Exploring potential therapeutic candidates against COVID-19: a molecular docking study
et al., Discover Molecules, doi:10.1007/s44345-024-00005-5, Nov 2024
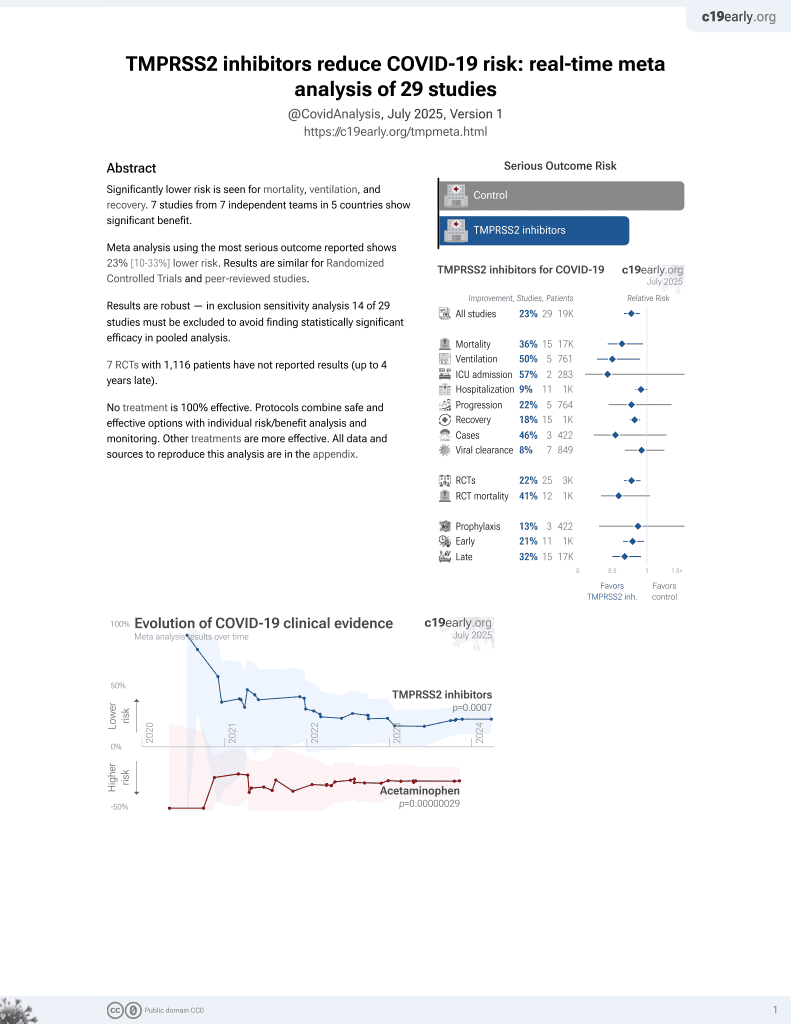
22nd treatment shown to reduce risk in
April 2021, now with p = 0.00063 from 29 studies.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
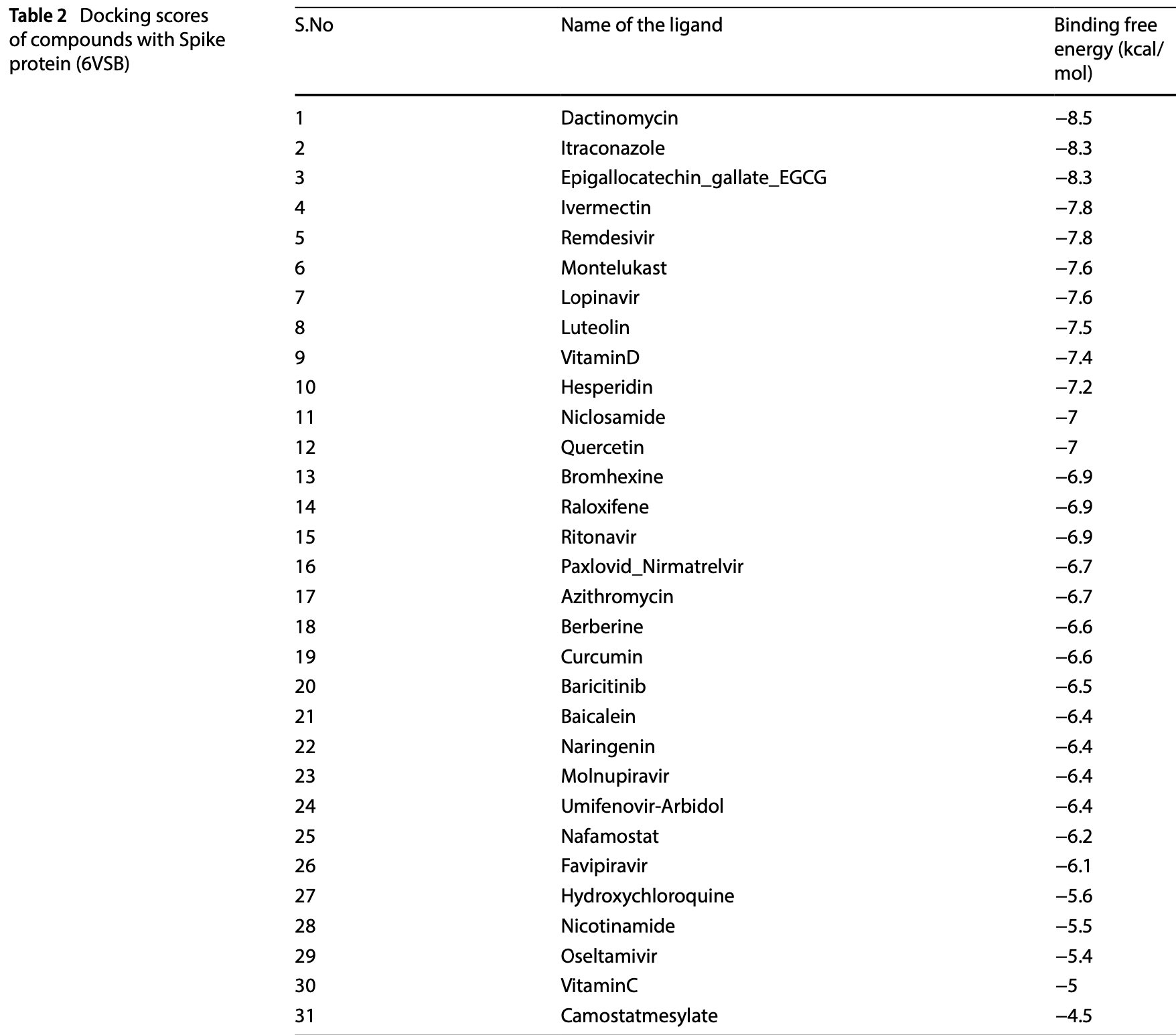
In silico study showing potential inhibition of SARS-CoV-2 proteins by various compounds including dactinomycin, itraconazole, ivermectin, vitamin D, quercetin, curcumin, montelukast, bromhexine, hesperidin, EGCG and raloxifene. Authors performed molecular docking of 31 compounds against 6 SARS-CoV-2 proteins: spike protein, main protease, RNA-dependent RNA polymerase, papain-like protease, helicase, and nucleocapsid protein. Dactinomycin and itraconazole showed the highest binding affinity for the spike protein, while EGCG, hesperidin, ivermectin and raloxifene showed strong binding to other viral proteins.
13 preclinical studies support the efficacy of TMPRSS2 inhibitors for COVID-19:
Study covers TMPRSS2 inhibitors, bromhexine, vitamin D, ivermectin, quercetin, curcumin, montelukast, and niclosamide.
1.
Haque et al., Exploring potential therapeutic candidates against COVID-19: a molecular docking study, Discover Molecules, doi:10.1007/s44345-024-00005-5.
2.
González-Paz et al., Biophysical Analysis of Potential Inhibitors of SARS-CoV-2 Cell Recognition and Their Effect on Viral Dynamics in Different Cell Types: A Computational Prediction from In Vitro Experimental Data, ACS Omega, doi:10.1021/acsomega.3c06968.
3.
Umar et al., Inhibitory potentials of ivermectin, nafamostat, and camostat on spike protein and some nonstructural proteins of SARS-CoV-2: Virtual screening approach, Jurnal Teknologi Laboratorium, doi:10.29238/teknolabjournal.v11i1.344.
4.
Unal et al., Favipiravir, umifenovir and camostat mesylate: a comparative study against SARS-CoV-2, bioRxiv, doi:10.1101/2022.01.11.475889.
5.
Sgrignani et al., Computational Identification of a Putative Allosteric Binding Pocket in TMPRSS2, Frontiers in Molecular Biosciences, doi:10.3389/fmolb.2021.666626.
6.
Yathindranath et al., Lipid Nanoparticle-Based Inhibitors for SARS-CoV-2 Host Cell Infection, International Journal of Nanomedicine, doi:10.2147/IJN.S448005.
7.
Martins et al., In Vitro Inhibition of SARS-CoV-2 Infection by Bromhexine hydrochloride, bioRxiv, doi:10.1101/2022.12.23.521817.
8.
Schultz et al., Pyrimidine inhibitors synergize with nucleoside analogues to block SARS-CoV-2, Nature, doi:10.1038/s41586-022-04482-x.
9.
Hempel et al., Synergistic inhibition of SARS-CoV-2 cell entry by otamixaban and covalent protease inhibitors: pre-clinical assessment of pharmacological and molecular properties, Chemical Science, doi:10.1039/D1SC01494C.
Haque et al., 21 Nov 2024, peer-reviewed, 3 authors.
Contact: sukanta999bhadra@gmail.com.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Exploring potential therapeutic candidates against COVID-19: a molecular docking study
Discover Molecules, doi:10.1007/s44345-024-00005-5
The COVID-19 pandemic, caused by SARS-CoV-2, has made it urgently necessary to develop effective therapeutic options, and in recent years, computational biological tool assisted to achieve this goal. We conducted MD simulations using six SARS-CoV-2 proteins and 31 ligands to identify possible inhibitors as well as evaluate the binding efficiency of them. Post simulation study showed that 6VSB-Dactinomycin complex reveals the most stable binding, protein flexibility, and robust structural integrity, making it a promising model for therapeutic studies. Our study assessed the therapeutic potential of antiviral candidates against covid-19 virus using computational and experimental method, including the flexibility and stability of twelve docked protein-ligand complexes using normal mode analysis (NMA) and molecular dynamics (MD) simulations. After MD simulation, Dactinomycin and Ivermectin showed significant deformability and high binding affinities for Spike protein and RNA-dependent RNA polymerase, respectively. Remarkably, Dactinomycin showed greater binding to the Helicase protein, but Hesperidin and Epigallocatechin gallate (EGCG) exhibited encouraging interactions with the Nucleocapsid protein and Main protease. Analysis of docking experiments and ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) showed the toxicity profiles and binding affinity of various drugs with key viral proteins. Toxicity of major drugs exhibited low to moderate although Dactinomycin, Ivermectin and vitamin-D exhibited higher degree of toxicity. Carcinogenicity observed in Quercetin, Baricitinib, Luteolin, Berberine, and Favipiravir while hepatotoxicity was not found in most case. Although EGCG and Hesperidin exhibit potential, more studies are required to evaluate effectiveness against approved drugs such as Remdesivir. This integrated approach shows that an integration of computational predictions with experimental data can help to support the development of antiviral drugs by adding novel perspectives in the safety profiles and efficiency of potential drugs.
Author contributions E.H. wrote the main manuscript text and prepared all figures. S. B. prepared figures, revised and supervised and N.K conceptualized the work. All authors reviewed the manuscript.
Declarations Competing Interests The authors declare no competing interests. Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http:// creat iveco mmons. org/ licen ses/ by-nc-nd/4. 0/. Publisher's Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Angkasekwinai, Rattanaumpawan, Chayakulkeeree, Phoompoung, Koomanachai et al., Safety and efficacy of ivermectin for the prevention and treatment of covid-19: a double-blinded randomized placebocontrolled study, Antibiotics, doi:10.3390/antibiotics11060796
Arévalo, Pagotto, Pórfido, Daghero, Segovia et al., Ivermectin reduces in vivo coronavirus infection in a mouse experimental model, Sci Rep, doi:10.1038/s41598-021-86679-0
Baldi, Computational approaches for drug design and discovery: an overview, Syst Rev Pharm, doi:10.4103/0975-8453.59519
Banerjee, Perera, Tillekeratne, Potential SARS-CoV-2 main protease inhibitors, Drug Discov Today, doi:10.1016/j.drudis.2020.12.005
Batalha, Forezi, Lima, Pauli, Boechat et al., Drug repurposing for the treatment of COVID-19: pharmacological aspects and synthetic approaches, Bioorg Chem, doi:10.1016/j.bioorg.2020.104488
Bellavite, Reappraisal of dietary phytochemicals for coronavirus infection focus on hesperidin and quercetin
Bioviads, Discovery Studio
Caly, Druce, Catton, Jans, Wagstaff, The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro, Antiviral Res, doi:10.1016/j.antiviral.2020.104787
Chen, Jia, Chen, Wang, Hu, The pharmacokinetics of raloxifene and its interaction with apigenin in rat, Molecules, doi:10.3390/molecules15118478
Cheng, Huynh, Yang, Hu, Shen et al., Hesperidin Is a Potential Inhibitor against SARS-CoV-2 Infection, Nutrients, doi:10.3390/nu13082800
Ciotti, Ciccozzi, Terrinoni, Jiang, Wang et al., The COVID-19 pandemic, Crit Rev Clin Lab Sci, doi:10.1080/10408363.2020.1783198
Costa, Vale, A review of repurposed cancer drugs in clinical trials for potential treatment of COVID-19, Pharmaceutics, doi:10.3390/pharmaceutics13060815
Das, Sarmah, Lyndem, Roy, An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study, J Biomol Struct Dyn, doi:10.1080/07391102.2020.1763201
Dearden, In silico prediction of drug toxicity, J Comput-Aided Mol Design, doi:10.1023/A:1025361621494
Drwal, Banerjee, Dunkel, Wettig, Preissner, ProTox: a web server for the in silico prediction of rodent oral toxicity, Nucleic Acids Res, doi:10.1093/nar/gku401
Dutta, Mazumdar, Gordy, The nucleocapsid protein of SARS-CoV-2: a target for vaccine development, J Virol, doi:10.1128/jvi.00647-20
Gaurav, Agrawal, Al-Nema, Gautam, Computational approaches in the discovery and development of therapeutic and prophylactic agents for viral diseases, Curr Top Med Chem, doi:10.2174/1568026623666221019110334
Gelderblom, Snyman, Abel, Lebepe-Mazur, Smuts et al., Hepatotoxicity and-carcinogenicity of the fumonisins in rats: a review regarding mechanistic implications for establishing risk in humans, Fumonisins Food, doi:10.1007/978-1-4899-1379-1_24
Ghazwani, Bakheit, Hakami, Alkahtani, Almehizia, Virtual screening and molecular docking studies for discovery of potential RNA-dependent RNA polymerase inhibitors, Crystals, doi:10.3390/cryst11050471
Gupta, Savytskyi, Coban, Venugopal, Pleqi et al., Protein structure-based in-silico approaches to drug discovery: guide to COVID-19 therapeutics, Mol Aspects Med, doi:10.1016/j.mam.2022.101151
Gupta, Singh, Kushwaha, Prajapati, Shuaib et al., Identification of potential natural inhibitors of SARS-CoV2 main protease by molecular docking and simulation studies, J Biomol Struct Dyn, doi:10.1080/07391102.2020.1776157
Habtemariam, Nabavi, Banach, Berindan-Neagoe, Sarkar et al., Should we try SARS-CoV-2 helicase inhibitors for COVID-19 therapy?, Arch Med Res, doi:10.1016/j.arcmed.2020.05.024
Huff, Kummetha, Tiwari, Huante, Clark et al., Discovery and mechanism of SARS-CoV-2 main protease inhibitors, J Med Chem, doi:10.1021/acs.jmedchem.1c00566
Humeau, Sauvat, Cerrato, Xie, Loos et al., Inhibition of transcription by dactinomycin reveals a new characteristic of immunogenic cell stress, EMBO Mol Med, doi:10.1525/emmm.201911622
Hurst, Dickinson, Hsu, Epigallocatechin-3-gallate (EGCG) Inhibits SARS-CoV-2 infection in primate epithelial cells: (a short communication), Microbiol Infect Dis, doi:10.3342/2639-9458.1116
Iaconis, Bordi, Matusali, Talarico, Manelfi et al., Characterization of raloxifene as a potential pharmacological agent against SARS-CoV-2 and its variants, Cell Death Dis, doi:10.1038/s41419-022-04961-z
Jean, Lee, Hsueh, Treatment options for COVID-19: the reality and challenges, J Microbiol Immunol Infect, doi:10.1016/j.jmii.2020.03.034v
Jia, Yan, Ren, Wu, Wang et al., Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis, Nucleic Acids Res, doi:10.1093/nar/gkz409
Jiang, Yin, Xu, RNA-dependent RNA polymerase: Structure, mechanism, and drug discovery for COVID-19, Biochem Biophys Res Commun, doi:10.1016/j.bbrc.2020.08.116
Kang, Yang, Hong, Zhang, Huang et al., Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites, Acta PharmaceuticaSinica B, doi:10.1016/j.apsb.2020.04.009
Kennedy, Cm, Inhibition of coronavirus 229E replication by actinomycin D, J Virol, doi:10.1128/JVI.29.1.401-404.1979
Klemm, Ebert, Calleja, Allison, Richardson et al., Mechanism and inhibition of the papain-like protease PLpro of SARS-CoV-2, EMBO J, doi:10.1525/embj.2020106275
Kowalczyk, Hesperidin, a potential antiviral agent against SARS-CoV-2: the influence of citrus consumption on COVID-19 incidence and severity in China, Medicina, doi:10.3390/medicina60060892
Langholz, Skolnik, Barrett, Renbarger, Seibel et al., Dactinomycin and vincristine toxicity in the treatment of childhood cancer: a retrospective study from the children's oncology group, Pediatr Blood Cancer, doi:10.1002/pbc.22882
Lestner, Hope, Itraconazole: an update on pharmacology and clinical use for treatment of invasive and allergic fungal infections, Expert Opin Drug Metab Toxicol, doi:10.1517/17425255.2013.794785
Li, Hilgenfeld, Whitley, Clercq, Therapeutic strategies for COVID-19: progress and lessons learned, Nat Rev Drug Discov
Li, Li, Li, Du, Zhang et al., In vivo pharmacokinetics of hesperidin are affected by treatment with glucosidase-like BglA protein isolated from yeasts, J Agric Food Chem, doi:10.1021/jf800105c
Li, Luan, Zhang, Molecular docking of potential SARS-CoV-2 papain-like protease inhibitors, Biochem Biophys Res Commun, doi:10.1016/j.bbrc.2020.11.083
Liesenborghs, Spriet, Jochmans, Belmans, Gyselinck et al., Itraconazole for COVID-19: preclinical studies and a proof-of-concept randomized clinical trial, EBioMedicine, doi:10.1016/j.ebiom.2021.103288
Liu, Bodnar, Meng, Khan, Wang et al., Epigallocatechingallate from green tea effectively blocks infection of SARS-CoV-2 and new variants by inhibiting spike binding to ACE2 receptor, Cell Biosci, doi:10.1186/s13578-021-00680-8
Madabhavi, Sarkar, Kadakol, COVID-19: a review, Monaldi Arch Chest Dis
Madhavisastry, Adzhigirey, Day, Annabhimoju, Sherman, Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments, J Comput-Aided Mol Design, doi:10.1007/s10822-013-9644-8
Mongia, Saha, Chouzenoux, Majumdar, A computational approach to aid clinicians in selecting anti-viral drugs for COVID-19 trials, Sci Rep, doi:10.1038/s41598-021-88153-3
Morris, Huey, Lindstrom, Sanner, Belew et al., Autodock4 and AutoDockTools4: automated docking with selective receptor flexiblity, J Comput Chem
Nicastri, Marinangeli, Pivetta, Reggiani, Fiorentino et al., A phase 2 randomized, double-blinded, placebo-controlled, multicenter trial evaluating the efficacy and safety of raloxifene for patients with mild to moderate COVID-19, EClinicalMedicine, doi:10.1016/j.eclinm.2022.101450
Noga, Michalska, Jurowski, Application of toxicology in silico methods for prediction of acute toxicity (LD50) for Novichoks, Arch Toxic, doi:10.1007/s00204-023-03507-2
Park, Park, Jang, Park, Therapeutic potential of EGCG, a green tea polyphenol, for treatment of coronavirus diseases, Life, doi:10.3390/life11030197
Parvez, Karim, Hasan, Jaman, Karim et al., Prediction of potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2 using comprehensive drug repurposing and molecular docking approach, Int J Biol Macromol, doi:10.1016/j.ijbiomac.2020.09.098
Peng, Du, Lei, Dorje, Qi et al., Structures of the SARS-CoV-2 nucleocapsid and their perspectives for drug design, EMBO J, doi:10.3390/v13061115
Pitsillou, Liang, Ververis, Hung, Karagiannis, Interaction of small molecules with the SARS-CoV-2 papain-like protease: In silico studies and in vitro validation of protease activity inhibition using an enzymatic inhibition assay, J Mol Graph Model, doi:10.1016/j.jmgm.2021.107851
Pitsillou, Liang, Ververis, Lim, Hung et al., Identification of small molecule inhibitors of the deubiquitinating activity of the SARS-CoV-2 papain-like protease: in silico molecular docking studies and in vitro enzymatic activity assay, Front Chem, doi:10.3389/fchem.2020.623971
Ravi, Kannabiran, A handbook on protein-ligand docking tool AutoDock 4, Innovare J Med Sci
Schlede, Mischke, Diener, Kayser, The international validation study of the acute toxic class method (oral), Arch Toxic, doi:10.1007/s002040050229
Schrödinger, Delano, None, PyMOL
Shi, Wang, Cai, Deng, Zheng et al., An overview of COVID-19, J Zhejiang Univ Sci B, doi:10.1631/jzus.B2000083
Skariyachan, Gopal, Chakrabarti, Kempanna, Uttarkar et al., Structural and molecular basis of the interaction mechanism of selected drugs towards multiple targets of SARS-CoV-2 by molecular docking and dynamic simulation studiesdeciphering the scope of repurposed drugs, Comput Biol Med, doi:10.1016/j.compbiomed.2020.104054
Soussi, Gargouri, Magné, Ben-Nasr, Kausar et al., -)-Epigallocatechingallate (EGCG) pharmacokinetics and molecular interactions towards amelioration of hyperglycemia, hyperlipidemia associated hepatorenal oxidative injury in alloxan induced diabetic mice, Chem Biol Interact, doi:10.1016/j.cbi.2022.110230
Sternberg, Naujokat, Structural features of coronavirus SARS-CoV-2 spike protein: targets for vaccination, Life Sci, doi:10.1016/j.lfs.2020.118056
Tzenios, Chahine, Tazanios, Better strategies for coronavirus (COVID-19) vaccination, Special J Med Acad Life Sci, doi:10.5867/sjmas.v1i1.11
Van Damme, Meyer, Bojkova, Ciesek, Cinatl et al., In vitro activity of itraconazole against SARS-CoV-2, J Med Virol, doi:10.1002/jmv.26917
Vivek-Ananth, Krishnaswamy, Samal, Potential phytochemical inhibitors of SARS-CoV-2 helicase Nsp13: a molecular docking and dynamic simulation study, Mol Divers, doi:10.1007/s11030-021-10251-1
White, Cheng, Discovery of COVID-19 inhibitors targeting the SARS-CoV-2 Nsp13 helicase, J Phys Chem Lett, doi:10.1021/acs.jpclett.0c02421
Xia, Domains and functions of spike protein in Sars-Cov-2 in the context of vaccine design, Viruses, doi:10.3390/v13010109
Yadav, Parihar, Dhiman, Dhamija, Upadhyay et al., Docking of fda approved drugs targeting nsp-16, n-protein and main protease of sars-cov-2 as dual inhibitors, Biointerface Res Appl Chem, doi:10.3326/BRIAC113.98489861
Yin, Mao, Luan, Shen, Shen et al., Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir, Science, doi:10.1126/science.abc1560
Yuki, Fujiogi, Koutsogiannaki, COVID-19 pathophysiology: a review, Clin Immunol, doi:10.1016/j.clim.2020.108427
Zhang, Xiao, Cai, Chen, Structure of SARS-CoV-2 spike protein, Curr Opin Virol, doi:10.1016/j.coviro.2021.08.010
Zhu, Chen, Gorshkov, Xu, Lo et al., RNA-dependent RNA polymerase as a target for COVID-19 drug discovery, SLAS Disco, doi:10.1177/2472555220942123
DOI record:
{
"DOI": "10.1007/s44345-024-00005-5",
"ISSN": [
"3004-9350"
],
"URL": "http://dx.doi.org/10.1007/s44345-024-00005-5",
"alternative-id": [
"5"
],
"article-number": "5",
"assertion": [
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Received",
"name": "received",
"order": 1,
"value": "9 May 2024"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "11 November 2024"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "First Online",
"name": "first_online",
"order": 3,
"value": "21 November 2024"
},
{
"group": {
"label": "Declarations",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 1
},
{
"group": {
"label": "Competing Interests",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 2,
"value": "The authors declare no competing interests."
}
],
"author": [
{
"affiliation": [],
"family": "Haque",
"given": "S. k. Erfanul",
"sequence": "first"
},
{
"affiliation": [],
"family": "Bhadra",
"given": "Sukanta",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Pal",
"given": "Nishith Kumar",
"sequence": "additional"
}
],
"container-title": "Discover Molecules",
"container-title-short": "Discov Mol",
"content-domain": {
"crossmark-restriction": false,
"domain": [
"link.springer.com"
]
},
"created": {
"date-parts": [
[
2024,
11,
21
]
],
"date-time": "2024-11-21T15:59:41Z",
"timestamp": 1732204781000
},
"deposited": {
"date-parts": [
[
2024,
11,
21
]
],
"date-time": "2024-11-21T16:14:53Z",
"timestamp": 1732205693000
},
"indexed": {
"date-parts": [
[
2024,
11,
22
]
],
"date-time": "2024-11-22T05:25:14Z",
"timestamp": 1732253114569,
"version": "3.28.0"
},
"is-referenced-by-count": 0,
"issue": "1",
"issued": {
"date-parts": [
[
2024,
11,
21
]
]
},
"journal-issue": {
"issue": "1",
"published-online": {
"date-parts": [
[
2024,
12
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by-nc-nd/4.0",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
11,
21
]
],
"date-time": "2024-11-21T00:00:00Z",
"timestamp": 1732147200000
}
},
{
"URL": "https://creativecommons.org/licenses/by-nc-nd/4.0",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
11,
21
]
],
"date-time": "2024-11-21T00:00:00Z",
"timestamp": 1732147200000
}
}
],
"link": [
{
"URL": "https://link.springer.com/content/pdf/10.1007/s44345-024-00005-5.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://link.springer.com/article/10.1007/s44345-024-00005-5/fulltext.html",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://link.springer.com/content/pdf/10.1007/s44345-024-00005-5.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "297",
"original-title": [],
"prefix": "10.1007",
"published": {
"date-parts": [
[
2024,
11,
21
]
]
},
"published-online": {
"date-parts": [
[
2024,
11,
21
]
]
},
"publisher": "Springer Science and Business Media LLC",
"reference": [
{
"DOI": "10.1016/j.clim.2020.108427",
"author": "K Yuki",
"doi-asserted-by": "publisher",
"journal-title": "Clin Immunol",
"key": "5_CR1",
"unstructured": "Yuki K, Fujiogi M, Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin Immunol. 2020;215: 108427. https://doi.org/10.1016/j.clim.2020.108427.",
"volume": "215",
"year": "2020"
},
{
"DOI": "10.1080/10408363.2020.1783198",
"author": "M Ciotti",
"doi-asserted-by": "publisher",
"first-page": "365",
"issue": "6",
"journal-title": "Crit Rev Clin Lab Sci",
"key": "5_CR2",
"unstructured": "Ciotti M, Ciccozzi M, Terrinoni A, Jiang WC, Wang CB, Bernardini S. The COVID-19 pandemic. Crit Rev Clin Lab Sci. 2020;57(6):365–88. https://doi.org/10.1080/10408363.2020.1783198.",
"volume": "57",
"year": "2020"
},
{
"DOI": "10.4081/monaldi.2020.1298",
"author": "I Madabhavi",
"doi-asserted-by": "publisher",
"first-page": "2",
"journal-title": "Monaldi Arch Chest Dis",
"key": "5_CR3",
"unstructured": "Madabhavi I, Sarkar M, Kadakol N. COVID-19: a review. Monaldi Arch Chest Dis. 2020;90:2.",
"volume": "90",
"year": "2020"
},
{
"DOI": "10.1631/jzus.B2000083",
"author": "Y Shi",
"doi-asserted-by": "publisher",
"first-page": "343",
"issue": "5",
"journal-title": "J Zhejiang Univ Sci B",
"key": "5_CR4",
"unstructured": "Shi Y, Wang G, Cai XP, Deng JW, Zheng L, Zhu HH, Zheng M, Yang B, Chen Z. An overview of COVID-19. J Zhejiang Univ Sci B. 2020;21(5):343. https://doi.org/10.1631/jzus.B2000083.",
"volume": "21",
"year": "2020"
},
{
"DOI": "10.1016/j.jmii.2020.03.034v",
"author": "SS Jean",
"doi-asserted-by": "publisher",
"first-page": "436",
"issue": "3",
"journal-title": "J Microbiol Immunol Infect",
"key": "5_CR5",
"unstructured": "Jean SS, Lee PI, Hsueh PR. Treatment options for COVID-19: the reality and challenges. J Microbiol Immunol Infect. 2020;53(3):436–43. https://doi.org/10.1016/j.jmii.2020.03.034v.",
"volume": "53",
"year": "2020"
},
{
"DOI": "10.5867/sjmas.v1i1.11",
"author": "N Tzenios",
"doi-asserted-by": "publisher",
"journal-title": "Special J Med Acad Life Sci",
"key": "5_CR6",
"unstructured": "Tzenios N, Chahine M, Tazanios M. Better strategies for coronavirus (COVID-19) vaccination. Special J Med Acad Life Sci. 2023. https://doi.org/10.5867/sjmas.v1i1.11.",
"year": "2023"
},
{
"DOI": "10.1038/s41573-023-00672-y",
"author": "G Li",
"doi-asserted-by": "publisher",
"first-page": "6",
"journal-title": "Nat Rev Drug Discov",
"key": "5_CR7",
"unstructured": "Li G, Hilgenfeld R, Whitley R, De Clercq E. Therapeutic strategies for COVID-19: progress and lessons learned. Nat Rev Drug Discov. 2023;22:6.",
"volume": "22",
"year": "2023"
},
{
"DOI": "10.1016/j.mam.2022.101151",
"author": "Y Gupta",
"doi-asserted-by": "publisher",
"journal-title": "Mol Aspects Med",
"key": "5_CR8",
"unstructured": "Gupta Y, Savytskyi OV, Coban M, Venugopal A, Pleqi V, Weber CA, Chitale R, Durvasula R, Hopkins C, Kempaiah P, Caulfield TR. Protein structure-based in-silico approaches to drug discovery: guide to COVID-19 therapeutics. Mol Aspects Med. 2023;91: 101151. https://doi.org/10.1016/j.mam.2022.101151.",
"volume": "91",
"year": "2023"
},
{
"DOI": "10.2174/1568026623666221019110334",
"author": "A Gaurav",
"doi-asserted-by": "publisher",
"first-page": "2190",
"issue": "26",
"journal-title": "Curr Top Med Chem",
"key": "5_CR9",
"unstructured": "Gaurav A, Agrawal N, Al-Nema M, Gautam V. Computational approaches in the discovery and development of therapeutic and prophylactic agents for viral diseases. Curr Top Med Chem. 2022;22(26):2190–206. https://doi.org/10.2174/1568026623666221019110334.",
"volume": "22",
"year": "2022"
},
{
"DOI": "10.1038/s41598-021-88153-3",
"author": "A Mongia",
"doi-asserted-by": "publisher",
"journal-title": "Sci Rep",
"key": "5_CR10",
"unstructured": "Mongia A, Saha SK, Chouzenoux E, Majumdar A. A computational approach to aid clinicians in selecting anti-viral drugs for COVID-19 trials. Sci Rep. 2021. https://doi.org/10.1038/s41598-021-88153-3.",
"year": "2021"
},
{
"DOI": "10.1016/j.lfs.2020.118056",
"author": "A Sternberg",
"doi-asserted-by": "publisher",
"journal-title": "Life Sci",
"key": "5_CR11",
"unstructured": "Sternberg A, Naujokat C. Structural features of coronavirus SARS-CoV-2 spike protein: targets for vaccination. Life Sci. 2020;257: 118056. https://doi.org/10.1016/j.lfs.2020.118056.",
"volume": "257",
"year": "2020"
},
{
"DOI": "10.3390/v13010109",
"author": "X Xia",
"doi-asserted-by": "publisher",
"first-page": "109",
"issue": "1",
"journal-title": "Viruses",
"key": "5_CR12",
"unstructured": "Xia X. Domains and functions of spike protein in Sars-Cov-2 in the context of vaccine design. Viruses. 2021;13(1):109. https://doi.org/10.3390/v13010109.",
"volume": "13",
"year": "2021"
},
{
"DOI": "10.1016/j.coviro.2021.08.010",
"author": "J Zhang",
"doi-asserted-by": "publisher",
"first-page": "173",
"journal-title": "Curr Opin Virol",
"key": "5_CR13",
"unstructured": "Zhang J, Xiao T, Cai Y, Chen B. Structure of SARS-CoV-2 spike protein. Curr Opin Virol. 2021;50:173–82. https://doi.org/10.1016/j.coviro.2021.08.010.",
"volume": "50",
"year": "2021"
},
{
"DOI": "10.1016/j.drudis.2020.12.005",
"author": "R Banerjee",
"doi-asserted-by": "publisher",
"first-page": "804",
"issue": "3",
"journal-title": "Drug Discov Today",
"key": "5_CR14",
"unstructured": "Banerjee R, Perera L, Tillekeratne LV. Potential SARS-CoV-2 main protease inhibitors. Drug Discov Today. 2021;26(3):804–16. https://doi.org/10.1016/j.drudis.2020.12.005.",
"volume": "26",
"year": "2021"
},
{
"DOI": "10.1021/acs.jmedchem.1c00566",
"author": "S Huff",
"doi-asserted-by": "publisher",
"first-page": "2866",
"issue": "4",
"journal-title": "J Med Chem",
"key": "5_CR15",
"unstructured": "Huff S, Kummetha IR, Tiwari SK, Huante MB, Clark AE, Wang S, Bray W, Smith D, Carlin AF, Endsley M, Rana TM. Discovery and mechanism of SARS-CoV-2 main protease inhibitors. J Med Chem. 2021;65(4):2866–79. https://doi.org/10.1021/acs.jmedchem.1c00566.",
"volume": "65",
"year": "2021"
},
{
"DOI": "10.1525/embj.2020106275",
"author": "T Klemm",
"doi-asserted-by": "publisher",
"journal-title": "EMBO J",
"key": "5_CR16",
"unstructured": "Klemm T, Ebert G, Calleja DJ, Allison CC, Richardson LW, Bernardini JP, Lu BG, Kuchel NW, Grohmann C, Shibata Y, Gan ZY. Mechanism and inhibition of the papain-like protease PLpro of SARS-CoV-2. EMBO J. 2020. https://doi.org/10.1525/embj.2020106275.",
"year": "2020"
},
{
"DOI": "10.1016/j.bbrc.2020.08.116",
"author": "Y Jiang",
"doi-asserted-by": "publisher",
"first-page": "47",
"journal-title": "Biochem Biophys Res Commun",
"key": "5_CR17",
"unstructured": "Jiang Y, Yin W, Xu HE. RNA-dependent RNA polymerase: Structure, mechanism, and drug discovery for COVID-19. Biochem Biophys Res Commun. 2021;538:47–53. https://doi.org/10.1016/j.bbrc.2020.08.116.",
"volume": "538",
"year": "2021"
},
{
"DOI": "10.1177/2472555220942123",
"author": "W Zhu",
"doi-asserted-by": "publisher",
"first-page": "1141",
"issue": "10",
"journal-title": "SLAS Disco",
"key": "5_CR18",
"unstructured": "Zhu W, Chen CZ, Gorshkov K, Xu M, Lo DC, Zheng W. RNA-dependent RNA polymerase as a target for COVID-19 drug discovery. SLAS Disco. 2020;25(10):1141–51. https://doi.org/10.1177/2472555220942123.",
"volume": "25",
"year": "2020"
},
{
"DOI": "10.1126/science.abc1560",
"author": "W Yin",
"doi-asserted-by": "publisher",
"first-page": "1499",
"issue": "6498",
"journal-title": "Science",
"key": "5_CR19",
"unstructured": "Yin W, Mao C, Luan X, Shen DD, Shen Q, Su H, Wang X, Zhou F, Zhao W, Gao M, Chang S. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368(6498):1499–504. https://doi.org/10.1126/science.abc1560.",
"volume": "368",
"year": "2020"
},
{
"DOI": "10.1021/acs.jpclett.0c02421",
"author": "MA White",
"doi-asserted-by": "publisher",
"first-page": "9144",
"issue": "21",
"journal-title": "J Phys Chem Lett",
"key": "5_CR20",
"unstructured": "White MA, Lin W, Cheng X. Discovery of COVID-19 inhibitors targeting the SARS-CoV-2 Nsp13 helicase. J Phys Chem Lett. 2020;11(21):9144–51. https://doi.org/10.1021/acs.jpclett.0c02421.",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1016/j.arcmed.2020.05.024",
"author": "S Habtemariam",
"doi-asserted-by": "publisher",
"first-page": "733",
"issue": "7",
"journal-title": "Arch Med Res",
"key": "5_CR21",
"unstructured": "Habtemariam S, Nabavi SF, Banach M, Berindan-Neagoe I, Sarkar K, Sil PC, Nabavi SM. Should we try SARS-CoV-2 helicase inhibitors for COVID-19 therapy? Arch Med Res. 2020;51(7):733–5. https://doi.org/10.1016/j.arcmed.2020.05.024.",
"volume": "51",
"year": "2020"
},
{
"DOI": "10.3390/v13061115",
"author": "Y Peng",
"doi-asserted-by": "publisher",
"issue": "20",
"journal-title": "EMBO J",
"key": "5_CR22",
"unstructured": "Peng Y, Du N, Lei Y, Dorje S, Qi J, Luo T, Gao GF, Song H. Structures of the SARS-CoV-2 nucleocapsid and their perspectives for drug design. EMBO J. 2020;39(20): e105938. https://doi.org/10.3390/v13061115.",
"volume": "39",
"year": "2020"
},
{
"DOI": "10.1128/jvi.00647-20",
"author": "NK Dutta",
"doi-asserted-by": "publisher",
"first-page": "10",
"issue": "13",
"journal-title": "J Virol",
"key": "5_CR23",
"unstructured": "Dutta NK, Mazumdar K, Gordy JT. The nucleocapsid protein of SARS–CoV-2: a target for vaccine development. J Virol. 2020;94(13):10–1128. https://doi.org/10.1128/jvi.00647-20.",
"volume": "94",
"year": "2020"
},
{
"DOI": "10.1016/j.apsb.2020.04.009",
"author": "S Kang",
"doi-asserted-by": "publisher",
"first-page": "1228",
"issue": "7",
"journal-title": "Acta PharmaceuticaSinica B",
"key": "5_CR24",
"unstructured": "Kang S, Yang M, Hong Z, Zhang L, Huang Z, Chen X, He S, Zhou Z, Zhou Z, Chen Q, Yan Y. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta PharmaceuticaSinica B. 2020;10(7):1228–38. https://doi.org/10.1016/j.apsb.2020.04.009.",
"volume": "10",
"year": "2020"
},
{
"DOI": "10.1093/nar/gku401",
"author": "MN Drwal",
"doi-asserted-by": "publisher",
"first-page": "W53",
"issue": "W1",
"journal-title": "Nucleic Acids Res",
"key": "5_CR25",
"unstructured": "Drwal MN, Banerjee P, Dunkel M, Wettig MR, Preissner R. ProTox: a web server for the in silico prediction of rodent oral toxicity. Nucleic Acids Res. 2014;42(W1):W53–8. https://doi.org/10.1093/nar/gku401.",
"volume": "42",
"year": "2014"
},
{
"DOI": "10.1023/A:1025361621494",
"author": "JC Dearden",
"doi-asserted-by": "publisher",
"first-page": "119",
"issue": "2",
"journal-title": "J Comput-Aided Mol Design",
"key": "5_CR26",
"unstructured": "Dearden JC. In silico prediction of drug toxicity. J Comput-Aided Mol Design. 2003;17(2):119–27. https://doi.org/10.1023/A:1025361621494.",
"volume": "17",
"year": "2003"
},
{
"DOI": "10.35693/2500-1388-2016-0-3-28-32",
"author": "L Ravi",
"doi-asserted-by": "crossref",
"first-page": "28",
"journal-title": "Innovare J Med Sci",
"key": "5_CR27",
"unstructured": "Ravi L, Kannabiran K. A handbook on protein-ligand docking tool AutoDock 4. Innovare J Med Sci. 2016;1:28–33.",
"volume": "1",
"year": "2016"
},
{
"DOI": "10.4103/0975-8453.59519",
"author": "A Baldi",
"doi-asserted-by": "publisher",
"first-page": "99",
"issue": "1",
"journal-title": "Syst Rev Pharm",
"key": "5_CR28",
"unstructured": "Baldi A. Computational approaches for drug design and discovery: an overview. Syst Rev Pharm. 2010;1(1):99. https://doi.org/10.4103/0975-8453.59519.",
"volume": "1",
"year": "2010"
},
{
"DOI": "10.1007/s10822-013-9644-8",
"author": "G MadhaviSastry",
"doi-asserted-by": "publisher",
"journal-title": "J Comput-Aided Mol Design",
"key": "5_CR29",
"unstructured": "MadhaviSastry G, Adzhigirey M, Day T, Annabhimoju R, Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput-Aided Mol Design. 2013. https://doi.org/10.1007/s10822-013-9644-8.",
"year": "2013"
},
{
"DOI": "10.1002/jcc.21256",
"author": "GM Morris",
"doi-asserted-by": "publisher",
"first-page": "2785",
"journal-title": "J Comput Chem",
"key": "5_CR30",
"unstructured": "Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. Autodock4 and AutoDockTools4: automated docking with selective receptor flexiblity. J Comput Chem. 2009;16:2785–91.",
"volume": "16",
"year": "2009"
},
{
"key": "5_CR31",
"unstructured": "Schrödinger L, DeLano W. PyMOL 2020. http://www.pymol.org/pymol"
},
{
"key": "5_CR32",
"unstructured": "BioviaDS.[Discovery Studio. San Diego: DassaultSystemes: 2021."
},
{
"DOI": "10.1007/s002040050229",
"author": "E Schlede",
"doi-asserted-by": "publisher",
"first-page": "659",
"journal-title": "Arch Toxic",
"key": "5_CR33",
"unstructured": "Schlede E, Mischke U, Diener W, Kayser D. The international validation study of the acute toxic class method (oral). Arch Toxic. 1995;69:659–70. https://doi.org/10.1007/s002040050229.",
"volume": "69",
"year": "1995"
},
{
"DOI": "10.1007/s00204-023-03507-2",
"author": "M Noga",
"doi-asserted-by": "publisher",
"first-page": "1691",
"issue": "6",
"journal-title": "Arch Toxic",
"key": "5_CR34",
"unstructured": "Noga M, Michalska A, Jurowski K. Application of toxicology in silico methods for prediction of acute toxicity (LD50) for Novichoks. Arch Toxic. 2023;97(6):1691–700. https://doi.org/10.1007/s00204-023-03507-2.",
"volume": "97",
"year": "2023"
},
{
"DOI": "10.1007/978-1-4899-1379-1_24",
"author": "WCA Gelderblom",
"doi-asserted-by": "publisher",
"journal-title": "Fumonisins Food",
"key": "5_CR35",
"unstructured": "Gelderblom WCA, Snyman SD, Abel S, Lebepe-Mazur S, Smuts CM, Van der Westhuizen L, Marasas WF, Victor TC, Knasmüller S, Huber W. Hepatotoxicity and-carcinogenicity of the fumonisins in rats: a review regarding mechanistic implications for establishing risk in humans. Fumonisins Food. 1996. https://doi.org/10.1007/978-1-4899-1379-1_24.",
"year": "1996"
},
{
"DOI": "10.1016/j.compbiomed.2020.104054",
"author": "S Skariyachan",
"doi-asserted-by": "publisher",
"journal-title": "Comput Biol Med",
"key": "5_CR36",
"unstructured": "Skariyachan S, Gopal D, Chakrabarti S, Kempanna P, Uttarkar A, Muddebihalkar AG, Niranjan V. Structural and molecular basis of the interaction mechanism of selected drugs towards multiple targets of SARS-CoV-2 by molecular docking and dynamic simulation studies-deciphering the scope of repurposed drugs. Comput Biol Med. 2020;126: 104054. https://doi.org/10.1016/j.compbiomed.2020.104054.",
"volume": "126",
"year": "2020"
},
{
"DOI": "10.1080/07391102.2020.1776157",
"author": "S Gupta",
"doi-asserted-by": "publisher",
"first-page": "4334",
"issue": "12",
"journal-title": "J Biomol Struct Dyn",
"key": "5_CR37",
"unstructured": "Gupta S, Singh AK, Kushwaha PP, Prajapati KS, Shuaib M, Senapati S, Kumar S. Identification of potential natural inhibitors of SARS-CoV2 main protease by molecular docking and simulation studies. J Biomol Struct Dyn. 2021;39(12):4334–45. https://doi.org/10.1080/07391102.2020.1776157.",
"volume": "39",
"year": "2021"
},
{
"DOI": "10.1080/07391102.2020.1763201",
"author": "S Das",
"doi-asserted-by": "publisher",
"first-page": "3347",
"issue": "9",
"journal-title": "J Biomol Struct Dyn",
"key": "5_CR38",
"unstructured": "Das S, Sarmah S, Lyndem S, Singha Roy A. An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. J Biomol Struct Dyn. 2021;39(9):3347–57. https://doi.org/10.1080/07391102.2020.1763201.",
"volume": "39",
"year": "2021"
},
{
"DOI": "10.3390/cryst11050471",
"author": "MY Ghazwani",
"doi-asserted-by": "publisher",
"first-page": "471",
"issue": "5",
"journal-title": "Crystals",
"key": "5_CR39",
"unstructured": "Ghazwani MY, Bakheit AH, Hakami AR, Alkahtani HM, Almehizia AA. Virtual screening and molecular docking studies for discovery of potential RNA-dependent RNA polymerase inhibitors. Crystals. 2021;11(5):471. https://doi.org/10.3390/cryst11050471.",
"volume": "11",
"year": "2021"
},
{
"DOI": "10.1016/j.ijbiomac.2020.09.098",
"author": "MSA Parvez",
"doi-asserted-by": "publisher",
"first-page": "1787",
"journal-title": "Int J Biol Macromol",
"key": "5_CR40",
"unstructured": "Parvez MSA, Karim MA, Hasan M, Jaman J, Karim Z, Tahsin T, Hasan MN, Hosen MJ. Prediction of potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2 using comprehensive drug repurposing and molecular docking approach. Int J Biol Macromol. 2020;163:1787–97. https://doi.org/10.1016/j.ijbiomac.2020.09.098.",
"volume": "163",
"year": "2020"
},
{
"DOI": "10.1016/j.jmgm.2021.107851",
"author": "E Pitsillou",
"doi-asserted-by": "publisher",
"journal-title": "J Mol Graph Model",
"key": "5_CR41",
"unstructured": "Pitsillou E, Liang J, Ververis K, Hung A, Karagiannis TC. Interaction of small molecules with the SARS-CoV-2 papain-like protease: In silico studies and in vitro validation of protease activity inhibition using an enzymatic inhibition assay. J Mol Graph Model. 2021;104: 107851. https://doi.org/10.1016/j.jmgm.2021.107851.",
"volume": "104",
"year": "2021"
},
{
"DOI": "10.3389/fchem.2020.623971",
"author": "E Pitsillou",
"doi-asserted-by": "publisher",
"journal-title": "Front Chem",
"key": "5_CR42",
"unstructured": "Pitsillou E, Liang J, Ververis K, Lim KW, Hung A, Karagiannis TC. Identification of small molecule inhibitors of the deubiquitinating activity of the SARS-CoV-2 papain-like protease: in silico molecular docking studies and in vitro enzymatic activity assay. Front Chem. 2020;8: 623971. https://doi.org/10.3389/fchem.2020.623971.",
"volume": "8",
"year": "2020"
},
{
"DOI": "10.1016/j.bbrc.2020.11.083",
"author": "D Li",
"doi-asserted-by": "publisher",
"first-page": "72",
"journal-title": "Biochem Biophys Res Commun",
"key": "5_CR43",
"unstructured": "Li D, Luan J, Zhang L. Molecular docking of potential SARS-CoV-2 papain-like protease inhibitors. Biochem Biophys Res Commun. 2021;538:72–9. https://doi.org/10.1016/j.bbrc.2020.11.083.",
"volume": "538",
"year": "2021"
},
{
"DOI": "10.1007/s11030-021-10251-1",
"author": "RP Vivek-Ananth",
"doi-asserted-by": "publisher",
"first-page": "429",
"issue": "1",
"journal-title": "Mol Divers",
"key": "5_CR44",
"unstructured": "Vivek-Ananth RP, Krishnaswamy S, Samal A. Potential phytochemical inhibitors of SARS-CoV-2 helicase Nsp13: a molecular docking and dynamic simulation study. Mol Divers. 2022;26(1):429–42. https://doi.org/10.1007/s11030-021-10251-1.",
"volume": "26",
"year": "2022"
},
{
"DOI": "10.1093/nar/gkz409",
"author": "Z Jia",
"doi-asserted-by": "publisher",
"first-page": "6538",
"issue": "12",
"journal-title": "Nucleic Acids Res",
"key": "5_CR45",
"unstructured": "Jia Z, Yan L, Ren Z, Wu L, Wang J, Guo J, Zheng L, Ming Z, Zhang L, Lou Z, Rao Z. Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019;47(12):6538–50. https://doi.org/10.1093/nar/gkz409.",
"volume": "47",
"year": "2019"
},
{
"DOI": "10.3326/BRIAC113.98489861",
"author": "R Yadav",
"doi-asserted-by": "publisher",
"journal-title": "Biointerface Res Appl Chem",
"key": "5_CR46",
"unstructured": "Yadav R, Parihar RD, Dhiman U, Dhamija P, Upadhyay SK, Imran M, Behera SK, Prasad TK. Docking of fda approved drugs targeting nsp-16, n-protein and main protease of sars-cov-2 as dual inhibitors. Biointerface Res Appl Chem. 2020. https://doi.org/10.3326/BRIAC113.98489861.",
"year": "2020"
},
{
"DOI": "10.1002/pbc.22882",
"author": "B Langholz",
"doi-asserted-by": "publisher",
"first-page": "252",
"issue": "2",
"journal-title": "Pediatr Blood Cancer",
"key": "5_CR47",
"unstructured": "Langholz B, Skolnik JM, Barrett JS, Renbarger J, Seibel NL, Zajicek A, Arndt CA. Dactinomycin and vincristine toxicity in the treatment of childhood cancer: a retrospective study from the children’s oncology group. Pediatr Blood Cancer. 2011;57(2):252–7. https://doi.org/10.1002/pbc.22882.",
"volume": "57",
"year": "2011"
},
{
"DOI": "10.1016/j.ebiom.2021.103288",
"author": "L Liesenborghs",
"doi-asserted-by": "publisher",
"journal-title": "EBioMedicine",
"key": "5_CR48",
"unstructured": "Liesenborghs L, Spriet I, Jochmans D, Belmans A, Gyselinck I, Teuwen LA, Ter Horst S, Dreesen E, Geukens T, Engelen MM, Landeloos E, Geldhof V, Ceunen H, Debaveye B, Vandenberk B, Van der Linden L, Jacobs S, Langendries L, Boudewijns R, Do TND, Verhamme P. Itraconazole for COVID-19: preclinical studies and a proof-of-concept randomized clinical trial. EBioMedicine. 2021. https://doi.org/10.1016/j.ebiom.2021.103288.",
"year": "2021"
},
{
"DOI": "10.1002/jmv.26917",
"author": "E Van Damme",
"doi-asserted-by": "publisher",
"first-page": "4454",
"issue": "7",
"journal-title": "J Med Virol",
"key": "5_CR49",
"unstructured": "Van Damme E, De Meyer S, Bojkova D, Ciesek S, Cinatl J, De Jonghe S, Jochmans D, Leyssen P, Buyck C, Neyts J, Van Loock M. In vitro activity of itraconazole against SARS-CoV-2. J Med Virol. 2021;93(7):4454–60. https://doi.org/10.1002/jmv.26917.",
"volume": "93",
"year": "2021"
},
{
"DOI": "10.1016/j.antiviral.2020.104787",
"author": "L Caly",
"doi-asserted-by": "publisher",
"journal-title": "Antiviral Res",
"key": "5_CR50",
"unstructured": "Caly L, Druce JD, Catton MG, Jans DA, Wagstaff KM. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020;178: 104787. https://doi.org/10.1016/j.antiviral.2020.104787.",
"volume": "178",
"year": "2020"
},
{
"DOI": "10.1038/s41598-021-86679-0",
"author": "AP Arévalo",
"doi-asserted-by": "publisher",
"first-page": "7132",
"issue": "1",
"journal-title": "Sci Rep",
"key": "5_CR51",
"unstructured": "Arévalo AP, Pagotto R, Pórfido JL, Daghero H, Segovia M, Yamasaki K, Varela B, Hill M, Verdes JM, Duhalde Vega M, Bollati-Fogolín M, Crispo M. Ivermectin reduces in vivo coronavirus infection in a mouse experimental model. Sci Rep. 2021;11(1):7132. https://doi.org/10.1038/s41598-021-86679-0.",
"volume": "11",
"year": "2021"
},
{
"DOI": "10.3390/medicina60060892",
"author": "A Kowalczyk",
"doi-asserted-by": "publisher",
"first-page": "892",
"issue": "6",
"journal-title": "Medicina",
"key": "5_CR52",
"unstructured": "Kowalczyk A. Hesperidin, a potential antiviral agent against SARS-CoV-2: the influence of citrus consumption on COVID-19 incidence and severity in China. Medicina. 2024;60(6):892. https://doi.org/10.3390/medicina60060892.",
"volume": "60",
"year": "2024"
},
{
"DOI": "10.3390/nu13082800",
"author": "FJ Cheng",
"doi-asserted-by": "publisher",
"first-page": "2800",
"issue": "8",
"journal-title": "Nutrients",
"key": "5_CR53",
"unstructured": "Cheng FJ, Huynh TK, Yang CS, Hu DW, Shen YC, Tu CY, Wu YC, Tang CH, Huang WC, Chen Y, Ho CY. Hesperidin Is a Potential Inhibitor against SARS-CoV-2 Infection. Nutrients. 2021;13(8):2800. https://doi.org/10.3390/nu13082800.",
"volume": "13",
"year": "2021"
},
{
"author": "P Bellavite",
"key": "5_CR54",
"unstructured": "Bellavite P. Reappraisal of dietary phytochemicals for coronavirus infection focus on hesperidin and quercetin. In: Waisundara Viduranga, editor. Antioxidants. London: IntechOpen; 2021.",
"volume-title": "Antioxidants",
"year": "2021"
},
{
"DOI": "10.1186/s13578-021-00680-8",
"author": "J Liu",
"doi-asserted-by": "publisher",
"first-page": "168",
"issue": "1",
"journal-title": "Cell Biosci",
"key": "5_CR55",
"unstructured": "Liu J, Bodnar BH, Meng F, Khan AI, Wang X, Saribas S, Wang T, Lohani SC, Wang P, Wei Z, Luo J, Zhou L, Wu J, Luo G, Li Q, Hu W, Ho W. Epigallocatechingallate from green tea effectively blocks infection of SARS-CoV-2 and new variants by inhibiting spike binding to ACE2 receptor. Cell Biosci. 2021;11(1):168. https://doi.org/10.1186/s13578-021-00680-8.",
"volume": "11",
"year": "2021"
},
{
"DOI": "10.3342/2639-9458.1116",
"author": "BL Hurst",
"doi-asserted-by": "publisher",
"journal-title": "Microbiol Infect Dis",
"key": "5_CR56",
"unstructured": "Hurst BL, Dickinson D, Hsu S. Epigallocatechin-3-gallate (EGCG) Inhibits SARS-CoV-2 infection in primate epithelial cells: (a short communication). Microbiol Infect Dis. 2021. https://doi.org/10.3342/2639-9458.1116.",
"year": "2021"
},
{
"DOI": "10.3390/life11030197",
"author": "J Park",
"doi-asserted-by": "publisher",
"first-page": "197",
"issue": "3",
"journal-title": "Life",
"key": "5_CR57",
"unstructured": "Park J, Park R, Jang M, Park Y-I. Therapeutic potential of EGCG, a green tea polyphenol, for treatment of coronavirus diseases. Life. 2021;11(3):197. https://doi.org/10.3390/life11030197.",
"volume": "11",
"year": "2021"
},
{
"DOI": "10.1038/s41419-022-04961-z",
"author": "D Iaconis",
"doi-asserted-by": "publisher",
"first-page": "498",
"issue": "5",
"journal-title": "Cell Death Dis",
"key": "5_CR58",
"unstructured": "Iaconis D, Bordi L, Matusali G, Talarico C, Manelfi C, Cesta MC, Zippoli M, Caccuri F, Bugatti A, Zani A, Filippini F, Scorzolini L, Gobbi M, Beeg M, Piotti A, Montopoli M, Cocetta V, Bressan S, Bucci EM, Caruso A, Beccari AR. Characterization of raloxifene as a potential pharmacological agent against SARS-CoV-2 and its variants. Cell Death Dis. 2022;13(5):498. https://doi.org/10.1038/s41419-022-04961-z.",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.1016/j.eclinm.2022.101450",
"author": "E Nicastri",
"doi-asserted-by": "publisher",
"journal-title": "EClinicalMedicine",
"key": "5_CR59",
"unstructured": "Nicastri E, Marinangeli F, Pivetta E, Torri E, Reggiani F, Fiorentino G, Scorzolini L, Vettori S, Marsiglia C, Gavioli EM, Beccari AR, Terpolilli G, De Pizzol M, Goisis G, Mantelli F, Vaia F, Allegretti M. A phase 2 randomized, double-blinded, placebo-controlled, multicenter trial evaluating the efficacy and safety of raloxifene for patients with mild to moderate COVID-19. EClinicalMedicine. 2022. https://doi.org/10.1016/j.eclinm.2022.101450.",
"year": "2022"
},
{
"DOI": "10.1016/j.cbi.2022.110230",
"author": "A Soussi",
"doi-asserted-by": "publisher",
"journal-title": "Chem Biol Interact",
"key": "5_CR60",
"unstructured": "Soussi A, Gargouri M, Magné C, Ben-Nasr H, Kausar MA, Siddiqui AJ, Saeed M, Snoussi M, Adnan M, El-Feki A, Chappard D, Badraoui R. (-)-Epigallocatechingallate (EGCG) pharmacokinetics and molecular interactions towards amelioration of hyperglycemia, hyperlipidemia associated hepatorenal oxidative injury in alloxan induced diabetic mice. Chem Biol Interact. 2022;368: 110230. https://doi.org/10.1016/j.cbi.2022.110230.",
"volume": "368",
"year": "2022"
},
{
"DOI": "10.1525/emmm.201911622",
"author": "J Humeau",
"doi-asserted-by": "publisher",
"journal-title": "EMBO Mol Med",
"key": "5_CR61",
"unstructured": "Humeau J, Sauvat A, Cerrato G, Xie W, Loos F, Iannantuoni F, Bezu L, Lévesque S, Paillet J, Pol J, Leduc M, Zitvogel L, de Thé H, Kepp O, Kroemer G. Inhibition of transcription by dactinomycin reveals a new characteristic of immunogenic cell stress. EMBO Mol Med. 2020. https://doi.org/10.1525/emmm.201911622.",
"year": "2020"
},
{
"DOI": "10.3390/antibiotics11060796",
"author": "N Angkasekwinai",
"doi-asserted-by": "publisher",
"first-page": "796",
"issue": "6",
"journal-title": "Antibiotics",
"key": "5_CR62",
"unstructured": "Angkasekwinai N, Rattanaumpawan P, Chayakulkeeree M, Phoompoung P, Koomanachai P, Chantarasut S, Wangchinda W, Srinonprasert V, Thamlikitkul V. Safety and efficacy of ivermectin for the prevention and treatment of covid-19: a double-blinded randomized placebo-controlled study. Antibiotics. 2022;11(6):796. https://doi.org/10.3390/antibiotics11060796.",
"volume": "11",
"year": "2022"
},
{
"DOI": "10.3390/molecules15118478",
"author": "Y Chen",
"doi-asserted-by": "publisher",
"first-page": "8478",
"issue": "11",
"journal-title": "Molecules",
"key": "5_CR63",
"unstructured": "Chen Y, Jia X, Chen J, Wang J, Hu M. The pharmacokinetics of raloxifene and its interaction with apigenin in rat. Molecules. 2010;15(11):8478–87. https://doi.org/10.3390/molecules15118478.",
"volume": "15",
"year": "2010"
},
{
"DOI": "10.1021/jf800105c",
"author": "YM Li",
"doi-asserted-by": "publisher",
"first-page": "5550",
"issue": "14",
"journal-title": "J Agric Food Chem",
"key": "5_CR64",
"unstructured": "Li YM, Li XM, Li GM, Du WC, Zhang J, Li WX, Xu J, Hu M, Zhu Z. In vivo pharmacokinetics of hesperidin are affected by treatment with glucosidase-like BglA protein isolated from yeasts. J Agric Food Chem. 2008;56(14):5550–7. https://doi.org/10.1021/jf800105c.",
"volume": "56",
"year": "2008"
},
{
"DOI": "10.1517/17425255.2013.794785",
"author": "J Lestner",
"doi-asserted-by": "publisher",
"first-page": "911",
"issue": "7",
"journal-title": "Expert Opin Drug Metab Toxicol",
"key": "5_CR65",
"unstructured": "Lestner J, Hope WW. Itraconazole: an update on pharmacology and clinical use for treatment of invasive and allergic fungal infections. Expert Opin Drug Metab Toxicol. 2013;9(7):911–26. https://doi.org/10.1517/17425255.2013.794785.",
"volume": "9",
"year": "2013"
},
{
"DOI": "10.3390/pharmaceutics13060815",
"author": "B Costa",
"doi-asserted-by": "publisher",
"first-page": "815",
"issue": "6",
"journal-title": "Pharmaceutics",
"key": "5_CR66",
"unstructured": "Costa B, Vale N. A review of repurposed cancer drugs in clinical trials for potential treatment of COVID-19. Pharmaceutics. 2021;13(6):815. https://doi.org/10.3390/pharmaceutics13060815.",
"volume": "13",
"year": "2021"
},
{
"DOI": "10.1128/JVI.29.1.401-404.1979",
"author": "DA Kennedy",
"doi-asserted-by": "publisher",
"first-page": "401",
"issue": "1",
"journal-title": "J Virol",
"key": "5_CR67",
"unstructured": "Kennedy DA, Johnson-Lussenburg CM. Inhibition of coronavirus 229E replication by actinomycin D. J Virol. 1979;29(1):401–4. https://doi.org/10.1128/JVI.29.1.401-404.1979.",
"volume": "29",
"year": "1979"
},
{
"DOI": "10.1016/j.bioorg.2020.104488",
"author": "PN Batalha",
"doi-asserted-by": "publisher",
"journal-title": "Bioorg Chem",
"key": "5_CR68",
"unstructured": "Batalha PN, Forezi LSM, Lima CGS, Pauli FP, Boechat FCS, de Souza MCBV, Cunha AC, Ferreira VF, da Silva FC. Drug repurposing for the treatment of COVID-19: pharmacological aspects and synthetic approaches. Bioorg Chem. 2021;106: 104488. https://doi.org/10.1016/j.bioorg.2020.104488.",
"volume": "106",
"year": "2021"
}
],
"reference-count": 68,
"references-count": 68,
"relation": {},
"resource": {
"primary": {
"URL": "https://link.springer.com/10.1007/s44345-024-00005-5"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Exploring potential therapeutic candidates against COVID-19: a molecular docking study",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1007/springer_crossmark_policy",
"volume": "1"
}