Pemivibart is less active against recent SARS-CoV-2 JN.1 sublineages
Qian Wang, Yicheng Guo, Jerren Ho, David D Ho
doi:10.1101/2024.08.12.607496
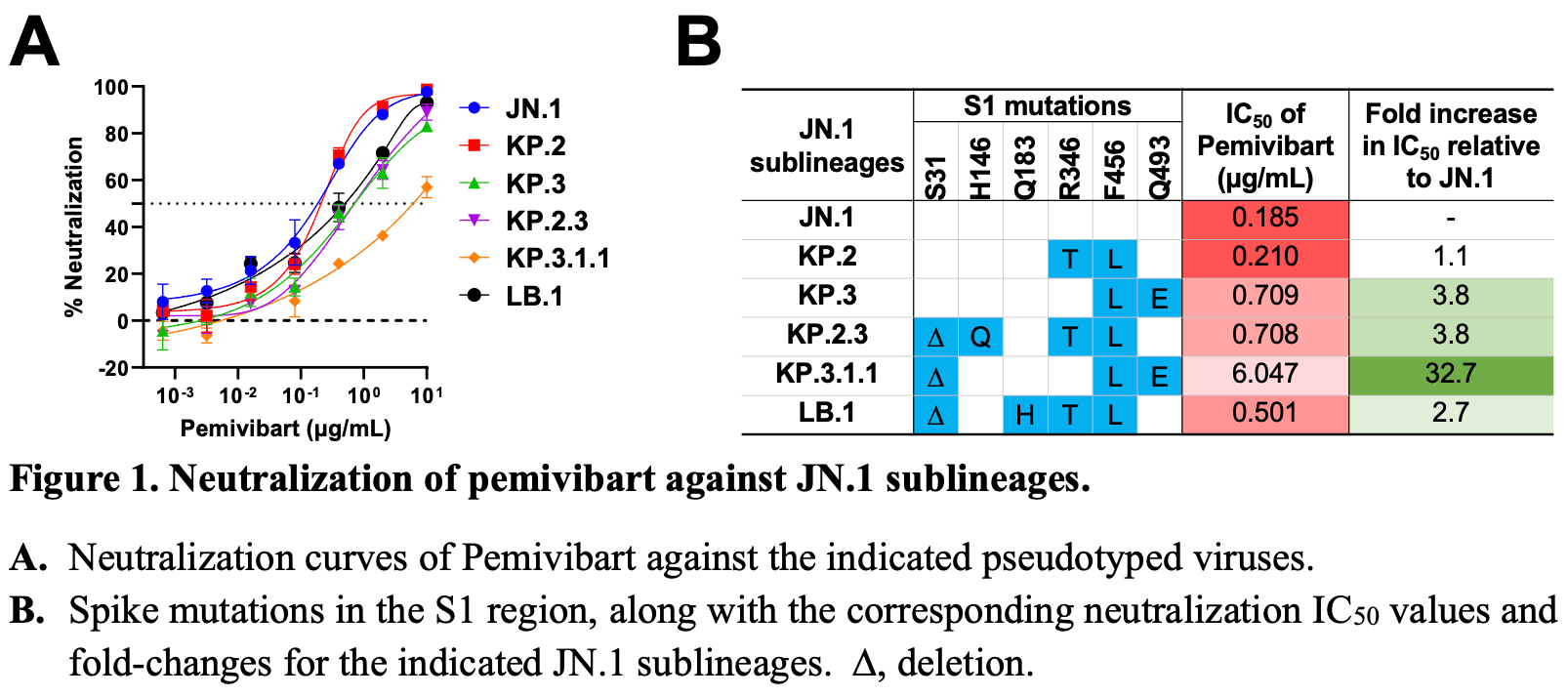
Protection from COVID-19 vaccination is suboptimal in many immunocompromised individuals. In March 2024, the Food and Drug Administration issued an Emergency Use Authorization for pemivibart (Permagard/VYD222), an engineered human monoclonal antibody, for pre-exposure prophylaxis in this vulnerable population. However, SARS-CoV-2 has since evolved extensively, resulting in multiple Omicron JN.1 sublineages. We therefore evaluated the in vitro neutralizing activity of pemivibart against the prevalent forms of JN.1, including KP.2, KP.3, KP.2.3, LB.1, and, importantly, KP.3.1.1, which is now expanding most rapidly. A panel of VSV-based pseudoviruses representing major JN.1 sublineages was generated to assess their susceptibility to pemivibart neutralization in vitro. Structural analyses were then conducted to understand the impact of specific spike mutations on the virus-neutralization results. Pemivibart neutralized both JN.1 and KP.2 in vitro with comparable activity, whereas its potency was decreased slightly against LB.1, KP.2.3, and KP.3 but substantially against KP.3.1.1. Critically, the 50% inhibitory concentration of pemivibart against KP.3.1.1 was ~6 µg/mL, or ~32.7 fold higher than that of JN.1. Structural analyses suggest that Q493E and the S31-deletion mutations in viral spike contribute to the antibody evasion, with the latter having a more pronounced effect. Our findings show that pemivibart has lost substantial neutralizing activity in vitro against KP.3.1.1, the most rapidly expanding lineage of SARS-CoV-2 today. Close monitoring of its clinical efficacy is therefore warranted. These results also highlight the imperative to expand our arsenal of preventive agents to protect millions of immunocompromised individuals who could not respond robustly to COVID-19 vaccines.
Supplementary Methods Cell lines Vero-E6 (CRL-1586) cells and HEK293T (CRL-3216) cells were obtained from ATCC and cultured at 37°C with 5% CO2 in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. Morphology of each cell line was confirmed visually before use. All cell lines tested mycoplasma negative. SARS-CoV-2 variant spike plasmids Spike-expressing plasmids for JN.1, KP.2, and KP.3 were previously generated 1 . To make the spike-expressing plasmids for KP.2.3, KP.3.1.1, and LB.1 for pseudovirus production, spike mutations were introduced to the JN.1 spike-expressing construct using the QuikChange II XL site-directed mutagenesis kit (Agilent). Spike mutations of each subvariant on top of JN.1 are shown in Figure S1A . Pseudovirus production SARS-CoV-2 pseudoviruses were produced in a vesicular stomatitis virus (VSV) background, in which the native VSV glycoprotein was replaced by SARS-CoV-2 variant spikes, as previously described 2 . Briefly, the spike-expressing construct was transfected into HEK293T cells using 1mg/mL PEI MAX (Polysciences Inc). 24 hours later, the transfected cells were infected with VSV-G pseudotyped ΔG-luciferase (G*ΔG-luciferase, Kerafast) for 2 hours, then washed three times with culture medium before being cultured in fresh medium for an additional 24 hours. Anti-VSVG (I1) antibody 3 was added to deplete non-pseudotyped viruses. Pseudoviruses were then harvested, clarified by..
References
Lefrancios, Lyles, The interactionof antiody with the major surface glycoprotein of vesicular stomatitis virus. I. Analysis of neutralizing epitopes with monoclonal antibodies, Virology
Liu, Wang, Nair, Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike, Nature
Rappazzo, Tse, Kaku, Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody, Science
Taylor, Starr, Deep mutational scanning of SARS-CoV-2 Omicron BA.2.86 and epistatic emergence of the KP.3 variant. bioRxiv
Wang, Mellis, Bowen, Recurrent SARS-CoV-2 spike mutations confer growth advantages to select JN.1 sublineages, bioRxiv
Wang, Mellis, Bowen, Recurrent SARS-CoV-2 spike mutations confer growth advantages to select JN.1 sublineages, bioRxiv
DOI record:
{
"DOI": "10.1101/2024.08.12.607496",
"URL": "http://dx.doi.org/10.1101/2024.08.12.607496",
"abstract": "<jats:title>Abstract</jats:title><jats:p>Protection from COVID-19 vaccination is suboptimal in many immunocompromised individuals. In March 2024, the Food and Drug Administration issued an Emergency Use Authorization for pemivibart (Permagard/VYD222), an engineered human monoclonal antibody, for pre-exposure prophylaxis in this vulnerable population. However, SARS-CoV-2 has since evolved extensively, resulting in multiple Omicron JN.1 sublineages. We therefore evaluated the in vitro neutralizing activity of pemivibart against the prevalent forms of JN.1, including KP.2, KP.3, KP.2.3, LB.1, and, importantly, KP.3.1.1, which is now expanding most rapidly. A panel of VSV-based pseudoviruses representing major JN.1 sublineages was generated to assess their susceptibility to pemivibart neutralization in vitro. Structural analyses were then conducted to understand the impact of specific spike mutations on the virus-neutralization results. Pemivibart neutralized both JN.1 and KP.2 in vitro with comparable activity, whereas its potency was decreased slightly against LB.1, KP.2.3, and KP.3 but substantially against KP.3.1.1. Critically, the 50% inhibitory concentration of pemivibart against KP.3.1.1 was ∼6 µg/mL, or ∼32.7 fold higher than that of JN.1. Structural analyses suggest that Q493E and the S31-deletion mutations in viral spike contribute to the antibody evasion, with the latter having a more pronounced effect. Our findings show that pemivibart has lost substantial neutralizing activity in vitro against KP.3.1.1, the most rapidly expanding lineage of SARS-CoV-2 today. Close monitoring of its clinical efficacy is therefore warranted. These results also highlight the imperative to expand our arsenal of preventive agents to protect millions of immunocompromised individuals who could not respond robustly to COVID-19 vaccines.</jats:p>",
"accepted": {
"date-parts": [
[
2024,
8,
13
]
]
},
"author": [
{
"ORCID": "http://orcid.org/0000-0001-9549-3140",
"affiliation": [],
"authenticated-orcid": false,
"family": "Wang",
"given": "Qian",
"sequence": "first"
},
{
"affiliation": [],
"family": "Guo",
"given": "Yicheng",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Ho",
"given": "Jerren",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Ho",
"given": "David D.",
"sequence": "additional"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2024,
8,
14
]
],
"date-time": "2024-08-14T03:40:15Z",
"timestamp": 1723606815000
},
"deposited": {
"date-parts": [
[
2024,
8,
16
]
],
"date-time": "2024-08-16T08:35:25Z",
"timestamp": 1723797325000
},
"group-title": "Immunology",
"indexed": {
"date-parts": [
[
2024,
8,
17
]
],
"date-time": "2024-08-17T00:19:12Z",
"timestamp": 1723853952373
},
"institution": [
{
"name": "bioRxiv"
}
],
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2024,
8,
13
]
]
},
"license": [
{
"URL": "https://www.biorxiv.org/about/FAQ#license",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
8,
13
]
],
"date-time": "2024-08-13T00:00:00Z",
"timestamp": 1723507200000
}
}
],
"link": [
{
"URL": "https://syndication.highwire.org/content/doi/10.1101/2024.08.12.607496",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "246",
"original-title": [],
"posted": {
"date-parts": [
[
2024,
8,
13
]
]
},
"prefix": "10.1101",
"published": {
"date-parts": [
[
2024,
8,
13
]
]
},
"publisher": "Cold Spring Harbor Laboratory",
"reference": [
{
"key": "2024081601350428000_2024.08.12.607496v1.1",
"unstructured": "FDA Roundup: March 22, 2024. 2024. (Accessed 07/25/2024, at https://www.fda.gov/news-events/press-announcements/fda-roundup-march-22-2024.)"
},
{
"DOI": "10.1126/science.abf4830",
"doi-asserted-by": "publisher",
"key": "2024081601350428000_2024.08.12.607496v1.2"
},
{
"key": "2024081601350428000_2024.08.12.607496v1.3",
"unstructured": "CDER Scientific Review Documents Supporting Emergency Use Authorizations for Drug and Biological Therapeutic Products | COVID-19. 2024. (Accessed 08/10/2024, at https://www.fda.gov/drugs/coronavirus-covid-19-drugs/cder-scientific-review-documents-supporting-emergency-use-authorizations-drug-and-biological.)"
},
{
"DOI": "10.1101/2024.05.29.596362",
"doi-asserted-by": "crossref",
"key": "2024081601350428000_2024.08.12.607496v1.4",
"unstructured": "Wang Q , Mellis IA , Bowen A , et al. Recurrent SARS-CoV-2 spike mutations confer growth advantages to select JN.1 sublineages. bioRxiv 2024:2024.05.29.596362."
},
{
"DOI": "10.1101/2024.07.23.604853",
"doi-asserted-by": "crossref",
"key": "2024081601350428000_2024.08.12.607496v1.5",
"unstructured": "Taylor AL , Starr TN . Deep mutational scanning of SARS-CoV-2 Omicron BA.2.86 and epistatic emergence of the KP.3 variant. bioRxiv 2024."
},
{
"DOI": "10.1101/2024.05.29.596362",
"doi-asserted-by": "crossref",
"key": "2024081601350428000_2024.08.12.607496v1.6",
"unstructured": "Wang Q , Mellis IA , Bowen A , et al. Recurrent SARS-CoV-2 spike mutations confer growth advantages to select JN.1 sublineages. bioRxiv 2024:2024.05.29.596362."
},
{
"DOI": "10.1038/s41586-020-2571-7",
"doi-asserted-by": "publisher",
"key": "2024081601350428000_2024.08.12.607496v1.7"
},
{
"DOI": "10.1016/0042-6822(82)90125-8",
"doi-asserted-by": "publisher",
"key": "2024081601350428000_2024.08.12.607496v1.8"
}
],
"reference-count": 8,
"references-count": 8,
"relation": {},
"resource": {
"primary": {
"URL": "http://biorxiv.org/lookup/doi/10.1101/2024.08.12.607496"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"subtype": "preprint",
"title": "Pemivibart is less active against recent SARS-CoV-2 JN.1 sublineages",
"type": "posted-content"
}