond, Lin, Cheung, Wong et al., Epidemiology of infections with SARS-CoV-2 Omicron BA.2 variant in Hong Kong, doi:10.2807/1560-7917.ES.2022.27.18.2200178
Nhcotpsro, Notice on printing and distributing the COVID-19 pneumonia diagnosis and treatment plan (
Rossler, Riepler, Bante, Laer, Kimpel, SARS-CoV-2 Omicron Variant Neutralization in Serum from Vaccinated and Convalescent Persons, N Engl J Med, doi:10.1056/NEJMc2119236
Runfeng, Yunlong, Jicheng, Weiqi, Qinhai et al., Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2), Pharmacol Res, doi:10.1016/j.phrs.2020.104761
Singhal, The Emergence of Omicron: Challenging Times Are Here Again!, Indian J Pediatr, doi:10.1007/s12098-022-04077-4
Sun, Li, Huang, Shi, Xing et al., Case Report of China/Tianjin's First Novel Coronavirus Variant Omicron, Iran J Immunol, doi:10.22034/IJI.2022.94050.2278
Team, Omicron) Variant -United States, December 1, MMWR Morb Mortal Wkly Rep, doi:10.15585/mmwr.mm7050e1
Thakur, Ratho, A new SARS-CoV-2 variant of concern mounting worldwide fear, J Med Virol, doi:10.1002/jmv.27541
Torjesen, Covid-19: Peak of viral shedding is later with omicron variant, Japanese data suggest, BMJ, doi:10.1136/bmj.o89
Wang, Prather, Sznitman, Jimenez, Lakdawala et al., Airborne transmission of respiratory viruses, Science, doi:10.1126/science.abd9149
Widders, Broom, Broom, SARS-CoV-2: The viral shedding vs infectivity dilemma, Infect Dis Health, doi:10.1016/j.idh.2020.05.002
Wu, Guo, Tang, Hong, Zhou et al., Prolonged presence of SARS-CoV-2 viral RNA in faecal samples, Lancet Gastroenterol Hepatol, doi:10.1016/S2468-1253(20)30083-2.130-175130.0(121.0-141.0)138.0(131.0-143.0)130.0
Yan, Zhang, Chen, Jiang, Liu et al., Characteristics of Viral Shedding Time in SARS-CoV-2 Infections: A Systematic Review and Meta-Analysis, Front Public Health, doi:10.3389/fpubh.2021.652842
Yuan, Hou, Lin, Chen, Ren, How China responds to Omicron, J Infect, doi:10.1016/j.jinf.2022.04.017
DOI record:
{
"DOI": "10.1101/2022.07.03.22277169",
"URL": "http://dx.doi.org/10.1101/2022.07.03.22277169",
"abstract": "<jats:p>Background\n\nStudies on the Omicron variant infection have generally been restricted to descriptions of its initial clinical and epidemiological characteristics, we investigated the timeline-related advancement in individuals with Omicron variant.\n\nMethods\n \nWe conducted a retrospective, single centered study including 226 laboratory confirmed cases with Omicron variant between April 6th and May 11th, 2022 in Shanghai, China. Final date of follow-up was May 30, 2022.\n\nResults\n\nAmong 226 enrolled patients, the median age was 52 years old, and 118 (52.2%) were female. The duration from onset of symptoms to hospitalization was 3 (IQR, 2-4) days for symptomatic patients. Cough occurred in 168 patients (74.3% ). The median interval to negative reverse-transcriptase PCR tests of nasopharynx swab was 10 days (interquartile range [IQR]: 8-13 days). No radiographic progressions were found in 196 patients on the 7th day after onset of symptoms. The median duration of fever in all participants was 5 days (IQR: 4-6 days). The median PCR conversion time of Paxlovid-treated patients was 8 days (IQR: 7-10 days) compared with that of Lianhuaqingwen (10 days, IQR: 8-13 days) (p=0.00056). Booster vaccination can significantly decrease the severity of Omicron infection when compared with unvaccinated, partially or fully vaccinated patients (p=0.009). The median time of PCR conversion in individuals aged under 14 recovered significantly faster than older patients (median 8 days versus 11 days, P < 0.0001). In multivariate logistic analysis, erythrocyte sedimentation rate (ESR) (OR=1.05) was independently related to the severity of the infection.\n \nConclusions\n\nThe majority clinical symptoms of Omicron infection were not severe. Early and aggressive administration of Paxlovid can significantly reduce the PCR conversion time. Booster vaccination should also be highly recommended in population over 14 years old.</jats:p>",
"accepted": {
"date-parts": [
[
2022,
7,
5
]
]
},
"author": [
{
"affiliation": [],
"family": "Shao",
"given": "Jiasheng",
"sequence": "first"
},
{
"affiliation": [],
"family": "Fan",
"given": "Rong",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Zhang",
"given": "Tiejun",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Lee",
"given": "Catherine",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Huang",
"given": "Xuyuan",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Wang",
"given": "Fei",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Liang",
"given": "Haiying",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Jin",
"given": "Ye",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Jiang",
"given": "Ying",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Gu",
"given": "Yanhua",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Hu",
"given": "Jianrong",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Huang",
"given": "Gang",
"sequence": "additional"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2022,
7,
6
]
],
"date-time": "2022-07-06T14:30:14Z",
"timestamp": 1657117814000
},
"deposited": {
"date-parts": [
[
2022,
7,
6
]
],
"date-time": "2022-07-06T14:30:15Z",
"timestamp": 1657117815000
},
"group-title": "Infectious Diseases (except HIV/AIDS)",
"indexed": {
"date-parts": [
[
2022,
7,
6
]
],
"date-time": "2022-07-06T15:17:16Z",
"timestamp": 1657120636413
},
"institution": [
{
"name": "medRxiv"
}
],
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2022,
7,
6
]
]
},
"link": [
{
"URL": "https://syndication.highwire.org/content/doi/10.1101/2022.07.03.22277169",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "246",
"original-title": [],
"posted": {
"date-parts": [
[
2022,
7,
6
]
]
},
"prefix": "10.1101",
"published": {
"date-parts": [
[
2022,
7,
6
]
]
},
"publisher": "Cold Spring Harbor Laboratory",
"reference-count": 0,
"references-count": 0,
"relation": {},
"resource": {
"primary": {
"URL": "http://medrxiv.org/lookup/doi/10.1101/2022.07.03.22277169"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subtitle": [],
"subtype": "preprint",
"title": "Clinical advancement of patients infected with SARS-CoV-2 Omicron variant in Shanghai, China",
"type": "posted-content"
}