In-Silico Molecular Docking, Validation, Drug-Likeness, and ADMET Studies of Antiandrogens to Use in the Fight against SARS-CoV-2
et al., Physical Chemistry Research, doi:10.22036/pcr.2022.324549.2016, May 2022
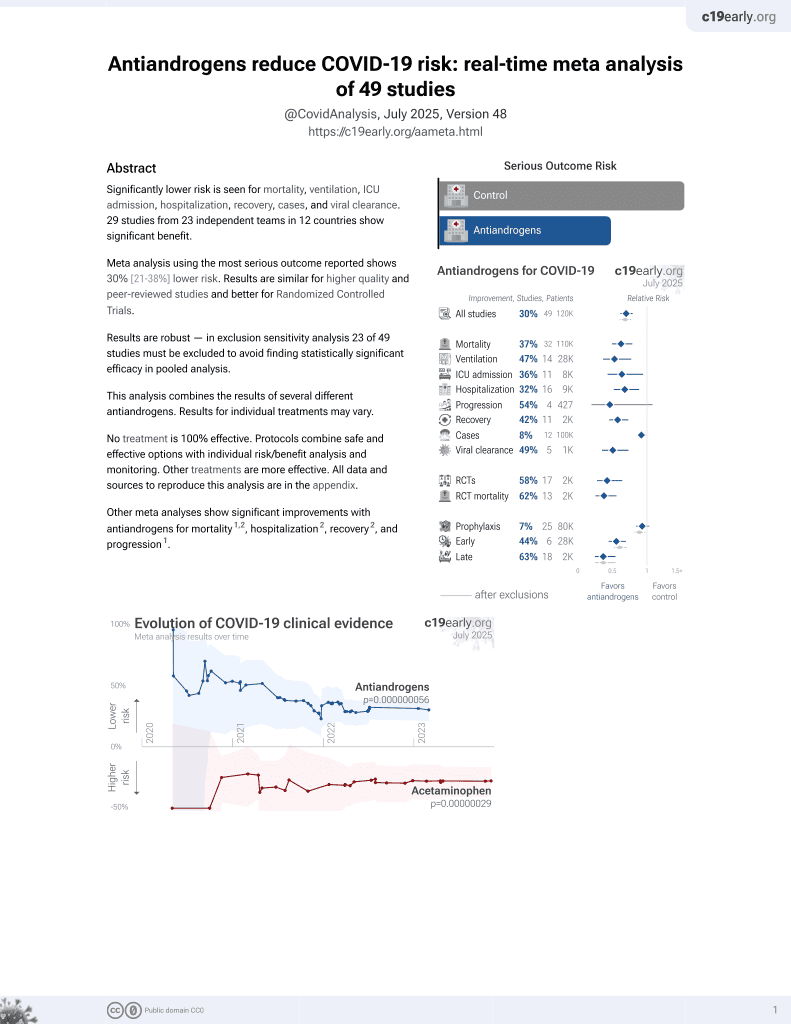
7th treatment shown to reduce risk in
September 2020, now with p = 0.000000056 from 49 studies.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
In silico study of several antiandrogens identifying strong candidates for inhibition of SARS-CoV-2. Apalutamide and bicalutamide showed the best binding affinity against TMPRSS2.
4 preclinical studies support the efficacy of antiandrogens for COVID-19:
1.
Saih et al., In-Silico Molecular Docking, Validation, Drug-Likeness, and ADMET Studies of Antiandrogens to Use in the Fight against SARS-CoV-2, Physical Chemistry Research, doi:10.22036/pcr.2022.324549.2016.
2.
Majidipur et al., Apalutamide Prevents SARS-CoV-2 Infection in Lung Epithelial Cells and in Human Nasal Epithelial Cells, International Journal of Molecular Sciences, doi:10.3390/ijms24043288.
Saih et al., 5 May 2022, Morocco, peer-reviewed, 11 authors.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
In-Silico Molecular Docking, Validation, Drug-Likeness, and ADMET Studies of Antiandrogens to Use in the Fight against SARS-CoV-2
doi:10.22036/PCR.2022.324549.2016
The SARS-CoV-2 is the novel coronavirus that causes the pandemic COVID-19, which has originated in Wuhan, China, in December 2019. Early studies have generally shown that human Angiotensin-Converting Enzyme 2 (ACE2) and transmembrane protease serine 2 (TMPRSS2) are responsible for the viral entry of SARS-CoV-2 into target cells. TMPRSS2 as androgen-regulated is highly expressed in the prostate and other tissues including the lung. We investigated the interaction between the TMPRSS2 protein and selected antiandrogens, namely bicalutamide, enzalutamide, apalutamide, flutamide, nilutamide, and darolutamide using in-silico molecular docking. The results showed that apalutamide (-8.8 Kcal mol -1 ) and bicalutamide (-8.6 Kcal mol -1 ) had the highest docking score. The molecular docking process was validated by re-docking the peptide-like-inhibitor-serine protease hepsin and superimposing them onto the reference complex. Last of all, the tested compounds have been evaluated for their pharmacokinetic and drug-likeness properties and concluded that these compounds except nilutamide (mutagenic) can be granted as potential inhibitors of SARS-CoV-2. This in-silico study result encourages its use as means for drug discovery of new COVID-19 treatment.
SUPPLEMENTARY MATERIALS Supplementary Table 1 : Molecular docking scores (in -Kcal mol -1 ) of TMPRSS2 protein with its selected inhibitors (ligands). Supplementary Table 2 : Identification of the active site of TMPRSS2 protein using the CASTp server Supplementary Table 3 : Drug likeness results of the six studied ligands. Supplementary Table 4 : pharmacokinetic parameters of the six tested compounds.
References
Binkowski, Naghibzadeh, Liang, CASTp: Computed atlas of surface topography of proteins, Nucleic Acids Res, doi:10.1093/nar/gkg512
Bohl, Gao, Miller, Bell, Dalton, Structural basis for antagonism and resistance of bicalutamide in prostate cancer, Proc. Natl. Acad. Sci. U S A, doi:10.1073/pnas.050038110
Burley, Berman, Bhikadiya, RCSB protein data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy, Nucleic Acids Res, doi:10.1093/nar/gky1004
Cava, Bertoli, Castiglioni, In silico discovery of candidate drugs against covid-19, Viruses, doi:10.3390/v12040404
Depfenhart, De Villiers, Lemperle, Meyer, Di Somma, Potential new treatment strategies for COVID-19: is there a role for bromhexine as addon therapy?, Intern. Emerg. Med, doi:10.1007/s11739-020-02383-3
Desai, Stadler, Vogelzang, Nilutamide: possible utility as a second-line hormonal agent, Urology, doi:10.1016/s0090-4295(01)01455-8
Diana, Michielin, Zoete, SwissADME: a free web tool to evaluate pharmacokinetics, druglikeness and medicinal chemistry friendliness of small molecules, Sci. Rep, doi:10.1038/srep42717
Eisenberg, Lüthy, Bowie, VERIFY3D: assessment of protein models with three-dimensional profiles, Methods Enzymol, doi:10.1016/s0076-6879(97)77022-8
Ertl, Molecular structure input on the web, J. Cheminform, doi:10.1186/1758-2946-2-1
Fizazi, Shore, Tammela, Nonmetastatic, castration-resistant prostate cancer and survival with darolutamide, N Engl. J. Med, doi:10.1056/NEJMoa2001342
Goldspiel, Kohler, Flutamide: an antiandrogen for advanced prostate cancer, DICP, doi:10.1177/106002809002400612
Hoffmann, Kleine-Weber, Schroeder, SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor, Cell, doi:10.1016/j.cell.2020.02.052
Hollingsworth, Karplus, A fresh look at the Ramachandran plot and the occurrence of standard structures in proteins, Biomol. Concepts, doi:10.1515/BMC.2010.022
Hussain, Jabeen, Amanullah, Molecular docking between human TMPRSS2 and SARS-CoV-2 spike protein: conformation and intermolecular interactions, AIMS Microbiol, doi:10.3934/microbiol.2020021
Kemmish, Fasnacht, Yan, Fully automated antibody structure prediction using BIOVIA tools: Validation study, PLoS One, doi:10.1371/journal.pone.0177923
Laskowski, Macarthur, Thornton, PROCHECK: A program to check the stereochemical quality of protein structures, J. App. Crystallography, doi:10.1107/S0021889892009944
Marvinsketch, None
Mckee, Sternberg, Stange, Laufer, Naujokat, Candidate drugs against SARS-CoV-2 and COVID-19, Pharmacol. Res, doi:10.1016/j.phrs.2020.104859
Montopoli, Zumerle, Vettor, Androgendeprivation therapies for prostate cancer and risk of infection by SARS-CoV-2: a population-based study (N = 4532), Ann. Oncol, doi:10.1016/j.annonc.2020.04.479
Morris, Huey, Lindstrom, AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility, J. Comput. Chem, doi:10.1002/jcc.21256
Pires, Blundell, Ascher, pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures, J. Med. Chem, doi:10.1021/ACS.JMEDCHEM.5B00104
Piva, Sabanovic, Cecati, Giulietti, Expression and co-expression analyses of TMPRSS2, a key element in COVID-19, Eur. J. Clin. Microbiol. Infect. Dis, doi:10.1007/s10096-020-04089-y
Ragia, Manolopoulos, Inhibition of SARS-CoV-2 entry through the ACE2/TMPRSS2 pathway: a promising approach for uncovering early COVID-19 drug therapies, Eur. J. Clin. Pharmacol, doi:10.1007/s00228-020-02963-4
Sanchez, Cathelineau, Pinto, Clinical and surgical assistance in prostate cancer during the COVID-19 pandemic: Implementation of assistance protocols, Int. Braz. J. Urol, doi:10.1590/S1677-5538.IBJU.2020.S106
Scott, Enzalutamide: A review in castrationresistant prostate cancer, Drugs, doi:10.1007/s40265-018-1029-9
Sobanska, Hekner, Brzezińska, RP-18 HPLC analysis of drugs' ability to cross the blood-brain barrier, J. Chem, doi:10.1155/2019/5795402
Squire, Park, Yoshimoto, Prostate cancer as a model system for genetic diversity in tumors, Adv. Cancer Res, doi:10.1016/B978-0-12-387688-1.00007-7
Strope, Pharmd, Figg, TMPRSS2: Potential biomarker for COVID-19 outcomes, J. Clin. Pharmacol, doi:10.1002/jcph.1641
Teli, Shah, Chhabria, In silico screening of natural compounds as potential inhibitors of SARS-CoV-2 main protease and spike RBD: Targets for COVID-19, Front. Mol. Biosci, doi:10.3389/fmolb.2020.599079
Trott, Olson, AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithread ding, J. Comput. Chem, doi:10.1002/jcc.21334
Uniprot, UniProt: a worldwide hub of protein knowledge, Nucleic Acids Res, doi::10.1093/nar/gky1049
Wambier, Goren, Vaño-Galván, Androgen sensitivity gateway to COVID-19 disease severity, Drug Dev Res, doi:10.1002/ddr.21688
Wang, Xiao, Suzek, Zhang, Wang et al., PubChem: a public information system for analyzing bioactivities of small molecules, Nucleic Acids Res, doi:10.1093/nar/gkp456
Waterhouse, Bertoni, Bienert, SWISS-MODEL: homology modelling of protein structures and complexes, Nucleic Acids Res, doi:10.1093/nar/gky427
Xu, Zhang, Improving the physical realism and structural accuracy of protein models by a twostep atomic-level energy minimization, Biophys. J, doi:10.1016/j.bpj.2011.10.024
Yuan, Stephen Chan, Zhenquan, Hu, Using PyMOL as a platform for computational drug design, WIREs Comput. Mol. Sci, doi:10.1002/wcms.1298