Identification of druggable host dependency factors shared by multiple SARS-CoV-2 variants of concern
et al., Journal of Molecular Cell Biology. doi:10.1093/jmcb/mjae004, Feb 2024
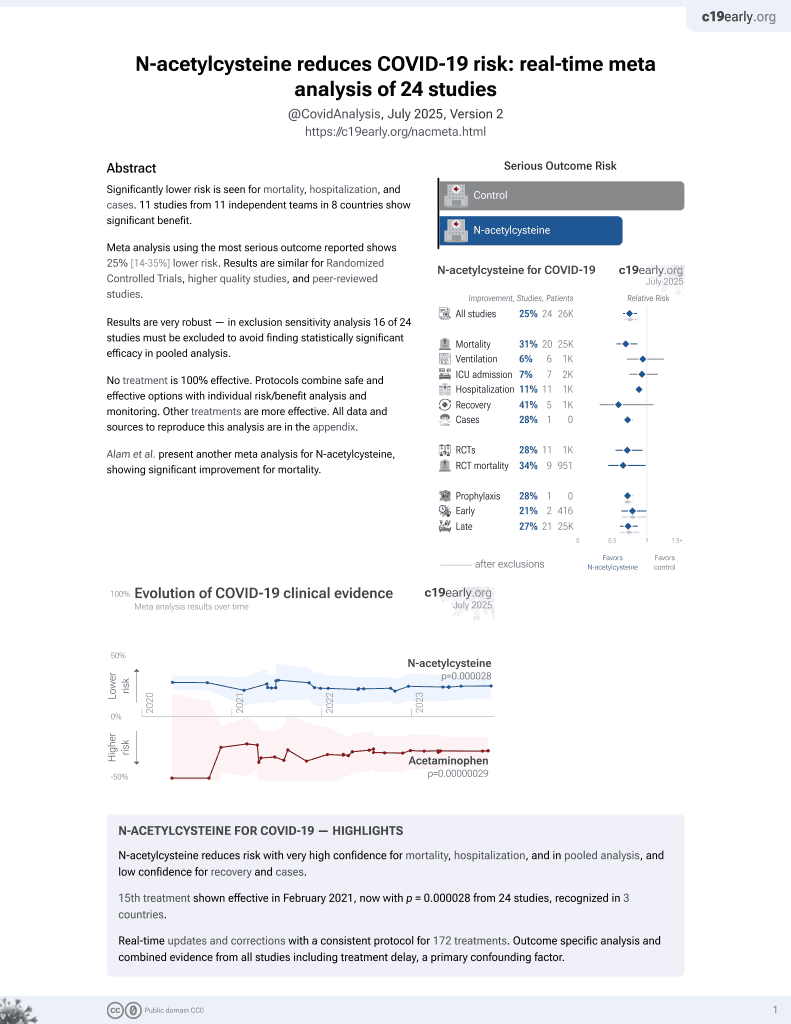
16th treatment shown to reduce risk in
February 2021, now with p = 0.0000032 from 25 studies, recognized in 3 countries.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
In vitro study showing that inhibition of host cell factors rather than viral elements may prevent emergence of SARS-CoV-2 variants resistant to current drugs. Authors identified host genes/proteins commonly required for infection by major variants using CRISPR screening of infected human lung cells. Targeting hits RIPK4, SLC7A11 or MASTL with existing drugs blocked variant replication and virus-induced cytotoxicity. Mechanistic studies revealed variants rapidly induce reactive oxygen species (ROS), while ROS suppression with antioxidants (NAC/GSH) potently blocked infection. NAC treatment in infected humanized mice reduced lung viral load.
14 preclinical studies support the efficacy of N-acetylcysteine for COVID-19:
Severe COVID-19 is marked by endotheliopathy with elevated von Willebrand factor (VWF) levels and platelet/VWF-rich microthrombi; N-acetylcysteine can reduce VWF multimers and lyse VWF-dependent clots in vivo, potentially helping to alleviate thrombosis associated with COVID-1910-12.
N-acetylcysteine shows dose-dependent inhibition of SARS-CoV-24,7,9 , shows anti-inflammatory and immunomodulatory effects against SARS-CoV-2-induced immune responses in combination with bromelain6, suppressed virus-induced reactive oxygen species and blocked viral replication in a humanized mouse model and in human lung cells5, may limit COVID-19 induced cardiac damage by boosting cellular antioxidant defenses and potentially mitigating the oxidative stress caused by spike protein-induced ROS production in cardiac fibroblasts3, and reduces disulfide bonds in proteins and exhibits antioxidant properties that may inhibit viral replication and modulate inflammatory responses2.
NAC may be beneficial for COVID-19 by replenishing glutathione stores and reinforcing the glutathione peroxidase-4 pathway to inhibit ferroptosis, an oxidative stress-induced cell death pathway implicated in COVID-1913.
NAC reinforces glutathione levels, reduces ROS, and minimizes ferroptosis and cytokine storm14.
1.
Agamah et al., Network-based multi-omics-disease-drug associations reveal drug repurposing candidates for COVID-19 disease phases, ScienceOpen, doi:10.58647/DRUGARXIV.PR000010.v1.
2.
Reis et al., Antiviral effect of Bromelain combined with acetylcysteine against SARS-CoV-2 Omicron variant, Scientific Reports, doi:10.1038/s41598-025-92242-y.
3.
Van Tin et al., Spike Protein of SARS-CoV-2 Activates Cardiac Fibrogenesis through NLRP3 Inflammasomes and NF-κB Signaling, Cells, doi:10.3390/cells13161331.
4.
Chaopreecha et al., Andrographolide attenuates SARS-CoV-2 infection via an up-regulation of glutamate-cysteine ligase catalytic subunit (GCLC), Phytomedicine, doi:10.1016/j.phymed.2024.156279.
5.
Frasson et al., Identification of druggable host dependency factors shared by multiple SARS-CoV-2 variants of concern, Journal of Molecular Cell Biology. doi:10.1093/jmcb/mjae004, academic.oup.com/jmcb/advance-article/doi/10.1093/jmcb/mjae004/7596546.
6.
Ferreira et al., Taming the SARS-CoV-2-mediated proinflammatory response with BromAc®, Frontiers in Immunology, doi:10.3389/fimmu.2023.1308477.
7.
La Maestra et al., Inhibition of the Cell Uptake of Delta and Omicron SARS-CoV-2 Pseudoviruses by N-Acetylcysteine Irrespective of the Oxidoreductive Environment, Cells, doi:10.3390/cells11203313.
8.
Goc et al., Inhibitory effects of specific combination of natural compounds against SARS-CoV-2 and its Alpha, Beta, Gamma, Delta, Kappa, and Mu variants, European Journal of Microbiology and Immunology, doi:10.1556/1886.2021.00022.
9.
Akhter et al., The Combination of Bromelain and Acetylcysteine (BromAc) Synergistically Inactivates SARS-CoV-2, Viruses, doi:10.3390/v13030425.
10.
Martinez de Lizarrondo et al., Potent Thrombolytic Effect of N-Acetylcysteine on Arterial Thrombi, Circulation, doi:10.1161/CIRCULATIONAHA.117.027290.
11.
Chen et al., N-acetylcysteine reduces the size and activity of von Willebrand factor in human plasma and mice, Journal of Clinical Investigation, doi:10.1172/JCI41062.
12.
Goshua et al., Endotheliopathy in COVID-19-associated coagulopathy: evidence from a single-centre, cross-sectional study, The Lancet Haematology, doi:10.1016/S2352-3026(20)30216-7.
Frasson et al., 1 Feb 2024, peer-reviewed, 16 authors.
Abstract: Identification of druggable host dependency factors shared by multiple
SARS-CoV-2 variants of concern
A
N
U
SC
RI
PT
1 Department of Molecular Medicine, University of Padua, Padua 35121, Italy
2 Department of Biology, Armenise/Harvard Pluripotent Stem Cell Biology Laboratory,
University of Padua, Padua 35131, Italy
3 Department. of Surgery, Oncology and Gastroenterology, University of Padua, Padua
35128, Italy
4 Veneto Institute of Oncology IOV-IRCCS, Padua 35128, Italy
5 Department of Comparative Biomedicine and Food Science, University of Padua, Padua
35020, Italy
6 Telethon Institute of Genetics and Medicine (TIGEM), Armenise/Harvard Laboratory of
Integrative Genomics, Pozzuoli 80078, Italy
7 Department of Translational Medicine, Federico II University, Naples 80138, Italy
8 School for Advanced Studies, Genomics and Experimental Medicine Program, University
of Naples “Federico II”, Naples 80138, Italy
9 Microbiology and Virology Unit, Padua University Hospital, Padua 35128, Italy
†
O
RI
G
IN
A
L
U
N
ED
IT
ED
M
These authors contributed equally to this work.
* Correspondence to: Sara N. Richter, E-mail: sara.richter@unipd.it; Marco Montagner, Email: marco.montagner@unipd.it; Graziano Martello, E-mail: graziano.martello@unipd.it
© The Author(s) 2024. Published by Oxford University Press on behalf of Institute of Biochemistry and Cell Biology, Shanghai Institutes for
Biological Sciences, Chinese Academy of Sciences. This is an Open Access article distributed under the terms of the Creative Commons Attribution
License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted reuse, distribution, and reproduction
in any medium, provided the original work is properly cited.
Ilaria Frasson1,†, Linda Diamante1,2,†, Manuela Zangrossi1,†, Elena Carbognin2, Anna
Dalla Pietà3, Alessandro Penna3, Antonio Rosato3,4, Ranieri Verin5, Filippo
Torrigiani5, Cristiano Salata1, Marìa Paula Dizanzo1, Lorenzo Vaccaro6,7, Davide
Cacchiarelli6,7,8, Sara N. Richter1,9,*, Marco Montagner1,*, Graziano Martello2,*
Abstract
The high mutation rate of SARS-CoV-2 leads to the emergence of multiple variants,
some of which are resistant to vaccines and drugs targeting viral elements. Targeting
host dependency factors, e.g. cellular proteins required for viral replication, would
variants induce conserved cellular responses and exploit the same core host factors.
To this end, we compared three variants of concern and found that the host
transcriptional response was conserved, differing only in kinetics and magnitude.
PT
Through CRISPR screening, we identified host genes required for infection by each
RI
variant. Most of the genes were shared by multiple variants. We validated our hits
with small molecules and repurposed Food and Drug Administration-approved drugs.
SC
All the drugs were highly active against all the variants tested, including new variants
that emerged during the study (Delta and Omicron). Mechanistically, we identified
U
reactive oxygen species production as a key step in early virus replication.
N
Antioxidants such as N-acetyl cysteine (NAC) were effective against all the variants
A
in both human lung cells and a humanised mouse model. Our study supports the use
M
of available antioxidant drugs, such as NAC, as a general and effective..
DOI record:
{
"DOI": "10.1093/jmcb/mjae004",
"ISSN": [
"1674-2788",
"1759-4685"
],
"URL": "http://dx.doi.org/10.1093/jmcb/mjae004",
"abstract": "<jats:title>Abstract</jats:title>\n <jats:p>The high mutation rate of SARS-CoV-2 leads to the emergence of multiple variants, some of which are resistant to vaccines and drugs targeting viral elements. Targeting host dependency factors, e.g. cellular proteins required for viral replication, would help prevent resistance. However, it remains unclear whether different SARS-CoV-2 variants induce conserved cellular responses and exploit the same core host factors. To this end, we compared three variants of concern and found that the host transcriptional response was conserved, differing only in kinetics and magnitude. Through CRISPR screening, we identified host genes required for infection by each variant. Most of the genes were shared by multiple variants. We validated our hits with small molecules and repurposed Food and Drug Administration-approved drugs. All the drugs were highly active against all the variants tested, including new variants that emerged during the study (Delta and Omicron). Mechanistically, we identified reactive oxygen species production as a key step in early virus replication. Antioxidants such as N-acetyl cysteine (NAC) were effective against all the variants in both human lung cells and a humanised mouse model. Our study supports the use of available antioxidant drugs, such as NAC, as a general and effective anti-COVID-19 approach.</jats:p>",
"author": [
{
"ORCID": "http://orcid.org/0000-0001-5135-5707",
"affiliation": [
{
"name": "Department of Molecular Medicine, University of Padua , Padua 35121 , Italy"
}
],
"authenticated-orcid": false,
"family": "Frasson",
"given": "Ilaria",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Department of Molecular Medicine, University of Padua , Padua 35121 , Italy"
},
{
"name": "Department of Biology, Armenise/Harvard Pluripotent Stem Cell Biology Laboratory, University of Padua , Padua 35131 , Italy"
}
],
"family": "Diamante",
"given": "Linda",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Molecular Medicine, University of Padua , Padua 35121 , Italy"
}
],
"family": "Zangrossi",
"given": "Manuela",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Biology, Armenise/Harvard Pluripotent Stem Cell Biology Laboratory, University of Padua , Padua 35131 , Italy"
}
],
"family": "Carbognin",
"given": "Elena",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-7875-7914",
"affiliation": [
{
"name": "Department. of Surgery, Oncology and Gastroenterology, University of Padua , Padua 35128 , Italy"
}
],
"authenticated-orcid": false,
"family": "Dalla Pietà",
"given": "Anna",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department. of Surgery, Oncology and Gastroenterology, University of Padua , Padua 35128 , Italy"
}
],
"family": "Penna",
"given": "Alessandro",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-5263-8386",
"affiliation": [
{
"name": "Department. of Surgery, Oncology and Gastroenterology, University of Padua , Padua 35128 , Italy"
},
{
"name": "Veneto Institute of Oncology IOV-IRCCS , Padua 35128 , Italy"
}
],
"authenticated-orcid": false,
"family": "Rosato",
"given": "Antonio",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Comparative Biomedicine and Food Science, University of Padua , Padua 35020 , Italy"
}
],
"family": "Verin",
"given": "Ranieri",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Comparative Biomedicine and Food Science, University of Padua , Padua 35020 , Italy"
}
],
"family": "Torrigiani",
"given": "Filippo",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-5136-7406",
"affiliation": [
{
"name": "Department of Molecular Medicine, University of Padua , Padua 35121 , Italy"
}
],
"authenticated-orcid": false,
"family": "Salata",
"given": "Cristiano",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Molecular Medicine, University of Padua , Padua 35121 , Italy"
}
],
"family": "Dizanzo",
"given": "Marìa Paula",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Telethon Institute of Genetics and Medicine (TIGEM), Armenise/Harvard Laboratory of Integrative Genomics , Pozzuoli 80078 , Italy"
},
{
"name": "Department of Translational Medicine, Federico II University , Naples 80138 , Italy"
}
],
"family": "Vaccaro",
"given": "Lorenzo",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Telethon Institute of Genetics and Medicine (TIGEM), Armenise/Harvard Laboratory of Integrative Genomics , Pozzuoli 80078 , Italy"
},
{
"name": "Department of Translational Medicine, Federico II University , Naples 80138 , Italy"
},
{
"name": "School for Advanced Studies, Genomics and Experimental Medicine Program, University of Naples “Federico II” , Naples 80138 , Italy"
}
],
"family": "Cacchiarelli",
"given": "Davide",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-5446-9029",
"affiliation": [
{
"name": "Department of Molecular Medicine, University of Padua , Padua 35121 , Italy"
},
{
"name": "Microbiology and Virology Unit, Padua University Hospital , Padua 35128 , Italy"
}
],
"authenticated-orcid": false,
"family": "Richter",
"given": "Sara N",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Molecular Medicine, University of Padua , Padua 35121 , Italy"
}
],
"family": "Montagner",
"given": "Marco",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-5520-085X",
"affiliation": [
{
"name": "Department of Biology, Armenise/Harvard Pluripotent Stem Cell Biology Laboratory, University of Padua , Padua 35131 , Italy"
}
],
"authenticated-orcid": false,
"family": "Martello",
"given": "Graziano",
"sequence": "additional"
}
],
"container-title": "Journal of Molecular Cell Biology",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2024,
2,
2
]
],
"date-time": "2024-02-02T05:44:42Z",
"timestamp": 1706852682000
},
"deposited": {
"date-parts": [
[
2024,
2,
2
]
],
"date-time": "2024-02-02T05:44:42Z",
"timestamp": 1706852682000
},
"indexed": {
"date-parts": [
[
2024,
2,
2
]
],
"date-time": "2024-02-02T21:42:02Z",
"timestamp": 1706910122902
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2024,
2,
1
]
]
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "am",
"delay-in-days": 1,
"start": {
"date-parts": [
[
2024,
2,
2
]
],
"date-time": "2024-02-02T00:00:00Z",
"timestamp": 1706832000000
}
}
],
"link": [
{
"URL": "https://academic.oup.com/jmcb/advance-article-pdf/doi/10.1093/jmcb/mjae004/56545101/mjae004.pdf",
"content-type": "application/pdf",
"content-version": "am",
"intended-application": "syndication"
},
{
"URL": "https://academic.oup.com/jmcb/advance-article-pdf/doi/10.1093/jmcb/mjae004/56545101/mjae004.pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "286",
"original-title": [],
"prefix": "10.1093",
"published": {
"date-parts": [
[
2024,
2,
1
]
]
},
"published-online": {
"date-parts": [
[
2024,
2,
1
]
]
},
"publisher": "Oxford University Press (OUP)",
"reference-count": 0,
"references-count": 0,
"relation": {},
"resource": {
"primary": {
"URL": "https://academic.oup.com/jmcb/advance-article/doi/10.1093/jmcb/mjae004/7596546"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"Cell Biology",
"Genetics",
"Molecular Biology",
"General Medicine"
],
"subtitle": [],
"title": "Identification of druggable host dependency factors shared by multiple SARS-CoV-2 variants of concern",
"type": "journal-article"
}