Escape of SARS-CoV-2 Variants KP.1.1, LB.1, and KP.3.3 From Approved Monoclonal Antibodies
et al., Pathogens and Immunity, doi:10.20411/pai.v10i1.752, Sep 2024
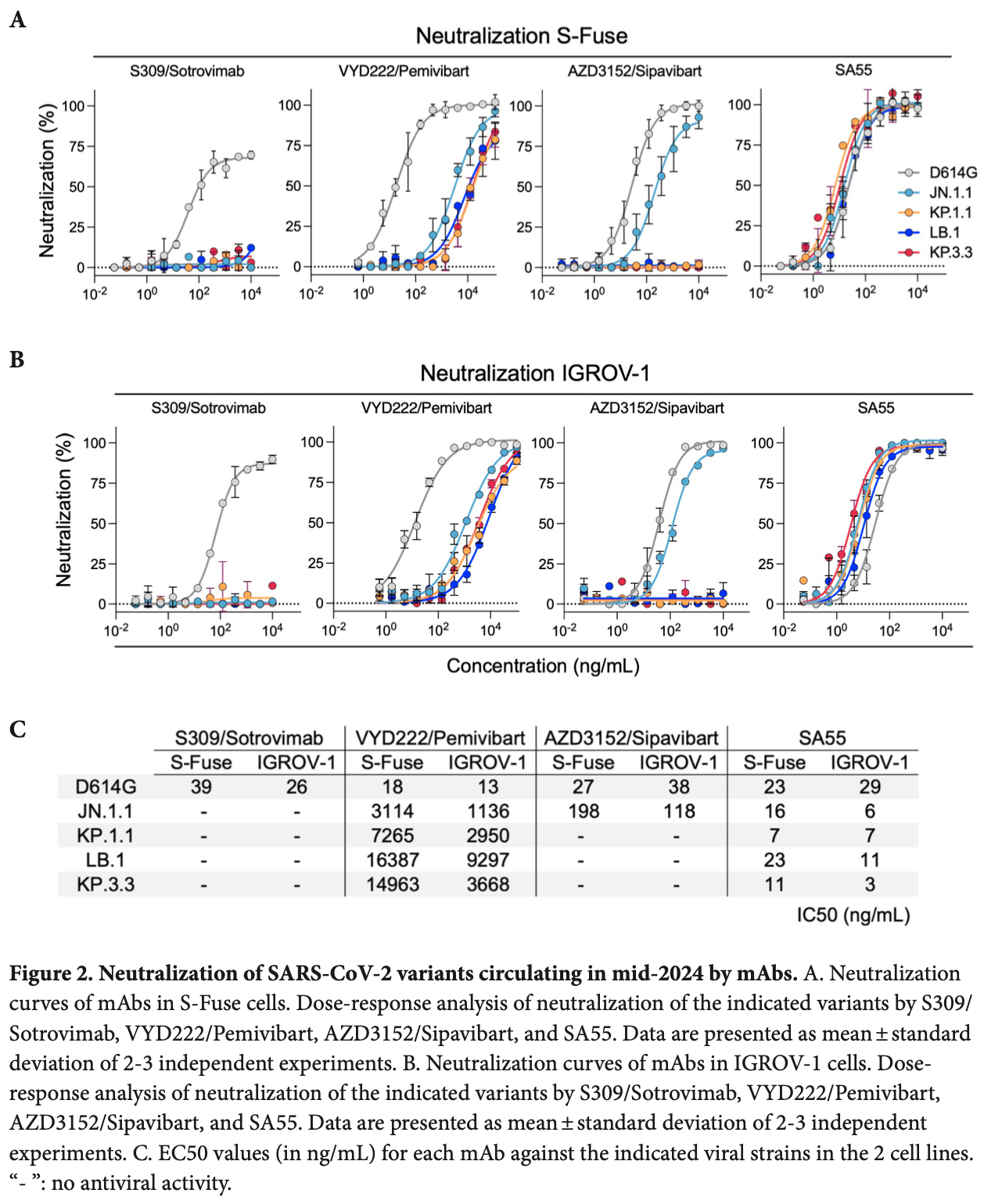
In vitro study showing significant escape of SARS-CoV-2 variants KP.1.1, LB.1, and KP.3.3 with monoclonal antibodies pemivibart (VYD222) and sipavibart (AZD3152). Sipavibart lost antiviral efficacy, while pemivibart maintained reduced activity. The mAb SA55 remained highly active against these variants. These results raise concerns about the clinical efficacy of pemivibart and sipavibart against JN.1 sublineages circulating in mid-2024.
Efficacy is variant dependent. In Vitro research shows reduced efficacy against KP.3.1.1, KP.1.1, LB.1, KP.3.3, and XEC variants1-4.
Study covers pemivibart and sipavibart.
1.
Xie et al., Molecular Basis of High-Blood-Pressure-Enhanced and High-Fever-Temperature-Weakened Receptor-Binding Domain/Peptidase Domain Binding: A Molecular Dynamics Simulation Study, International Journal of Molecular Sciences, doi:10.3390/ijms26073250.
2.
Wang et al., Activity of Research-Grade Pemivibart against Recent SARS-CoV-2 JN.1 Sublineages, New England Journal of Medicine, doi:10.1056/NEJMc2410203.
3.
Yao et al., Neutralizing Activity and Viral Escape of Pemivibart by SARS-CoV-2 JN.1 sublineages, bioRxiv, doi:10.1101/2024.11.08.622746.
4.
Planas et al., Escape of SARS-CoV-2 Variants KP.1.1, LB.1, and KP.3.3 From Approved Monoclonal Antibodies, Pathogens and Immunity, doi:10.20411/pai.v10i1.752.
Planas et al., 30 Sep 2024, France, peer-reviewed, 14 authors, study period 1 January, 2024 - 4 August, 2024.
Contact: olivier.schwartz@pasteur.fr.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Escape of SARS-CoV-2 Variants KP.1.1, LB.1, and KP.3.3 From Approved Monoclonal Antibodies
Pathogens and Immunity, doi:10.20411/pai.v10i1.752
Background: First-generation anti-SARS-CoV-2 monoclonal antibodies (mAbs) used for prophylaxis or therapeutic purposes in immunocompromised patients have been withdrawn because of the emergence of resistant Omicron variants. In 2024, 2 novel mAbs, VYD222/Pemivibart and AZD3152/Sipavibart, were approved by health authorities, but their activity against contemporary JN.1 sublineages is poorly characterized.
Methods: We isolated authentic JN.1.1, KP.1.1, LB.1, and KP.3.3 viruses and evaluated their sensitivity to neutralization by these mAbs in 2 target cell lines. Results: Compared to ancestral strains, VYD222/Pemivibart remained moderately active against JN.1 subvariants, with a strong increase of 50% Inhibitory Concentration (IC50), reaching up to 3 to 15 µg/mL for KP.3.3. AZD3152/Sipavibart neutralized JN.1.1 but lost antiviral efficacy against KP.1.1, LB.1, and KP.3.3.
Conclusions: Our results highlight the need for a close clinical monitoring of VYD222/Pemivibart and raise concerns about the clinical efficacy of AZD3152/Sipavibart.
AUTHOR CONTRIBUTIONS Experimental strategy design, experiments: DP, IS, CP, FG-B, ES-L, HM, M-ARW, OS. Vital materials: CP, EY, BJ, YR, VE, HM, M-ARW. Phylogenetic analysis: ES-L. Viral sequencing: EY, BJ, YR, VE, MP, FL, ES-L, M-ARW. Manuscript writing and editing: DP, ES-L, HM, M-ARW, OS.
References
Astrazeneca, Sipavibart EMA regulatory submission accepted under accelerated assessment for COVID-19 prevention
Boren, Efficacy and Safety of the Anti-COVID-19 Antibody SA55 for Injection in Patients With Hematological Disorders Who Are Persistently Positive for COVID-19
Bruel, Hadjadj, Maes, Planas, Seve et al., Serum neutralization of SARS-CoV-2 Omicron sublineages BA.1 and BA.2 in patients receiving monoclonal antibodies, Nat Med, doi:10.1038/s41591-022-01792-5
Buchrieser, Dufloo, Hubert, Monel, Planas et al., Syncytia formation by SARS-CoV-2-infected cells, EMBO J, doi:10.15252/embj.2020106267
Cai, Diallo, Rosenthal, Ren, Flores et al., AZD3152 neutralizes SARS-CoV-2 historical and contemporary variants and is protective in hamsters and well tolerated in adults, Sci Transl Med, doi:10.1126/scitranslmed.ado2817
Cao, Jian, Zhang, Yisimayi, Hao et al., Rational identification of potent and broad sarbecovirus-neutralizing antibody cocktails from SARS convalescents, Cell Rep, doi:10.1016/j.celrep.2022.111845
Elbe, Buckland-Merrett, Data, disease and diplomacy: GISAID's innovative contribution to global health, Glob Chall, doi:10.1002/gch2.1018
Focosi, Franchini, Casadevall, Maggi, An update on the anti-spike monoclonal antibody pipeline for SARS-CoV-2, Clin Microbiol Infect, doi:10.1016/j.cmi.2024.04.012
Kaku, Uriu, Kosugi, Okumura, Yamasoba et al., Virological characteristics of the SARS-CoV-2 KP.2 variant, Lancet Infect Dis, doi:10.1016/S1473-3099(24)00298-6
Kaku, Yo, Tolentino, Uriu, Okumura et al., Virological characteristics of the SARS-CoV-2 KP.3, LB.1, and KP.2.3 variants, Lancet Infect Dis, doi:10.1016/S1473-3099(24)00415-8
Planas, Bruel, Grzelak, Guivel-Benhassine, Staropoli et al., Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies, Nat Med, doi:10.1038/s41591-021-01318-5
Planas, Saunders, Maes, Guivel-Benhassine, Planchais et al., Considerable escape of SARS-CoV-2 Omicron to antibody neutralization, Nature, doi:10.1038/s41586-021-04389-z
Planas, Staropoli, Michel, Lemoine, Donati et al., Distinct evolution of SARS-CoV-2 Omicron XBB and BA.2.86/JN.1 lineages combining increased fitness and antibody evasion, Nat Commun, doi:http://dx.doi.org.10.1038/s41467-024-46490-7
Planchais, Fernandez, Bruel, De Melo, Prot et al., Potent human broadly SARS-CoV-2-neutralizing IgA and IgG antibodies effective against Omicron BA.1 and BA.2, J Exp Med, doi:10.1084/jem.20220638
Shu, Mccauley, GISAID: Global initiative on sharing all influenza datafrom vision to reality, Euro Surveill, doi:10.2807/1560-7917.ES.2017.22.13.30494101
Tsueng, Mullen, Alkuzweny, Cano, Rush et al., Outbreak.info Research Library: a standardized, searchable platform to discover and explore COVID-19 resources, Nat Methods, doi:10.1038/s41592-023-01770-w
Wang, Guo, Ho, Ho, Pemivibart is less active against recent SARS-CoV-2 JN.1 sublineages, bioRxiv, doi:10.1101/2024.08.12.607496
Yang, Yu, Xu, Jian, Song et al., Fast evolution of SARS-CoV-2 BA.2.86 to JN.1 under heavy immune pressure, Lancet Infect Dis, doi:10.1016/S1473-3099(23)00744-2
DOI record:
{
"DOI": "10.20411/pai.v10i1.752",
"ISSN": [
"2469-2964"
],
"URL": "http://dx.doi.org/10.20411/pai.v10i1.752",
"abstract": "<jats:p>Background: First-generation anti-SARS-CoV-2 monoclonal antibodies (mAbs) used for prophylaxis or therapeutic purposes in immunocompromised patients have been withdrawn because of the emergence of resistant Omicron variants. In 2024, 2 novel mAbs, VYD222/Pemivibart and AZD3152/Sipavibart, were approved by health authorities, but their activity against contemporary JN.1 sublineages is poorly characterized. \nMethods: We isolated authentic JN.1.1, KP.1.1, LB.1, and KP.3.3 viruses and evaluated their sensitivity to neutralization by these mAbs in 2 target cell lines. \nResults: Compared to ancestral strains, VYD222/Pemivibart remained moderately active against JN.1 subvariants, with a strong increase of 50% Inhibitory Concentration (IC50), reaching up to 3 to 15 µg/mL for KP.3.3. AZD3152/Sipavibart neutralized JN.1.1 but lost antiviral efficacy against KP.1.1, LB.1, and KP.3.3. \nConclusions: Our results highlight the need for a close clinical monitoring of VYD222/Pemivibart and raise concerns about the clinical efficacy of AZD3152/Sipavibart.</jats:p>",
"author": [
{
"affiliation": [],
"family": "Planas",
"given": "Delphine",
"sequence": "first"
},
{
"affiliation": [],
"family": "Staropoli",
"given": "Isabelle",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Planchais",
"given": "Cyril",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Yab",
"given": "Emilie",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Jeyarajah",
"given": "Banujaa",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Rahou",
"given": "Yannis",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Prot",
"given": "Matthieu",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Guivel-Benhassine",
"given": "Florence",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Lemoine",
"given": "Frederic",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Enouf",
"given": "Vincent",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Simon-Loriere",
"given": "Etienne",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Mouquet",
"given": "Hugo",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Rameix-Welti",
"given": "Marie-Anne",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Schwartz",
"given": "Olivier",
"sequence": "additional"
}
],
"container-title": "Pathogens and Immunity",
"container-title-short": "PAI",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2024,
10,
2
]
],
"date-time": "2024-10-02T18:01:49Z",
"timestamp": 1727892109000
},
"deposited": {
"date-parts": [
[
2024,
10,
2
]
],
"date-time": "2024-10-02T18:02:01Z",
"timestamp": 1727892121000
},
"indexed": {
"date-parts": [
[
2024,
10,
3
]
],
"date-time": "2024-10-03T04:16:20Z",
"timestamp": 1727928980964
},
"is-referenced-by-count": 0,
"issue": "1",
"issued": {
"date-parts": [
[
2024,
9,
30
]
]
},
"journal-issue": {
"issue": "1",
"published-online": {
"date-parts": [
[
2024,
9,
30
]
]
}
},
"license": [
{
"URL": "http://creativecommons.org/licenses/by/4.0",
"content-version": "unspecified",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
9,
30
]
],
"date-time": "2024-09-30T00:00:00Z",
"timestamp": 1727654400000
}
}
],
"link": [
{
"URL": "https://www.paijournal.com/index.php/paijournal/article/download/752/833",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.paijournal.com/index.php/paijournal/article/download/752/830",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.paijournal.com/index.php/paijournal/article/download/752/830",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "7530",
"original-title": [],
"page": "1-11",
"prefix": "10.20411",
"published": {
"date-parts": [
[
2024,
9,
30
]
]
},
"published-online": {
"date-parts": [
[
2024,
9,
30
]
]
},
"publisher": "Case Western Reserve University",
"reference": [
{
"DOI": "10.1038/s41467-024-46490-7",
"doi-asserted-by": "crossref",
"key": "6623",
"unstructured": "<p>1.	Planas D, Staropoli I, Michel V, Lemoine F, Donati F, Prot M, Porrot F, Guivel-Benhassine F, Jeyarajah B, Brisebarre A, Dehan O, Avon L, Bolland WH, Hubert M, Buchrieser J, Vanhoucke T, Rosenbaum P, Veyer D, Pere H, Lina B, Trouillet-Assant S, Hocqueloux L, Prazuck T, Simon-Loriere E, Schwartz O. Distinct evolution of SARS-CoV-2 Omicron XBB and BA.2.86/JN.1 lineages combining increased fitness and antibody evasion. <em>Nat Commun</em>. 2024;15(1):2254. doi: <a href=\"http://dx.doi.org.10.1038/s41467-024-46490-7\" target=\"_blank\"><span>10.1038/s41467-024-46490-7</span></a>. PubMed PMID: 38480689; PMCID: PMC10938001.</p>"
},
{
"DOI": "10.1016/S1473-3099(24)00415-8",
"doi-asserted-by": "crossref",
"key": "6624",
"unstructured": "<p>2.	Kaku Y, Yo MS, Tolentino JE, Uriu K, Okumura K, Genotype to Phenotype Japan C, Ito J, Sato K. Virological characteristics of the SARS-CoV-2 KP.3, LB.1, and KP.2.3 variants. <em>Lancet Infect Dis</em>. 2024;24(8):e482-e3. doi: <a href=\"http://dx.doi.org/10.1016/S1473-3099(24)00415-8\"><span>10.1016/S1473-3099(24)00415-8</span></a>. PubMed PMID: 38945150.</p>"
},
{
"DOI": "10.1016/S1473-3099(24)00298-6",
"doi-asserted-by": "crossref",
"key": "6625",
"unstructured": "<p>3.	Kaku Y, Uriu K, Kosugi Y, Okumura K, Yamasoba D, Uwamino Y, Kuramochi J, Sadamasu K, Yoshimura K, Asakura H, Nagashima M, Genotype to Phenotype Japan C, Ito J, Sato K. Virological characteristics of the SARS-CoV-2 KP.2 variant. <em>Lancet Infect Dis</em>. 2024;24(7):e416. doi: <a href=\"http://dx.doi.org/10.1016/S1473-3099(24)00298-6\" target=\"_blank\"><span>10.1016/S1473-3099(24)00298-6</span></a>. PubMed PMID: 38782005.</p>"
},
{
"DOI": "10.1016/j.cmi.2024.04.012",
"doi-asserted-by": "crossref",
"key": "6626",
"unstructured": "<p>4.	Focosi D, Franchini M, Casadevall A, Maggi F. An update on the anti-spike monoclonal antibody pipeline for SARS-CoV-2. <em>Clin Microbiol Infect</em>. 2024;30(8):999-1006. doi: <a href=\"http://dx.doi.org/10.1016/j.cmi.2024.04.012\" target=\"_blank\"><span>10.1016/j.cmi.2024.04.012</span></a>. PubMed PMID: 38663655.</p>"
},
{
"DOI": "10.1126/scitranslmed.ado2817",
"doi-asserted-by": "crossref",
"key": "6627",
"unstructured": "<p>5.	Cai Y, Diallo S, Rosenthal K, Ren K, Flores DJ, Dippel A, Oganesyan V, van Dyk N, Chen X, Cantu E, Choudhary R, Sulikowski M, Adissu H, Chawla B, Kar S, Liu C, Dijokaite-Guraliuc A, Mongkolsapaya J, Rajan S, Loo YM, Beavon R, Webber C, Chang LJ, Thomas S, Clegg L, Zhang H, Screaton GR, Philbin N, Harre M, Selim A, Martinez-Alier N, Uriel A, Cohen TS, Perez JL, Esser MT, Blair W, Francica JR. AZD3152 neutralizes SARS-CoV-2 historical and contemporary variants and is protective in hamsters and well tolerated in adults. <em>Sci Transl Med</em>. 2024;16(753):eado2817. doi: <a href=\"http://dx.doi.org/10.1126/scitranslmed.ado2817\" target=\"_blank\"><span>10.1126/scitranslmed.ado2817</span></a>. PubMed PMID: 38924429.</p>"
},
{
"key": "6628",
"unstructured": "<p>6.	AstraZeneca. Sipavibart EMA regulatory submission accepted under accelerated assessment for COVID-19 prevention. 2024.</p>"
},
{
"key": "6629",
"unstructured": "<p>7.	Fact sheet for healthcare providers: Emergency Use Authorization of Pemgarda (pemivibart). Invivyd; 2024. Accessed March 25, 2024. In: Pemgarda, editor.: FDA.gov; 2024.</p>"
},
{
"DOI": "10.1016/j.celrep.2022.111845",
"doi-asserted-by": "crossref",
"key": "6630",
"unstructured": "<p>8.	Cao Y, Jian F, Zhang Z, Yisimayi A, Hao X, Bao L, Yuan F, Yu Y, Du S, Wang J, Xiao T, Song W, Zhang Y, Liu P, An R, Wang P, Wang Y, Yang S, Niu X, Zhang Y, Gu Q, Shao F, Hu Y, Yin W, Zheng A, Wang Y, Qin C, Jin R, Xiao J, Xie XS. Rational identification of potent and broad sarbecovirus-neutralizing antibody cocktails from SARS convalescents. <em>Cell Rep</em>. 2022;41(12):111845. doi: <a href=\"http://dx.doi.org/10.1016/j.celrep.2022.111845\" target=\"_blank\"><span>10.1016/j.celrep.2022.111845</span></a>. PubMed PMID: 36493787; PMCID: PMC9712074.</p>"
},
{
"DOI": "10.1016/S1473-3099(23)00744-2",
"doi-asserted-by": "crossref",
"key": "6631",
"unstructured": "<p>9.	Yang S, Yu Y, Xu Y, Jian F, Song W, Yisimayi A, Wang P, Wang J, Liu J, Yu L, Niu X, Wang J, Wang Y, Shao F, Jin R, Wang Y, Cao Y. Fast evolution of SARS-CoV-2 BA.2.86 to JN.1 under heavy immune pressure. <em>Lancet Infect Dis</em>. 2024;24(2):e70-e2. doi: <a href=\"http://dx.doi.org/10.1016/S1473-3099(23)00744-2\" target=\"_blank\"><span>10.1016/S1473-3099(23)00744-2</span></a>. PubMed PMID: 38109919.</p>"
},
{
"key": "6632",
"unstructured": "<p>10.	Beijing Boren Hospital. Efficacy and Safety of the Anti-COVID-19 Antibody SA55 for Injection in Patients With Hematological Disorders Who Are Persistently Positive for COVID-19 [Internet]. ClinicalTrials.gov identifier: NCT123456. Available from: <a href=\"https://clinicaltrials.gov/study/NCT05675943\" target=\"_blank\"><span>https://clinicaltrials.gov/study/NCT05675943</span></a>.</p>"
},
{
"DOI": "10.15252/embj.2020106267",
"doi-asserted-by": "crossref",
"key": "6633",
"unstructured": "<p>11.	Buchrieser J, Dufloo J, Hubert M, Monel B, Planas D, Rajah MM, Planchais C, Porrot F, Guivel-Benhassine F, Van der Werf S, Casartelli N, Mouquet H, Bruel T, Schwartz O. Syncytia formation by SARS-CoV-2-infected cells. <em>EMBO J</em>. 2020;39(23):e106267. doi: <a href=\"http://dx.doi.org/10.15252/embj.2020106267\" target=\"_blank\"><span>10.15252/embj.2020106267</span></a>. PubMed PMID: 33051876; PMCID: PMC7646020.</p>"
},
{
"DOI": "10.1038/s41586-021-04389-z",
"doi-asserted-by": "crossref",
"key": "6634",
"unstructured": "<p>12.	Planas D, Saunders N, Maes P, Guivel-Benhassine F, Planchais C, Buchrieser J, Bolland WH, Porrot F, Staropoli I, Lemoine F, Pere H, Veyer D, Puech J, Rodary J, Baele G, Dellicour S, Raymenants J, Gorissen S, Geenen C, Vanmechelen B, Wawina-Bokalanga T, Marti-Carreras J, Cuypers L, Seve A, Hocqueloux L, Prazuck T, Rey FA, Simon-Loriere E, Bruel T, Mouquet H, Andre E, Schwartz O. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. <em>Nature</em>. 2022;602(7898):671-5. doi: <a href=\"http://dx.doi.org/10.1038/s41586-021-04389-z\" target=\"_blank\"><span>10.1038/s41586-021-04389-z</span></a>. PubMed PMID: 35016199.</p>"
},
{
"DOI": "10.1038/s41591-021-01318-5",
"doi-asserted-by": "crossref",
"key": "6635",
"unstructured": "<p>13.	Planas D, Bruel T, Grzelak L, Guivel-Benhassine F, Staropoli I, Porrot F, Planchais C, Buchrieser J, Rajah MM, Bishop E, Albert M, Donati F, Prot M, Behillil S, Enouf V, Maquart M, Smati-Lafarge M, Varon E, Schortgen F, Yahyaoui L, Gonzalez M, De Seze J, Pere H, Veyer D, Seve A, Simon-Loriere E, Fafi-Kremer S, Stefic K, Mouquet H, Hocqueloux L, van der Werf S, Prazuck T, Schwartz O. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. <em>Nat Med</em>. 2021;27(5):917-24. doi: <a href=\"http://dx.doi.org/10.1038/s41591-021-01318-5\" target=\"_blank\"><span>10.1038/s41591-021-01318-5</span></a>. PubMed PMID: 33772244.</p>"
},
{
"DOI": "10.1038/s41591-022-01792-5",
"doi-asserted-by": "crossref",
"key": "6636",
"unstructured": "<p>14.	Bruel T, Hadjadj J, Maes P, Planas D, Seve A, Staropoli I, Guivel-Benhassine F, Porrot F, Bolland WH, Nguyen Y, Casadevall M, Charre C, Pere H, Veyer D, Prot M, Baidaliuk A, Cuypers L, Planchais C, Mouquet H, Baele G, Mouthon L, Hocqueloux L, Simon-Loriere E, Andre E, Terrier B, Prazuck T, Schwartz O. Serum neutralization of SARS-CoV-2 Omicron sublineages BA.1 and BA.2 in patients receiving monoclonal antibodies. <em>Nat Med</em>. 2022;28(6):1297-302. doi: <a href=\"http://dx.doi.org/10.1038/s41591-022-01792-5\" target=\"_blank\"><span>10.1038/s41591-022-01792-5</span></a>. PubMed PMID: 35322239; PMCID: 35322239.</p>"
},
{
"DOI": "10.1084/jem.20220638",
"doi-asserted-by": "crossref",
"key": "6637",
"unstructured": "<p>15.	Planchais C, Fernandez I, Bruel T, de Melo GD, Prot M, Beretta M, Guardado-Calvo P, Dufloo J, Molinos-Albert LM, Backovic M, Chiaravalli J, Giraud E, Vesin B, Conquet L, Grzelak L, Planas D, Staropoli I, Guivel-Benhassine F, Hieu T, Boulle M, Cervantes-Gonzalez M, Ungeheuer MN, Charneau P, van der Werf S, Agou F, French CCSG, Group CS, Dimitrov JD, Simon-Loriere E, Bourhy H, Montagutelli X, Rey FA, Schwartz O, Mouquet H. Potent human broadly SARS-CoV-2-neutralizing IgA and IgG antibodies effective against Omicron BA.1 and BA.2. <em>J Exp Med</em>. 2022;219(7). doi: <a href=\"http://dx.doi.org/10.1084/jem.20220638\" target=\"_blank\"><span>10.1084/jem.20220638</span></a>. PubMed PMID: 35704748; PMCID: PMC9206116.</p>"
},
{
"DOI": "10.1002/gch2.1018",
"doi-asserted-by": "crossref",
"key": "6638",
"unstructured": "<p>16.	Elbe S, Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. <em>Glob Chall</em>. 2017;1(1):33-46. doi: <a href=\"http://dx.doi.org/10.1002/gch2.1018\" target=\"_blank\"><span>10.1002/gch2.1018</span></a>. PubMed PMID: 31565258; PMCID: PMC6607375.</p>"
},
{
"key": "6639",
"unstructured": "<p>17.	Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. <em>Euro Surveill</em>. 2017;22(13):30494. doi: <a href=\"http://dx.doi.org/10.2807/1560-7917.ES.2017.22.13.30494101\" target=\"_blank\"><span>10.2807/1560-7917.ES.2017.22.13.30494</span></a>. PubMed PMID: 28382917; PMCID: PMC5388101.</p>"
},
{
"DOI": "10.1038/s41592-023-01770-w",
"doi-asserted-by": "crossref",
"key": "6640",
"unstructured": "<p>18.	Tsueng G, Mullen JL, Alkuzweny M, Cano M, Rush B, Haag E, Lin J, Welzel DJ, Zhou X, Qian Z, Latif AA, Hufbauer E, Zeller M, Andersen KG, Wu C, Su AI, Gangavarapu K, Hughes LD. Outbreak.info Research Library: a standardized, searchable platform to discover and explore COVID-19 resources. <em>Nat Methods</em>. 2023;20(4):536-40. doi: <a href=\"http://dx.doi.org/10.1038/s41592-023-01770-w\" target=\"_blank\"><span>10.1038/s41592-023-01770-w</span></a>. PubMed PMID: 36823331; PMCID: PMC10393269.</p>"
},
{
"key": "6641",
"unstructured": "<p>19.	Wang Q, Guo Y, Ho J, Ho DD. Pemivibart is less active against recent SARS-CoV-2 JN.1 sublineages. <em>bioRxiv</em>. 2024:2024.08.12.607496. doi: <a href=\"http://dx.doi.org/10.1101/2024.08.12.607496\" target=\"_blank\"><span>10.1101/2024.08.12.607496</span></a>.</p>"
}
],
"reference-count": 19,
"references-count": 19,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.paijournal.com/index.php/paijournal/article/view/752"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Escape of SARS-CoV-2 Variants KP.1.1, LB.1, and KP.3.3 From Approved Monoclonal Antibodies",
"type": "journal-article",
"volume": "10"
}