Dynamics of SARS-CoV-2 variants and mutations in Central Sweden between 2023 and 2024 and their potential implications on monoclonal antibodies pemivibart and sipavibart as PrEP in the region
et al., Infectious Diseases, doi:10.1080/23744235.2025.2509011, May 2025
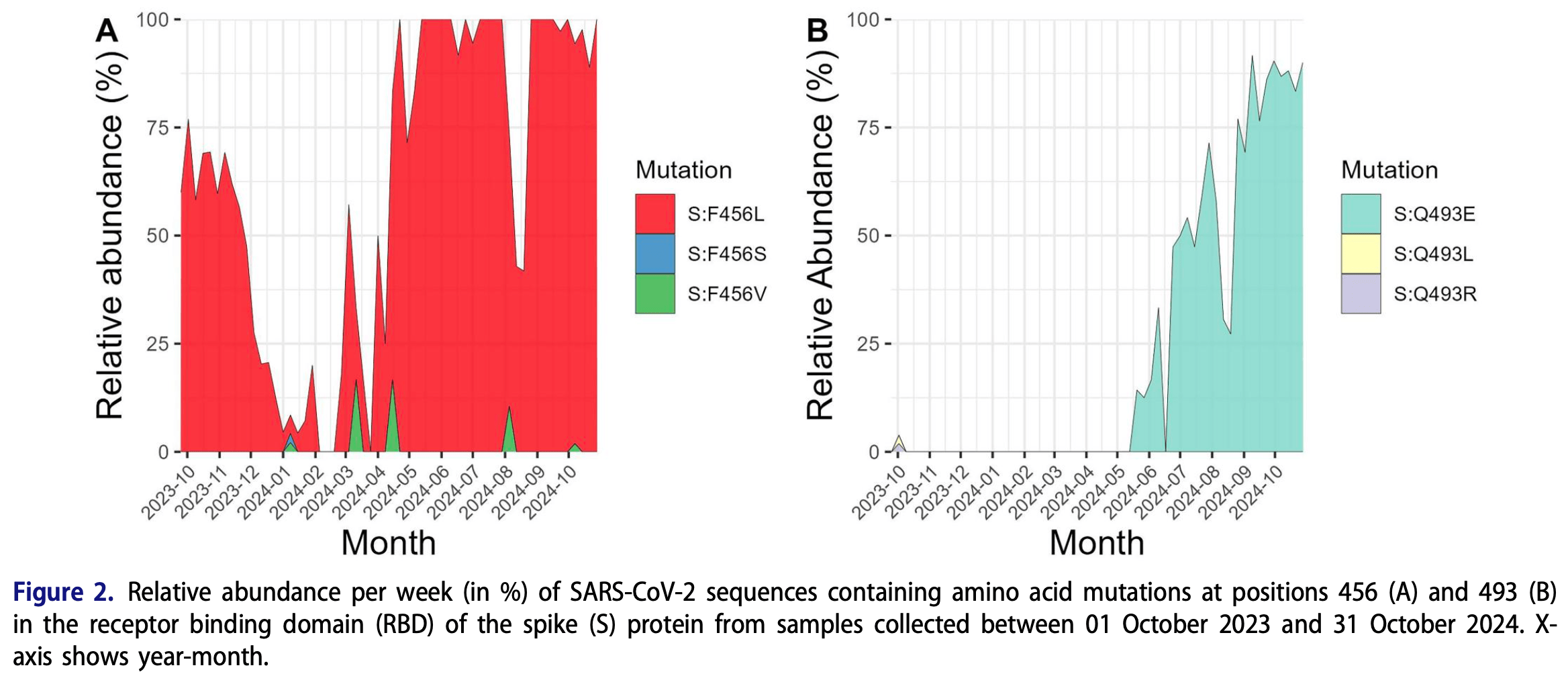
Analysis of SARS-CoV-2 variants and mutations in central Sweden from October 2023 to October 2024, showing the rise of resistance mutations that likely render monoclonal antibodies sipavibart and pemivibart ineffective.
Efficacy is variant dependent. In Vitro research shows reduced efficacy against KP.3.1.1, KP.1.1, LB.1, KP.3.3, and XEC variants1-4.
Study covers sipavibart and pemivibart.
1.
Xie et al., Molecular Basis of High-Blood-Pressure-Enhanced and High-Fever-Temperature-Weakened Receptor-Binding Domain/Peptidase Domain Binding: A Molecular Dynamics Simulation Study, International Journal of Molecular Sciences, doi:10.3390/ijms26073250.
2.
Wang et al., Activity of Research-Grade Pemivibart against Recent SARS-CoV-2 JN.1 Sublineages, New England Journal of Medicine, doi:10.1056/NEJMc2410203.
Haars et al., 26 May 2025, Sweden, peer-reviewed, 12 authors.
Contact: jonathan.haars@uu.se, johan.lennerstrand@medsci.uu.se, n.palanisamy@chester.ac.uk.
Dynamics of SARS-CoV-2 variants and mutations in Central Sweden between 2023 and 2024 and their potential implications on monoclonal antibodies pemivibart and sipavibart as PrEP in the region
Infectious Diseases, doi:10.1080/23744235.2025.2509011
Background: Monoclonal antibodies (mAbs) are an important option against SARS-CoV-2, especially as pre-exposure prophylaxis (PrEP) for patients with immune system impairment. PrEP mAbs like sipavibart and pemivibart have been approved for limited use in several countries. Certain SARS-CoV-2 variants carry mutations in the spike (S) protein, conferring resistance to these mAbs. Objectives: We aimed to examine the relative abundance of different circulating SARS-CoV-2 variants/ mutations in central Sweden between 2023 and 2024, and to predict the effectiveness of sipavibart and pemivibart. Methods: An amplicon-based Nanopore sequencing method was used for sequencing SARS-CoV-2 samples. Coronapp was used to identify mutations in these sequences. Using the published in vitro resistance data for sipavibart and pemivibart, the effectiveness of these mAbs was inferred.
Results: We have observed that the relative abundance of the KP.3.1.1 variant and the Q493E mutation started to increase in the later part of 2024 in the region. Also, since April 2024, the relative abundance of the F456L mutation reached 100% during many weeks until the end of the study period. The KP.3.1.1 variant is significantly resistant to pemivibart. Further, the presence of the F456L mutation in the Omicron subvariants confers high fold resistance towards sipavibart.
Conclusion: The use of sipavibart or pemivibart as PrEP for COVID-19 in the region may currently not be effective unless new SARS-CoV-2 variants appear not containing these resistance mutations. Further, new mAbs under development as PrEP for COVID-19 can be effectively used by routinely sequencing SARS-CoV-2 in patients to identify variants and resistance mutations.
Author contributions Conceptualisation, J.H., N.P. and J.Le.; methodology, J.H. and J.Le.; software, J.H. and R.K.; formal analysis, J.H.; investigation, J.H., F.W., K.E., P.M., P.E., R.K. and J.Le.; resources, J.Li., H.Y., M.S. and R.K.; data curation, J.H. and F.W.; writing-original draft preparation, J.H.; writing-review and editing, J.H., N.P., K.E., F.W., P.M., P.E., J.Li., M.S., R.K. and J.Le.; visualisation, J.H.; supervision, R.K. and J.Le.; funding acquisition, J.Le. All authors have read and agreed to the published version of the manuscript.
Disclosure statement Johan Lennerstrand received an unrestricted research grant from AstraZeneca AB, Sweden, but this company was not involved in any part of this study. All other authors declare no conflict of interest.
References
Akinosoglou, Rigopoulos, Kaiafa, Tixagevimab/Cilgavimab in SARS-CoV-2 prophylaxis and therapy: a comprehensive review of clinical experience, Viruses, doi:10.3390/v15010118
Aksamentov, Roemer, Hodcroft, Nextclade: clade assignment, mutation calling and quality control for viral genomes, JOSS, doi:10.21105/joss.03773
Amani, Amani, Efficacy and safety of sotrovimab in patients with COVID-19: a rapid review and meta-analysis, Rev Med Virol, doi:10.1002/rmv.2402
Astrazeneca, Phase, /III Randomized, Double Blind Study to Evaluate the Safety, Efficacy and Neutralizing Activity of AZD5156/AZD3152 for Pre Exposure Prophylaxis of COVID 19 in Participants With Conditions Causing Immune Impairment. Sub-Study: phase II Open Label Sub-Study to Evaluate the Safety, PK, and Neutralizing Activity of AZD3152 for Pre-Exposure Prophylaxis of COVID-19
Benotmane, Velay, Gautier-Vargas, Pre-exposure prophylaxis with 300 Mg Evusheld elicits limited neutralizing activity against the omicron variant, Kidney Int, doi:10.1016/j.kint.2022.05.008
Bhimraj, Falck-Ytter, Kim, clinical practice guideline update by the infectious diseases society of america on the management of COVID-19: anti-SARS-CoV-2 neutralizing antibody pemivibart for pre-exposure prophylaxis, Clin Infect Dis, doi:10.1093/cid/ciae435
Cai, Diallo, Rosenthal, AZD3152 Neutralizes SARS-CoV-2 historical and contemporary variants and is protective in hamsters and well tolerated in adults, Sci Transl Med, doi:10.1126/scitranslmed.ado2817
Calabrese, The future of COVID-19 for patients with immune-mediated inflammatory diseases: who is at risk?, J Rheumatol, doi:10.3899/jrheum.2023-1186
Cao, Jian, Wang, Imprinted SARS-CoV-2 humoral immunity induces convergent omicron RBD evolution, Nature, doi:10.1038/s41586-022-05644-7
Cdc Kp, 3.1.1 Is the Predominant Variant as COVID-19 Activity Increases
Cox, Peacock, Harvey, SARS-CoV-2 variant evasion of monoclonal antibodies based on in vitro studies, Nat Rev Microbiol, doi:10.1038/s41579-022-00809-7
Cumlin, Karlsson, Haars, From SARS-CoV-2 to global preparedness: a graphical interface for standardised high-throughput bioinformatics analysis in pandemic scenarios and surveillance of drug resistance, Int J Mol Sci, doi:10.3390/ijms25126645
Espinosa-Gongora, Berg, Rehn, Early detection of the emerging SARS-CoV-2 BA.2.86 lineage through integrated genomic surveillance of wastewater and COVID-19 cases in Sweden, weeks 31 to 38, Euro Surveill, doi:10.2807/1560-7917.ES.2023.28.46.2300595
Evusheld, None
Focosi, Casadevall, A critical analysis of the use of cilgavimab plus tixagevimab monoclonal antibody cocktail (Evusheld TM ) for COVID-19 prophylaxis and treatment, Viruses, doi:10.3390/v14091999
Focosi, Franchini, Casadevall, An update on the anti-spike monoclonal antibody pipeline for SARS-CoV-2, Clin Microbiol Infect, doi:10.1016/j.cmi.2024.04.012
Focosi, Mcconnell, Casadevall, Monoclonal antibody therapies against SARS-CoV-2, Lancet Infect Dis, doi:10.1016/S1473-3099(22)00311-5
Focosi, Quiroga, Mcconnell, Convergent evolution in SARS-CoV-2 spike creates a variant soup from which new COVID-19 waves emerge, Int J Mol Sci, doi:10.3390/ijms24032264
Focosi, Spezia, Gueli, The era of the flips: how spike mutations L455F and F456L (and A475V) are shaping SARS-CoV-2 evolution, Viruses, doi:10.3390/v16010003
Francica, Cai, Diallo, The SARS-CoV-2 monoclonal antibody AZD3152 potently neutralizes historical and emerging variants and is being developed for the prevention and treatment of COVID-19 in high-risk individuals j AZ RIA -QR Tool
Francica, Cai, Diallo, The SARS-CoV-2 monoclonal antibody AZD3152 potently neutralizes historical and emerging variants and is being developed for the prevention and treatment of COVID-19 in high-risk individuals, Open Forum Infect Dis, doi:10.1093/ofid/ofad500.1192
Garnier, Ross, Rudis, Viridis: colorblind-friendly color maps for R, doi:10.32614/CRAN.package.viridis
Gisaid -Gisaid, None, Org
Gliga, L€ Ubke, Killer, Rapid selection of sotrovimab escape variants in severe acute respiratory syndrome coronavirus 2 omicron-infected immunocompromised patients, Clin Infect Dis, doi:10.1093/cid/ciac802
Haars, Palanisamy, Wallin, Prevalence of SARS-CoV-2 omicron sublineages and spike protein mutations conferring resistance against monoclonal antibodies in a Swedish cohort during 2022-2023, Microorganisms, doi:10.3390/microorganisms11102417
Hemo, Islam, Jn, 1 as a New Variant of COVID-19 -Editorial, Ann Med Surg (Lond), doi:10.1097/MS9.0000000000001876
Heo, Sotrovimab: first approval, Drugs, doi:10.1007/s40265-022-01690-7
Jackson, Farzan, Chen, Mechanisms of SARS-CoV-2 entry into cells, Nat Rev Mol Cell Biol, doi:10.1038/s41580-021-00418-x
Jian, Feng, Yang, Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding, PLoS Pathog, doi:10.1371/journal.ppat.1011868
Koskela Von Sydow, Lindqvist, Asghar, Comparison of SARS-CoV-2 whole genome sequencing using tiled amplicon enrichment and bait hybridization, Sci Rep, doi:10.1038/s41598-023-33168-1
Kumar, Wu, Stosor, Real-world experience of bamlanivimab for coronavirus disease 2019 (COVID-19): a case-control study, Clin Infect Dis, doi:10.1093/cid/ciab305
Lee, Lee, Ko, Effectiveness of regdanvimab treatment in high-risk COVID-19 patients to prevent progression to severe disease, Front Immunol, doi:10.3389/fimmu.2021.772320
Li, Faraone, Hsu, Neutralization and Stability of JN.1-Derived LB.1, KP.2, doi:10.1101/2024.09.04.611219
Liu, Wei, Zhang, 501Y.V2 and 501Y.V3 Variants of SARS-CoV-2 lose binding to bamlanivimab in vitro, MAbs, doi:10.1080/19420862.2021.1919285
Mannsverk, Bergholm, Palanisamy, SARS-CoV-2 variants of concern and spike protein mutational dynamics in a Swedish cohort during 2021, studied by nanopore sequencing, Virol J, doi:10.1186/s12985-022-01896-x
Martinell, Andersson, Mannsverk, In-flight transmission of a SARS-CoV-2 Lineage B.1.617.2 harbouring the rare S:E484Q immune escape mutation, Viruses, doi:10.3390/v14030504
Mercatelli, Triboli, Fornasari, Coronapp: A web application to annotate and monitor SARS-CoV-2 mutations, J Med Virol, doi:10.1002/jmv.26678
Neuwirth, RColorBrewer: colorbrewer palettes
O'brien, Forleo-Neto, Sarkar, Effect of subcutaneous casirivimab and imdevimab antibody combination vs placebo on development of symptomatic COVID-19 in early asymptomatic SARS-CoV-2 infection: a randomized clinical trial, JAMA, doi:10.1001/jama.2021.24939
O'toole � A, Scher, Underwood, Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool, Virus Evol, doi:10.1093/ve/veab064
Outbreak, Info SARS-CoV-2 Data Explorer
Planas, Staropoli, Planchais, Escape of SARS-CoV-2 Variants KP1.1, LB.1, and KP3.3 from approved monoclonal antibodies, Pathog Immun, doi:10.20411/pai.v10i1.752
Research, FDA Announces Bebtelovimab Is Not Currently Authorized in Any US Region
Research, FDA Announces Evusheld Is Not Currently Authorized for Emergency Use in the U.S
Rosen, Tortorici, Marco, A potent pansarbecovirus neutralizing antibody resilient to epitope diversification, Cell, doi:10.1016/j.cell.2024.09.026
Somersan-Karakaya, Mylonakis, Menon, Casirivimab and imdevimab for the treatment of hospitalized patients with COVID-19, J Infect Dis, doi:10.1093/infdis/jiac320
Tai, He, Zhang, Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine, Cell Mol Immunol, doi:10.1038/s41423-020-0400-4
Team, A Language and Environment for Statistical Computing
Vasta-Ainehoito, AZD3152) Covid-19-Infektion Ehk€ aisyyn Kantasolu-Ja Elinsiirtopotilailla
Wang, Guo, Ho, Pemivibart is less active against recent SARS-CoV-2 JN.1 Sublineages, bioRxiv, doi:10.1101/2024.08.12.607496
Wang, Iketani, Li, Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants, Cell, doi:10.1016/j.cell.2022.12.018
Wickham, Franc¸ois, Henry, PBC Dplyr: A grammar of data manipulation, doi:10.32614/CRAN.package.dplyr
Wickham, Ggplot2: elegant graphics for data analysis
Wickham, Pedersen, Seidel, Scales: scale functions for visualization, doi:10.32614/CRAN.package.scales
Wickham, RStudio forcats: tools for working with categorical variables (factors), doi:10.32614/CRAN.package.forcats
Wickham, Software, Stringr: simple, consistent wrappers for common string operations, doi:10.32614/CRAN.package.stringr
Wickham, Vaughan, Girlich, Tidyr: tidy Messy Data, doi:10.32614/CRAN.package.tidyr
DOI record:
{
"DOI": "10.1080/23744235.2025.2509011",
"ISSN": [
"2374-4235",
"2374-4243"
],
"URL": "http://dx.doi.org/10.1080/23744235.2025.2509011",
"alternative-id": [
"10.1080/23744235.2025.2509011"
],
"assertion": [
{
"label": "Peer Review Statement",
"name": "peerreview_statement",
"order": 1,
"value": "The publishing and review policy for this title is described in its Aims & Scope."
},
{
"URL": "http://www.tandfonline.com/action/journalInformation?show=aimsScope&journalCode=infd20",
"label": "Aim & Scope",
"name": "aims_and_scope_url",
"order": 2,
"value": "http://www.tandfonline.com/action/journalInformation?show=aimsScope&journalCode=infd20"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Received",
"name": "received",
"order": 0,
"value": "2025-02-07"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Revised",
"name": "revised",
"order": 1,
"value": "2025-05-14"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "2025-05-15"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Published",
"name": "published",
"order": 3,
"value": "2025-05-26"
}
],
"author": [
{
"affiliation": [
{
"name": "Department of Medical Sciences, Uppsala University",
"place": [
"Uppsala, Sweden"
]
}
],
"family": "Haars",
"given": "Jonathan",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Department of Laboratory Medicine, Clinical Microbiology, Örebro University Hospital, Södra Grev Rosengatan",
"place": [
"Örebro, Sweden"
]
}
],
"family": "Wallin",
"given": "Frans",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Clinical Microbiology, Laboratory Medicine, Falu Hospital",
"place": [
"Falun, Sweden"
]
}
],
"family": "Elfving",
"given": "Karin",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Clinical Microbiology, Laboratory Medicine, Falu Hospital",
"place": [
"Falun, Sweden"
]
}
],
"family": "Jonsson",
"given": "Anna-Karin",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Medical Sciences, Uppsala University",
"place": [
"Uppsala, Sweden"
]
}
],
"family": "Ellström",
"given": "Patrik",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Laboratory Medicine, Clinical Microbiology, Faculty of Medicine and Health, Örebro University",
"place": [
"Örebro, Sweden"
]
}
],
"family": "Mölling",
"given": "Paula",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Medical Sciences, Uppsala University",
"place": [
"Uppsala, Sweden"
]
}
],
"family": "Lindh",
"given": "Johan",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Clinical Microbiology, Laboratory Medicine, Falu Hospital",
"place": [
"Falun, Sweden"
]
}
],
"family": "Yin",
"given": "Hong",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Laboratory Medicine, Clinical Microbiology, Faculty of Medicine and Health, Örebro University",
"place": [
"Örebro, Sweden"
]
}
],
"family": "Sundqvist",
"given": "Martin",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Medical Sciences, Uppsala University",
"place": [
"Uppsala, Sweden"
]
},
{
"name": "Clinical Genomics Uppsala, SciLifeLab",
"place": [
"Uppsala, Sweden"
]
}
],
"family": "Kaden",
"given": "René",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Chester Medical School, University of Chester",
"place": [
"Chester, United Kingdom"
]
}
],
"family": "Palanisamy",
"given": "Navaneethan",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Medical Sciences, Uppsala University",
"place": [
"Uppsala, Sweden"
]
}
],
"family": "Lennerstrand",
"given": "Johan",
"sequence": "additional"
}
],
"container-title": "Infectious Diseases",
"container-title-short": "Infectious Diseases",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"www.tandfonline.com"
]
},
"created": {
"date-parts": [
[
2025,
5,
26
]
],
"date-time": "2025-05-26T13:49:35Z",
"timestamp": 1748267375000
},
"deposited": {
"date-parts": [
[
2025,
5,
26
]
],
"date-time": "2025-05-26T13:49:38Z",
"timestamp": 1748267378000
},
"funder": [
{
"award": [
"RFR-994199"
],
"name": "Regional Research Council Mid Sweden"
}
],
"indexed": {
"date-parts": [
[
2025,
5,
27
]
],
"date-time": "2025-05-27T04:12:36Z",
"timestamp": 1748319156142,
"version": "3.41.0"
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2025,
5,
26
]
]
},
"language": "en",
"license": [
{
"URL": "http://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
5,
26
]
],
"date-time": "2025-05-26T00:00:00Z",
"timestamp": 1748217600000
}
}
],
"link": [
{
"URL": "https://www.tandfonline.com/doi/pdf/10.1080/23744235.2025.2509011",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "301",
"original-title": [],
"page": "1-10",
"prefix": "10.1080",
"published": {
"date-parts": [
[
2025,
5,
26
]
]
},
"published-online": {
"date-parts": [
[
2025,
5,
26
]
]
},
"publisher": "Informa UK Limited",
"reference": [
{
"DOI": "10.1016/S1473-3099(22)00311-5",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_2_1"
},
{
"DOI": "10.1093/cid/ciab305",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_3_1"
},
{
"DOI": "10.1093/infdis/jiac320",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_4_1"
},
{
"DOI": "10.1001/jama.2021.24939",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_5_1"
},
{
"DOI": "10.3389/fimmu.2021.772320",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_6_1"
},
{
"DOI": "10.1007/s40265-022-01690-7",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_7_1"
},
{
"DOI": "10.1002/rmv.2402",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_8_1"
},
{
"key": "e_1_3_5_9_1",
"unstructured": "EMA. Evusheld. [accessed 2023 August 22]. Available at: https://www.ema.europa.eu/en/medicines/human/EPAR/evusheld."
},
{
"DOI": "10.1016/j.kint.2022.05.008",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_10_1"
},
{
"DOI": "10.3390/v15010118",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_11_1"
},
{
"key": "e_1_3_5_12_1",
"unstructured": "Research C. for D.E. and FDA Announces Bebtelovimab Is Not Currently Authorized in Any US Region. [accessed 2023 August 22]. Available at: https://www.fda.gov/drugs/drug-safety-and-availability/fda-announces-bebtelovimab-not-currently-authorized-any-us-region."
},
{
"DOI": "10.1080/19420862.2021.1919285",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_13_1"
},
{
"DOI": "10.1093/cid/ciac802",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_14_1"
},
{
"DOI": "10.1038/s41423-020-0400-4",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_15_1"
},
{
"DOI": "10.1371/journal.ppat.1011868",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_16_1"
},
{
"DOI": "10.3390/ijms24032264",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_17_1"
},
{
"DOI": "10.1038/s41580-021-00418-x",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_18_1"
},
{
"DOI": "10.1038/s41579-022-00809-7",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_19_1"
},
{
"DOI": "10.3390/v14091999",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_20_1"
},
{
"DOI": "10.1016/j.cell.2022.12.018",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_21_1"
},
{
"DOI": "10.1038/s41586-022-05644-7",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_22_1"
},
{
"DOI": "10.3390/microorganisms11102417",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_23_1"
},
{
"key": "e_1_3_5_24_1",
"unstructured": "Research C. for D.E. and FDA Announces Evusheld Is Not Currently Authorized for Emergency Use in the U.S. [accessed 2023 August 22]. Available at: https://www.fda.gov/drugs/drug-safety-and-availability/fda-announces-evusheld-not-currently-authorized-emergency-use-us."
},
{
"DOI": "10.3899/jrheum.2023-1186",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_25_1"
},
{
"key": "e_1_3_5_26_1",
"unstructured": "ECCMID Poster: P2636 The SARS-CoV-2 Monoclonal Antibody AZD3152 Potently Neutralises Historical and Currently Circulating Variants. 2023."
},
{
"DOI": "10.1093/ofid/ofad500.1192",
"doi-asserted-by": "crossref",
"key": "e_1_3_5_27_1",
"unstructured": "Francica JR Cai Y Diallo S et al. The SARS-CoV-2 monoclonal antibody AZD3152 potently neutralizes historical and emerging variants and is being developed for the prevention and treatment of COVID-19 in high-risk individuals | AZ RIA - QR Tool. [accessed 2024 May 31]. Available at: https://congresspublication.com/fbkgfb?utm_term=Congress&utm_medium=Poster&utm_campaign=ID-Week-2023&utm_content=IDWeek-2023-AZD3152-PK-tox-poster&utm_source=82&utm_contentcategory=PDF#Congress."
},
{
"DOI": "10.1093/ofid/ofad500.1192",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_28_1"
},
{
"key": "e_1_3_5_29_1",
"unstructured": "AstraZeneca A Phase I/III Randomized Double Blind Study to Evaluate the Safety Efficacy and Neutralizing Activity of AZD5156/AZD3152 for Pre Exposure Prophylaxis of COVID 19 in Participants With Conditions Causing Immune Impairment. Sub-Study: phase II Open Label Sub-Study to Evaluate the Safety PK and Neutralizing Activity of AZD3152 for Pre-Exposure Prophylaxis of COVID-19; clinicaltrials.gov; 2023."
},
{
"key": "e_1_3_5_30_1",
"unstructured": "SUPERNOVA Phase III Trial of Sipavibart Long-Acting Antibody Met Primary Endpoints in Preventing COVID-19 in Immunocompromised Patient Population (Cision). [accessed 2024 May 31]. Available at: https://www.placera.se/placera/pressmeddelanden/2024/05/16/astrazeneca-supernova-phase-iii-trial-of-sipavibart-long-acting-antibody-met-primary-endpoints-in-preventing-covid-19-in-immunocompromised-patient-population.html."
},
{
"key": "e_1_3_5_31_1",
"unstructured": "Vasta-Ainehoito (AZD3152) Covid-19-Infektion Ehkäisyyn Kantasolu- Ja Elinsiirtopotilailla. [accessed 2024 May 31]. Available at: https://hoito-ohjeet.fi/fi/Ohjepankki/VSSHP/Vasta-ainehoito%20%28AZD3152%29%20Covid-19-infektion%20ehkäisyyn%20kantasolu-%20ja%20elinsiirtopotilailla.pdf."
},
{
"key": "e_1_3_5_32_1",
"unstructured": "Liste Des Spécialités En Accès Dérogatoire - AZD3152 - ANSM. [accessed 2024 May 31]. Available at: https://ansm.sante.fr/tableau-acces-derogatoire/azd3152#."
},
{
"key": "e_1_3_5_33_1",
"unstructured": "Norwegian Medical Products Agency Compassionate Use Program (CUP) Approved by The Norwegian Medicinal Products Agency. [accessed 2024 May 31]. Available at: https://www.dmp.no/contentassets/b71d6f2f34264dd7a22822deab47c201/compassionate-use-program_eng_2024-04-30.pdf."
},
{
"key": "e_1_3_5_34_1",
"unstructured": "Kavigale | European Medicines Agency (EMA). [accessed 2025 February 4]. Available at: https://www.ema.europa.eu/en/medicines/human/EPAR/kavigale."
},
{
"DOI": "10.1126/scitranslmed.ado2817",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_35_1"
},
{
"DOI": "10.3390/v16010003",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_36_1"
},
{
"DOI": "10.20411/pai.v10i1.752",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_37_1"
},
{
"DOI": "10.1093/ve/veab064",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_38_1"
},
{
"key": "e_1_3_5_39_1",
"unstructured": "Invivyd Announces FDA Authorization for Emergency Use of PEMGARDATM (Formerly VYD222) for Pre-Exposure Prophylaxis (PrEP) of COVID-19 – Invivyd. [accessed 2024 April 4]. Available at: https://investors.invivyd.com/news-releases/news-release-details/invivyd-announces-fda-authorization-emergency-use-pemgardatm/."
},
{
"DOI": "10.1093/cid/ciae435",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_40_1"
},
{
"DOI": "10.1016/j.cmi.2024.04.012",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_41_1"
},
{
"DOI": "10.1101/2024.08.12.607496",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_42_1"
},
{
"DOI": "10.1186/s12985-022-01896-x",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_43_1"
},
{
"DOI": "10.3390/v14030504",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_44_1"
},
{
"DOI": "10.3390/ijms25126645",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_45_1"
},
{
"DOI": "10.1038/s41598-023-33168-1",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_46_1"
},
{
"key": "e_1_3_5_47_1",
"unstructured": "Geneious | Bioinformatics Software for Sequence Data Analysis. [accessed 2023 August 22]. Available at: https://www.geneious.com/."
},
{
"key": "e_1_3_5_48_1",
"unstructured": "Epi2me-Labs/Wf-Artic: ARTIC SARS-CoV-2 Workflow and Reporting. [accessed 2023 September 8]. Available at: https://github.com/epi2me-labs/wf-artic."
},
{
"key": "e_1_3_5_49_1",
"unstructured": "Geneious Wrapper Plugin for Pangolin. [accessed 2023 August 29]. Available at: https://github.com/clinical-genomics-uppsala/Geneious_pangolin_wrapper."
},
{
"DOI": "10.1002/jmv.26678",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_50_1"
},
{
"key": "e_1_3_5_51_1",
"unstructured": "R Core Team R: A Language and Environment for Statistical Computing. Vienna, Austria; R Foundation for Statistical Computing; 2021. https://www.R-project.org/.",
"volume-title": "R Core Team R: A Language and Environment for Statistical Computing. Vienna, Austria; R Foundation for Statistical Computing",
"year": "2021"
},
{
"DOI": "10.21105/joss.03773",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_52_1"
},
{
"DOI": "10.32614/CRAN.package.dplyr",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_53_1"
},
{
"DOI": "10.32614/CRAN.package.stringr",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_54_1"
},
{
"DOI": "10.32614/CRAN.package.tidyr",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_55_1"
},
{
"DOI": "10.1007/978-3-319-24277-4",
"author": "Wickham H.",
"doi-asserted-by": "crossref",
"key": "e_1_3_5_56_1",
"unstructured": "Wickham H. Ggplot2: elegant graphics for data analysis. New York: Springer-Verlag; 2016.",
"volume-title": "Ggplot2: elegant graphics for data analysis",
"year": "2016"
},
{
"DOI": "10.32614/CRAN.package.forcats",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_57_1"
},
{
"author": "Neuwirth E.",
"journal-title": "RColorBrewer: colorbrewer palettes",
"key": "e_1_3_5_58_1",
"unstructured": "Neuwirth E. RColorBrewer: colorbrewer palettes. 2022.",
"year": "2022"
},
{
"DOI": "10.32614/CRAN.package.scales",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_59_1"
},
{
"DOI": "10.32614/CRAN.package.viridis",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_60_1"
},
{
"key": "e_1_3_5_61_1",
"unstructured": "WHO Chief Declares End to COVID-19 as a Global Health Emergency | UN News. [accessed 2025 January 22]. Available at: https://news.un.org/en/story/2023/05/1136367."
},
{
"DOI": "10.1101/2024.09.04.611219",
"doi-asserted-by": "crossref",
"key": "e_1_3_5_62_1",
"unstructured": "Li P Faraone JN Hsu CC et al. Neutralization and Stability of JN.1-Derived LB.1 KP.2.3 KP.3 and KP.3.1.1 Subvariants. bioRxiv. 2024. doi: 10.1101/2024.09.04.611219."
},
{
"DOI": "10.1097/MS9.0000000000001876",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_63_1"
},
{
"DOI": "10.2807/1560-7917.ES.2023.28.46.2300595",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_64_1"
},
{
"key": "e_1_3_5_65_1",
"unstructured": "CDC KP.3.1.1 Is the Predominant Variant as COVID-19 Activity Increases. [accessed 2025 January 22]. Available at: https://www.cdc.gov/ncird/whats-new/kp-3-1-1-is-the-predominant-variant.html."
},
{
"key": "e_1_3_5_66_1",
"unstructured": "Outbreak.Info SARS-CoV-2 Data Explorer. [accessed 2025 January 25]. Available at: https://outbreak.info/."
},
{
"key": "e_1_3_5_67_1",
"unstructured": "GISAID - Gisaid.Org. [accessed 2025 January 20]. Available at: https://gisaid.org/."
},
{
"DOI": "10.1016/j.cell.2024.09.026",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_68_1"
}
],
"reference-count": 67,
"references-count": 67,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.tandfonline.com/doi/full/10.1080/23744235.2025.2509011"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Dynamics of SARS-CoV-2 variants and mutations in Central Sweden between 2023 and 2024 and their potential implications on monoclonal antibodies pemivibart and sipavibart as PrEP in the region",
"type": "journal-article",
"update-policy": "https://doi.org/10.1080/tandf_crossmark_01"
}
haars2