Exploring Therapeutic Potential of Nutraceutical Compounds from Propolis on MAPK1 Protein Using Bioinformatics Approaches as Anti-Coronavirus Disease 2019 (COVID-19)
et al., BIO Web of Conferences, doi:10.1051/bioconf/20248800007, Jan 2024
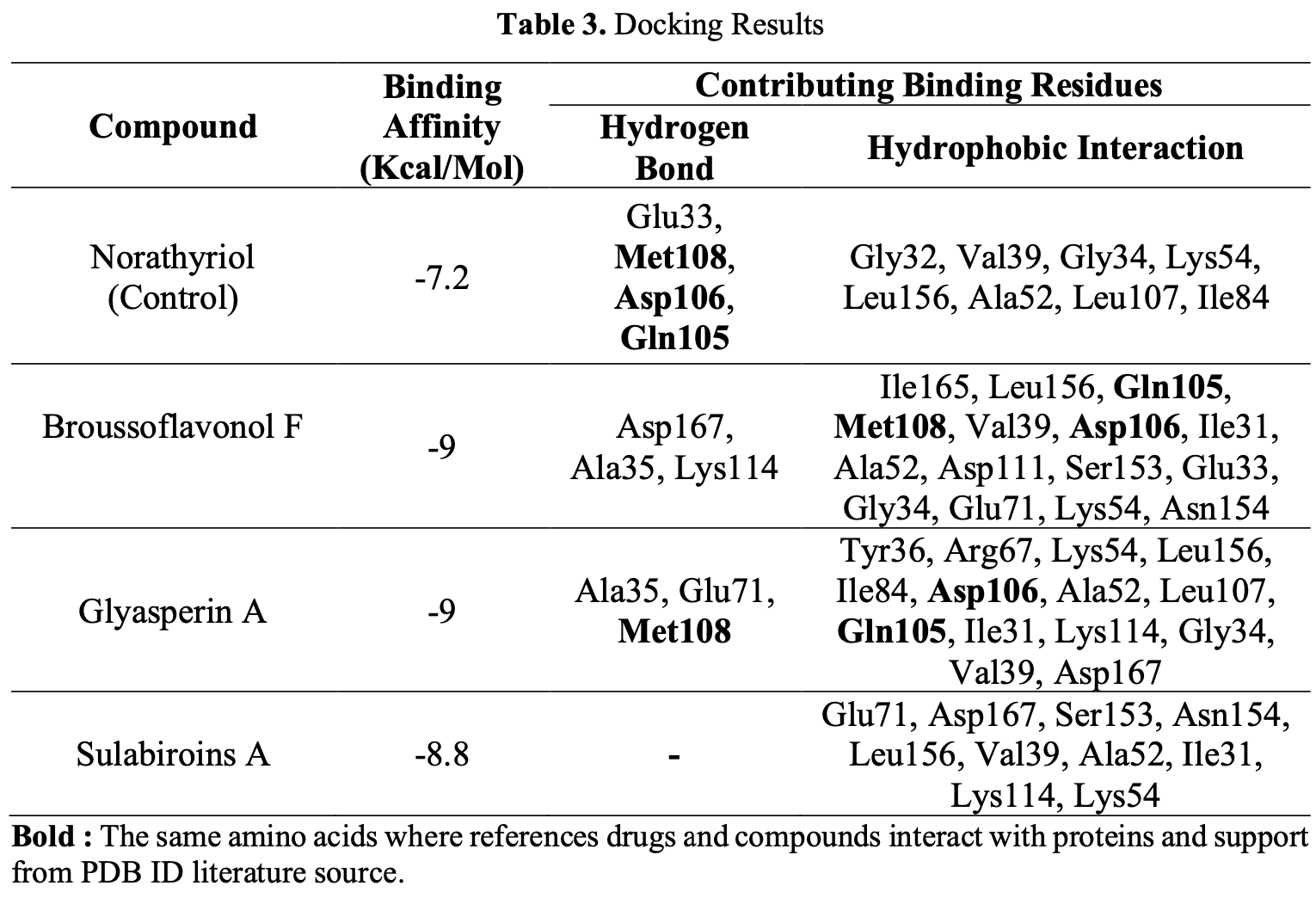
In silico study suggesting that propolis compounds broussoflavonol F, glyasperin A, and sulabiroins A may inhibit SARS-CoV-2 by targeting the MAPK1 protein. Using network pharmacology, authors identified 74 overlapping targets between propolis and COVID-19, with MAPK1 emerging as a key target from protein-protein interaction analysis. Molecular docking showed broussoflavonol F, glyasperin A, and sulabiroins A had higher binding affinities to MAPK1 (-9.0, -9.0, and -8.8 kcal/mol respectively) than the native ligand norathyriol (-7.2 kcal/mol). Molecular dynamics simulations over 20ns confirmed the stability of these complexes.
12 preclinical studies support the efficacy of propolis for COVID-19:
In silico studies predict inhibition of SARS-CoV-2 with propolis or metabolites via binding to the spikeA,1,2 , MproB,2, RNA-dependent RNA polymeraseC,1,2 , PLproD,1, ACE2E,6, and TMPRSS2F,6 proteins.
In vitro studies demonstrate inhibition of the ACE2E,6 and TMPRSS2F,6 proteins.
Propolis may inhibit spike protein and ACE2 interaction8, may inhibit SARS-CoV-2 through interactions with MAPK15, inhibited SARS-CoV-2 in Vero E6 cells at a concentration comparable to a combination of four antiviral components9, may mitigate hyperinflammation via STAT1, NOS2, and BTK targeting3, may inhibit SARS-CoV-2 entry by interfering with ACE2/TMPRSS2 interaction7, components of propolis show ACE2 downregulation in human cells6, modulates inflammatory responses by reducing pro-inflammatory cytokines IL-1β, IL-6, and TNF-α7, and may suppress Epstein-Barr Virus reactivation3.
1.
Ferreira Júnior et al., An In Silico Investigation of Brazilian Green Propolis Extracts as Potential Treatment for COVID-19, ACS Omega, doi:10.1021/acsomega.5c02121.
2.
Al balawi et al., Assessing multi-target antiviral and antioxidant activities of natural compounds against SARS-CoV-2: an integrated in vitro and in silico study, Bioresources and Bioprocessing, doi:10.1186/s40643-024-00822-z.
3.
Anshori et al., Uncovering the Therapeutic Potential of Propolis Extract in Managing Hyperinflammation and Long COVID‐19: A Comprehensive Bioinformatics Study, Chemistry & Biodiversity, doi:10.1002/cbdv.202401947.
4.
Ay et al., Investigation of The Inhibition of SARS-CoV-2 Spike RBD and ACE-2 Interaction by Phenolics of Propolis Extracts, Journal of Apitherapy and Nature, doi:10.35206/jan.1471090.
5.
Siregar et al., Exploring Therapeutic Potential of Nutraceutical Compounds from Propolis on MAPK1 Protein Using Bioinformatics Approaches as Anti-Coronavirus Disease 2019 (COVID-19), BIO Web of Conferences, doi:10.1051/bioconf/20248800007.
6.
Kumar et al., Computational and experimental evidence of the anti‐COVID‐19 potential of honeybee propolis ingredients, caffeic acid phenethyl ester and artepillin c, Phytotherapy Research, doi:10.1002/ptr.7717.
7.
Ferreira et al., Antiviral and anti-inflammatory efficacy of nanoencapsulated brazilian green propolis against SARS-CoV-2, Scientific Reports, doi:10.1038/s41598-025-05683-w.
a.
The trimeric spike (S) protein is a glycoprotein that mediates viral entry by binding to the host ACE2 receptor, is critical for SARS-CoV-2's ability to infect host cells, and is a target of neutralizing antibodies. Inhibition of the spike protein prevents viral attachment, halting infection at the earliest stage.
b.
The main protease or Mpro, also known as 3CLpro or nsp5, is a cysteine protease that cleaves viral polyproteins into functional units needed for replication. Inhibiting Mpro disrupts the SARS-CoV-2 lifecycle within the host cell, preventing the creation of new copies.
c.
RNA-dependent RNA polymerase (RdRp), also called nsp12, is the core enzyme of the viral replicase-transcriptase complex that copies the positive-sense viral RNA genome into negative-sense templates for progeny RNA synthesis. Inhibiting RdRp blocks viral genome replication and transcription.
d.
The papain-like protease (PLpro) has multiple functions including cleaving viral polyproteins and suppressing the host immune response by deubiquitination and deISGylation of host proteins. Inhibiting PLpro may block viral replication and help restore normal immune responses.
e.
The angiotensin converting enzyme 2 (ACE2) protein is a host cell transmembrane protein that serves as the cellular receptor for the SARS-CoV-2 spike protein. ACE2 is expressed on many cell types, including epithelial cells in the lungs, and allows the virus to enter and infect host cells. Inhibition may affect ACE2's physiological function in blood pressure control.
f.
Transmembrane protease serine 2 (TMPRSS2) is a host cell protease that primes the spike protein, facilitating cellular entry. TMPRSS2 activity helps enable cleavage of the spike protein required for membrane fusion and virus entry. Inhibition may especially protect respiratory epithelial cells, buy may have physiological effects.
Siregar et al., 22 Jan 2024, peer-reviewed, 4 authors.
Contact: 1911102415125@umkt.ac.id, pmk195@umkt.ac.id.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Exploring Therapeutic Potential of Nutraceutical Compounds from Propolis on MAPK1 Protein Using Bioinformatics Approaches as Anti-Coronavirus Disease 2019 (COVID-19)
BIO Web of Conferences, doi:10.1051/bioconf/20248800007
This study explores the potential of propolis, a natural substance, as a gene therapy for treating COVID-19. Despite the advent of COVID-19 vaccines, their side effects pose new health challenges. Utilizing network pharmacology, this research identifies propolis compounds through various databases and assesses their ability to target proteins associated with COVID-19. MAPK1 emerges as a potential therapeutic target, and molecular docking reveals Broussoflavonol F, Glyasperin A, and Sulabiroins as promising compounds with strong binding affinities, i.e., -9.0, -9.0, and -8.8 kcal/mol, respectively, exceeding the native ligand (-7.2 kcal/mol). Molecular Dynamics displays stable complex behavior, with backbone RMSD values consistently below 4 Angstroms and RMSF simulations showing minimal fluctuations within ±2 Angstroms error. Moreover, MM-PBSA analysis further supports the strong binding of Broussoflavonol F, Glyasperin A, and Sulabiroins A, with relative binding energies of -122.82±89.65, 131.48±95.39, and -155.97±111,37 kJ/mol, respectively. These results indicate that propolis has potential as an anti-COVID-19 agent, primarily through inhibiting the MAPK1 pathway. However, further research is needed to validate these results and develop practical applications for COVID-19 therapy. This study underscores the significance of network pharmacology and computational models in understanding propolis mechanisms, offering potential directions for future research and treatment strategies against COVID-19.
Bold : The same amino acids where references drugs and compounds interact with proteins and support from PDB ID literature source.
Molecular Dynamics The potential compound selected as a result of molecular docking is reported to have better interaction stability (RMSD, RMSF, and Binding Energy) than Norathyriol, based on molecular dynamics results. The Root-Mean-Square Deviation (RMSD) value of the backbone structure shows that the MAPK1 complex with Sulabiroins A has better stability than the Native Ligand. Similarly, RMSD of the ligand structure also confirmed that Sulalabiroins A had better structural stability than Native Ligand. The average RMSF values obtained for Norathyriol (Control), Broussoflavonol F, Glyasperin A and Sulabiroins A were 1.51 Å, 1.65 Å and 1.48 Å respectively, indicating relatively stable behavior. The stability of the Active Sites distributed in each complex is reported to confirm the Complex Docking and Conformation results obtained from dynamic simulations. Based on the structural arrangement of the MAPK1 complex and its interactions with the active site residues of several selected compounds, it shows consistent stability below 2 Å during the simulation. Based on a comprehensive analysis of root mean square fluctuations (RMSF), it can be concluded that no statistically significant fluctuations were observed in the investigated system. Thus, the results of binding energy calculations found that Sulabiroins A, Glyasperin A and Broussoflavonol F..
References
Al-Sadi, None, Front. Biosci
Ashraf, None, Phyther. Res
Berlin, Gulick, Martinez, None, N. Engl. J. Med
Burley, None, Nucleic Acids Res
Capraru, None, Medicina (B. Aires)
Che, None, Int. J. Chron. Obstruct. Pulmon. Dis
Chen, None, Hematology
Chin, None, BMC Syst. Biol
Chu, None, Transl. Cancer Res
Daina, Michielin, Zoete, None, Nucleic Acids Res
Daina, Michielin, Zoete, None, Sci. Rep
Dallakyan, Olson, None, Chemical Biology, doi:10.1007/978-1-4939-2269-7_19
Demeke, Woldeyohan, Kifle, None, Metab. Open
Dewi, None, Int. J. Appl. Pharm
Dilokthornsakul, Kosiyaporn, Wuttipongwaragon, Dilokthornsakul, None, J. Integr. Med
Fatriansyah, Rizqillah, Yandi, Fadilah, Sahlan, None, J. King Saud Univ. -Sci
Gentleman, Carey, Huber, Irizarry, Dudoit, Bioinformatics and Computational Biology Solutions Using R and Bioconductor, doi:10.1007/0-387-29362-0
Gentleman, None, Genome Biol
Hariri, Hardin, None, N. Engl. J. Med
Harisna, None, Biochem. Biophys. Reports
Hidayat, Susilo, Awwaly, Cahyati, None, J. Ilmu dan Teknol. Has. Ternak
Hopkins, None, Nat. Chem. Biol
Hossain, None, Chin. Med
Ibrahim, El-Banna, None, RSC Adv
Kanehisa, Furumichi, Tanabe, Sato, Morishima, None, Nucleic Acids Res
Kashyap, None, Molecules
Khayrani, None, J. King Saud Univ. -Sci
Kim, None, Nucleic Acids Res
Krieger, Vriend, None, J. Comput. Chem
Kuhn, Mering, Campillos, Jensen, Bork, None, Nucleic Acids Res
Kumar, None, Phyther. Res, doi:10.1002/ptr.7717
Laskowski, Swindells, None, J. Chem. Inf. Model
Lassi, Naseem, Salam, Siddiqui, Das, None, Int. J. Environ. Res. Public Health
Li, None, Cancer Res
Ma, . -H. Chan, Leung, None, Chem. Sci
Mahroum, None, Front. Immunol
Maier, None, J. Chem. Theory Comput
Meylani, None, Results Chem
Miller, None, J. Chem. Theory Comput
Pan, Zhou, Su, Li, None, Med. Sci. Monit
Papadopoulou, None, Pathogens
Pinzi, Rastelli, None, Int. J. Mol. Sci
Rabaan, None, Ann. Clin. Microbiol. Antimicrob
Rivas, Porritt, Cheng, Bahar, Arditi, None, J. Allergy Clin. Immunol
Rizzi, None, Int. J. Mol. Sci
Safran, Practical Guide to Life Science Databases, doi:10.1007/978-981-16-5812-9_2
Sahlan, None, J. King Saud Univ. -Sci
Shang, None, J. Ethnopharmacol
Shannon, None, Genome Res
Shi, None, Front. Genet
Siregar, Hanifa, None, J. Public Heal. Trop. Coast. Reg
Stelzer, None, Curr. Protoc. Bioinforma
Syaban, None, J. Trop. Life Sci
Szklarczyk, None, Nucleic Acids Res
Tsai, None, J. Taiwan Inst. Chem. Eng
Vaghasia, Sakaria, Prajapati, Saraf, None, Comput. Biol. Med
Velázquez-Libera, Durán-Verdugo, Valdés-Jiménez, Núñez-Vivanco, Caballero, None, Bioinformatics
Vidal-Limon, Aguilar-Toala, Liceaga, None, J. Agric. Food Chem
Wallace, Laskowski, Thornton, None, Protein Eng. Des. Sel
Xinyi, None, Digit. Chinese Med
Yao, None, J. Comput. Aided. Mol. Des
Yosri, None, Foods
Zhang, None, Int. J. Med. Sci
Zheng, Ji, Li, Feng, None, PeerJ
Zheng, Xue, Wang, Guo, Liu, None, Front. Bioinforma
DOI record:
{
"DOI": "10.1051/bioconf/20248800007",
"ISSN": [
"2117-4458"
],
"URL": "http://dx.doi.org/10.1051/bioconf/20248800007",
"abstract": "<jats:p>This study explores the potential of propolis, a natural substance, as a gene therapy for treating COVID-19. Despite the advent of COVID-19 vaccines, their side effects pose new health challenges. Utilizing network pharmacology, this research identifies propolis compounds through various databases and assesses their ability to target proteins associated with COVID-19. MAPK1 emerges as a potential therapeutic target, and molecular docking reveals Broussoflavonol F, Glyasperin A, and Sulabiroins as promising compounds with strong binding affinities, i.e.,- 9.0, -9.0, and -8.8 kcal/mol, respectively, exceeding the native ligand (-7.2 kcal/mol). Molecular Dynamics displays stable complex behavior, with backbone RMSD values consistently below 4 Angstroms and RMSF simulations showing minimal fluctuations within ±2 Angstroms error. Moreover, MM-PBSA analysis further supports the strong binding of Broussoflavonol F, Glyasperin A, and Sulabiroins A, with relative binding energies of -122.82±89.65, 131.48±95.39, and -155.97±111,37 kJ/mol, respectively. These results indicate that propolis has potential as an anti-COVID-19 agent, primarily through inhibiting the MAPK1 pathway. However, further research is needed to validate these results and develop practical applications for COVID-19 therapy. This study underscores the significance of network pharmacology and computational models in understanding propolis mechanisms, offering potential directions for future research and treatment strategies against COVID-19.</jats:p>",
"alternative-id": [
"bioconf_icias2024_00007"
],
"author": [
{
"affiliation": [],
"family": "Siregar",
"given": "Khalish Arsy Al Khairy",
"sequence": "first"
},
{
"affiliation": [],
"family": "Kustiawan",
"given": "Paula Mariana",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Sari",
"given": "Anissa Nofita",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Hermanto",
"given": "Feri Eko",
"sequence": "additional"
}
],
"container-title": "BIO Web of Conferences",
"container-title-short": "BIO Web Conf.",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2024,
1,
22
]
],
"date-time": "2024-01-22T08:52:51Z",
"timestamp": 1705913571000
},
"deposited": {
"date-parts": [
[
2024,
1,
22
]
],
"date-time": "2024-01-22T08:54:52Z",
"timestamp": 1705913692000
},
"editor": [
{
"affiliation": [],
"family": "Widodo",
"given": "E.",
"sequence": "first"
},
{
"affiliation": [],
"family": "Loh",
"given": "T.C.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Louton",
"given": "H.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Alimon",
"given": "A.R.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Khatib",
"given": "A.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Le Feuvre",
"given": "D.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Muchlas",
"given": "M.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Satria",
"given": "A.T.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Hermanto",
"given": "F.E.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Andri",
"given": "F.",
"sequence": "additional"
}
],
"indexed": {
"date-parts": [
[
2024,
1,
23
]
],
"date-time": "2024-01-23T00:18:18Z",
"timestamp": 1705969098246
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2024
]
]
},
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 21,
"start": {
"date-parts": [
[
2024,
1,
22
]
],
"date-time": "2024-01-22T00:00:00Z",
"timestamp": 1705881600000
}
}
],
"link": [
{
"URL": "https://www.bio-conferences.org/10.1051/bioconf/20248800007/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "250",
"original-title": [],
"page": "00007",
"prefix": "10.1051",
"published": {
"date-parts": [
[
2024
]
]
},
"published-online": {
"date-parts": [
[
2024,
1,
22
]
]
},
"published-print": {
"date-parts": [
[
2024
]
]
},
"publisher": "EDP Sciences",
"reference": [
{
"DOI": "10.14710/jphtcr.v4i1.10569",
"author": "Siregar",
"doi-asserted-by": "crossref",
"first-page": "28",
"journal-title": "J. Public Heal. Trop. Coast. Reg.",
"key": "R1",
"volume": "4",
"year": "2021"
},
{
"DOI": "10.3389/fendo.2023.1238981",
"doi-asserted-by": "crossref",
"key": "R2",
"unstructured": "Wu W. et al, Front. Endocrinol. (Lausanne). 14, (2023)."
},
{
"DOI": "10.3390/jcm9061753",
"author": "Baj",
"doi-asserted-by": "crossref",
"first-page": "1753",
"journal-title": "J. Clin. Med.",
"key": "R3",
"volume": "9",
"year": "2020"
},
{
"DOI": "10.1056/NEJMcp2009575",
"author": "Berlin",
"doi-asserted-by": "crossref",
"first-page": "2451",
"journal-title": "N. Engl. J. Med.",
"key": "R4",
"volume": "383",
"year": "2020"
},
{
"DOI": "10.1056/NEJMe2018629",
"author": "Hariri",
"doi-asserted-by": "crossref",
"first-page": "182",
"journal-title": "N. Engl. J. Med.",
"key": "R5",
"volume": "383",
"year": "2020"
},
{
"DOI": "10.1016/j.jaci.2020.10.008",
"author": "Rivas",
"doi-asserted-by": "crossref",
"first-page": "57",
"journal-title": "J. Allergy Clin. Immunol.",
"key": "R6",
"volume": "147",
"year": "2021"
},
{
"DOI": "10.1186/s12941-020-00384-w",
"author": "Rabaan",
"doi-asserted-by": "crossref",
"first-page": "40",
"journal-title": "Ann. Clin. Microbiol. Antimicrob.",
"key": "R7",
"volume": "19",
"year": "2020"
},
{
"DOI": "10.3390/ijerph18030988",
"author": "Lassi",
"doi-asserted-by": "crossref",
"first-page": "988",
"journal-title": "Int. J. Environ. Res. Public Health",
"key": "R8",
"volume": "18",
"year": "2021"
},
{
"DOI": "10.3389/fimmu.2022.872683",
"doi-asserted-by": "crossref",
"key": "R9",
"unstructured": "Mahroum N.. et al, Front. Immunol. 13, (2022)."
},
{
"DOI": "10.1016/j.joim.2022.01.008",
"author": "Dilokthornsakul",
"doi-asserted-by": "crossref",
"first-page": "114",
"journal-title": "J. Integr. Med.",
"key": "R10",
"volume": "20",
"year": "2022"
},
{
"DOI": "10.1016/j.metop.2021.100141",
"author": "Demeke",
"doi-asserted-by": "crossref",
"first-page": "100141",
"journal-title": "Metab. Open",
"key": "R11",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.3390/molecules27196374",
"author": "Kashyap",
"doi-asserted-by": "crossref",
"first-page": "6374",
"journal-title": "Molecules",
"key": "R12",
"volume": "27",
"year": "2022"
},
{
"DOI": "10.1186/s13020-022-00651-2",
"author": "Hossain",
"doi-asserted-by": "crossref",
"first-page": "100",
"journal-title": "Chin. Med.",
"key": "R13",
"volume": "17",
"year": "2022"
},
{
"DOI": "10.3390/foods10081776",
"author": "Yosri",
"doi-asserted-by": "crossref",
"first-page": "1776",
"journal-title": "Foods",
"key": "R14",
"volume": "10",
"year": "2021"
},
{
"DOI": "10.1002/ptr.7640",
"author": "Ashraf",
"doi-asserted-by": "crossref",
"first-page": "627",
"journal-title": "Phyther. Res.",
"key": "R15",
"volume": "37",
"year": "2023"
},
{
"DOI": "10.3390/pathogens11030311",
"author": "Papadopoulou",
"doi-asserted-by": "crossref",
"first-page": "311",
"journal-title": "Pathogens",
"key": "R16",
"volume": "11",
"year": "2022"
},
{
"DOI": "10.3390/medicina59050877",
"author": "Capraru",
"doi-asserted-by": "crossref",
"first-page": "877",
"journal-title": "Medicina (B. Aires).",
"key": "R17",
"volume": "59",
"year": "2023"
},
{
"DOI": "10.3390/ijms24087099",
"author": "Rizzi",
"doi-asserted-by": "crossref",
"first-page": "7099",
"journal-title": "Int. J. Mol. Sci.",
"key": "R18",
"volume": "24",
"year": "2023"
},
{
"DOI": "10.1016/j.jtice.2023.104898",
"author": "Tsai",
"doi-asserted-by": "crossref",
"first-page": "104898",
"journal-title": "J. Taiwan Inst. Chem. Eng.",
"key": "R19",
"volume": "147",
"year": "2023"
},
{
"DOI": "10.3389/fbinf.2022.928116",
"doi-asserted-by": "crossref",
"key": "R20",
"unstructured": "Zheng S.., Xue T.., Wang B.., Guo H. & Liu Q., Front. Bioinforma. 2, (2022)."
},
{
"DOI": "10.1038/nchembio.118",
"author": "Hopkins",
"doi-asserted-by": "crossref",
"first-page": "682",
"journal-title": "Nat. Chem. Biol.",
"key": "R21",
"volume": "4",
"year": "2008"
},
{
"DOI": "10.21037/tcr-20-2596",
"author": "Chu",
"doi-asserted-by": "crossref",
"first-page": "681",
"journal-title": "Transl. Cancer Res.",
"key": "R22",
"volume": "10",
"year": "2021"
},
{
"DOI": "10.3389/fphar.2020.01194",
"doi-asserted-by": "crossref",
"key": "R23",
"unstructured": "Lai X. et al, Front. Pharmacol. 11, (2020)."
},
{
"DOI": "10.7717/peerj.13737",
"author": "Zheng",
"doi-asserted-by": "crossref",
"first-page": "e13737",
"journal-title": "PeerJ",
"key": "R24",
"volume": "10",
"year": "2022"
},
{
"DOI": "10.1039/C1SC00152C",
"author": "Ma",
"doi-asserted-by": "crossref",
"first-page": "1656",
"journal-title": "Chem. Sci.",
"key": "R25",
"volume": "2",
"year": "2011"
},
{
"DOI": "10.3390/ijms20184331",
"author": "Pinzi",
"doi-asserted-by": "crossref",
"first-page": "4331",
"journal-title": "Int. J. Mol. Sci.",
"key": "R26",
"volume": "20",
"year": "2019"
},
{
"DOI": "10.1021/acs.jafc.1c06110",
"author": "Vidal-Limon",
"doi-asserted-by": "crossref",
"first-page": "934",
"journal-title": "J. Agric. Food Chem.",
"key": "R27",
"volume": "70",
"year": "2022"
},
{
"DOI": "10.1039/D1RA01390D",
"author": "Ibrahim",
"doi-asserted-by": "crossref",
"first-page": "11610",
"journal-title": "RSC Adv.",
"key": "R28",
"volume": "11",
"year": "2021"
},
{
"DOI": "10.1016/j.bbrep.2021.100969",
"author": "Harisna",
"doi-asserted-by": "crossref",
"first-page": "100969",
"journal-title": "Biochem. Biophys. Reports",
"key": "R29",
"volume": "26",
"year": "2021"
},
{
"DOI": "10.22159/ijap.2021.v13s2.20",
"author": "Dewi",
"doi-asserted-by": "crossref",
"first-page": "103",
"journal-title": "Int. J. Appl. Pharm.",
"key": "R30",
"volume": "13",
"year": "2021"
},
{
"DOI": "10.11594/jtls.12.02.08",
"author": "Syaban",
"doi-asserted-by": "crossref",
"first-page": "219",
"journal-title": "J. Trop. Life Sci.",
"key": "R31",
"volume": "12",
"year": "2022"
},
{
"DOI": "10.1016/j.jksus.2020.101297",
"author": "Khayrani",
"doi-asserted-by": "crossref",
"first-page": "101297",
"journal-title": "J. King Saud Univ. Sci.",
"key": "R32",
"volume": "33",
"year": "2021"
},
{
"DOI": "10.21776/ub.jitek.2022.017.02.7",
"author": "Hidayat",
"doi-asserted-by": "crossref",
"first-page": "123",
"journal-title": "J. Ilmu dan Teknol. Has. Ternak",
"key": "R33",
"volume": "17",
"year": "2022"
},
{
"DOI": "10.1016/j.jksus.2020.101234",
"author": "Sahlan",
"doi-asserted-by": "crossref",
"first-page": "101234",
"journal-title": "J. King Saud Univ. Sci.",
"key": "R34",
"volume": "33",
"year": "2021"
},
{
"DOI": "10.1016/j.jksus.2021.101707",
"author": "Fatriansyah",
"doi-asserted-by": "crossref",
"first-page": "101707",
"journal-title": "J. King Saud Univ. Sci.",
"key": "R35",
"volume": "34",
"year": "2022"
},
{
"DOI": "10.1002/ptr.7717",
"doi-asserted-by": "crossref",
"key": "R36",
"unstructured": "Kumar V.. et al, Phyther. Res. (2023) doi:10.1002/ptr.7717."
},
{
"DOI": "10.1038/srep42717",
"author": "Daina",
"doi-asserted-by": "crossref",
"first-page": "42717",
"journal-title": "Sci. Rep.",
"key": "R37",
"volume": "7",
"year": "2017"
},
{
"DOI": "10.1093/nar/gkm795",
"author": "Kuhn",
"doi-asserted-by": "crossref",
"first-page": "D684",
"journal-title": "Nucleic Acids Res.",
"key": "R38",
"volume": "36",
"year": "2007"
},
{
"DOI": "10.1007/s10822-016-9915-2",
"author": "Yao",
"doi-asserted-by": "crossref",
"first-page": "413",
"journal-title": "J. Comput. Aided. Mol. Des.",
"key": "R39",
"volume": "30",
"year": "2016"
},
{
"DOI": "10.1093/nar/gkz382",
"author": "Daina",
"doi-asserted-by": "crossref",
"first-page": "W357",
"journal-title": "Nucleic Acids Res.",
"key": "R40",
"volume": "47",
"year": "2019"
},
{
"DOI": "10.1007/978-981-16-5812-9_2",
"doi-asserted-by": "crossref",
"key": "R41",
"unstructured": "Safran M.. et al, in Practical Guide to Life Science Databases 27–56 (Springer Nature Singapore, 2021). doi:10.1007/978-981-16-5812-9_2."
},
{
"DOI": "10.1093/nar/gkw1092",
"author": "Kanehisa",
"doi-asserted-by": "crossref",
"first-page": "D353",
"journal-title": "Nucleic Acids Res.",
"key": "R42",
"volume": "45",
"year": "2017"
},
{
"DOI": "10.1179/1607845414Y.0000000209",
"author": "Chen",
"doi-asserted-by": "crossref",
"first-page": "336",
"journal-title": "Hematology",
"key": "R43",
"volume": "20",
"year": "2015"
},
{
"DOI": "10.12659/MSM.917240",
"author": "Pan",
"doi-asserted-by": "crossref",
"first-page": "4648",
"journal-title": "Med. Sci. Monit.",
"key": "R44",
"volume": "25",
"year": "2019"
},
{
"key": "R45",
"unstructured": "Gentleman R. C.. et al, Genome Biol. 5, (2004)."
},
{
"DOI": "10.1016/j.jep.2022.115876",
"author": "Shang",
"doi-asserted-by": "crossref",
"first-page": "115876",
"journal-title": "J. Ethnopharmacol.",
"key": "R46",
"volume": "302",
"year": "2023"
},
{
"DOI": "10.2147/COPD.S321877",
"author": "Che",
"doi-asserted-by": "crossref",
"first-page": "2503",
"journal-title": "Int. J. Chron. Obstruct. Pulmon. Dis.",
"key": "R47",
"volume": "16",
"year": "2021"
},
{
"DOI": "10.3389/fgene.2022.870640",
"doi-asserted-by": "crossref",
"key": "R48",
"unstructured": "Shi L.. et al, Front. Genet. 13, (2022)."
},
{
"DOI": "10.1093/nar/gku1003",
"author": "Szklarczyk",
"doi-asserted-by": "crossref",
"first-page": "D447",
"journal-title": "Nucleic Acids Res.",
"key": "R49",
"volume": "43",
"year": "2015"
},
{
"key": "R50",
"unstructured": "Stelzer G.. et al, Curr. Protoc. Bioinforma. 54, (2016)."
},
{
"DOI": "10.1101/gr.1239303",
"author": "Shannon",
"doi-asserted-by": "crossref",
"first-page": "2498",
"journal-title": "Genome Res.",
"key": "R51",
"volume": "13",
"year": "2003"
},
{
"DOI": "10.1007/0-387-29362-0",
"doi-asserted-by": "crossref",
"key": "R52",
"unstructured": "Gentleman R.., Carey V. J.., Huber W.., Irizarry R. A. & Dudoit S., Bioinformatics and Computational Biology Solutions Using R and Bioconductor. (Springer New York, 2005). doi:10.1007/0-387-29362-0."
},
{
"DOI": "10.1186/1752-0509-8-S4-S11",
"author": "Chin",
"doi-asserted-by": "crossref",
"first-page": "S11",
"journal-title": "BMC Syst. Biol.",
"key": "R53",
"volume": "8",
"year": "2014"
},
{
"DOI": "10.1016/j.compbiomed.2022.105994",
"author": "Vaghasia",
"doi-asserted-by": "crossref",
"first-page": "105994",
"journal-title": "Comput. Biol. Med.",
"key": "R54",
"volume": "149",
"year": "2022"
},
{
"DOI": "10.1007/978-1-4939-2269-7_19",
"doi-asserted-by": "crossref",
"key": "R55",
"unstructured": "Dallakyan S. & Olson A. J., in Chemical Biology 243–250 (Humana Press, 2015). doi:10.1007/978-1-4939-2269-7_19."
},
{
"DOI": "10.1158/0008-5472.CAN-11-2596",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "260",
"journal-title": "Cancer Res.",
"key": "R56",
"volume": "72",
"year": "2012"
},
{
"DOI": "10.1093/nar/gkaa1038",
"doi-asserted-by": "crossref",
"key": "R57",
"unstructured": "Burley S. K.. et al, Nucleic Acids Res. 49, D437–D451 (2021)."
},
{
"DOI": "10.1093/nar/gkac956",
"doi-asserted-by": "crossref",
"key": "R58",
"unstructured": "Kim S.. et al, Nucleic Acids Res. 51, D1373–D1380 (2023)."
},
{
"DOI": "10.1093/bioinformatics/btaa018",
"author": "Velázquez-Libera",
"doi-asserted-by": "crossref",
"first-page": "2912",
"journal-title": "Bioinformatics",
"key": "R59",
"volume": "36",
"year": "2020"
},
{
"DOI": "10.1016/j.dcmed.2022.03.003",
"author": "Xinyi",
"doi-asserted-by": "crossref",
"first-page": "18",
"journal-title": "Digit. Chinese Med.",
"key": "R60",
"volume": "5",
"year": "2022"
},
{
"DOI": "10.1093/protein/8.2.127",
"author": "Wallace",
"doi-asserted-by": "crossref",
"first-page": "127",
"journal-title": "Protein Eng. Des. Sel.",
"key": "R61",
"volume": "8",
"year": "1995"
},
{
"DOI": "10.1021/ci200227u",
"author": "Laskowski",
"doi-asserted-by": "crossref",
"first-page": "2778",
"journal-title": "J. Chem. Inf. Model.",
"key": "R62",
"volume": "51",
"year": "2011"
},
{
"DOI": "10.1002/jcc.23899",
"author": "Krieger",
"doi-asserted-by": "crossref",
"first-page": "996",
"journal-title": "J. Comput. Chem.",
"key": "R63",
"volume": "36",
"year": "2015"
},
{
"DOI": "10.1021/acs.jctc.5b00255",
"author": "Maier",
"doi-asserted-by": "crossref",
"first-page": "3696",
"journal-title": "J. Chem. Theory Comput.",
"key": "R64",
"volume": "11",
"year": "2015"
},
{
"DOI": "10.1021/ct300418h",
"author": "Miller",
"doi-asserted-by": "crossref",
"first-page": "3314",
"journal-title": "J. Chem. Theory Comput.",
"key": "R65",
"volume": "8",
"year": "2012"
},
{
"DOI": "10.1016/j.rechem.2022.100721",
"author": "Meylani",
"doi-asserted-by": "crossref",
"first-page": "100721",
"journal-title": "Results Chem.",
"key": "R66",
"volume": "5",
"year": "2023"
},
{
"DOI": "10.2741/3413",
"doi-asserted-by": "crossref",
"key": "R67",
"unstructured": "Al-Sadi R., Front. Biosci. Volume, 2765 (2009)."
},
{
"DOI": "10.7150/ijms.53685",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "1866",
"journal-title": "Int. J. Med. Sci.",
"key": "R68",
"volume": "18",
"year": "2021"
}
],
"reference-count": 68,
"references-count": 68,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.bio-conferences.org/10.1051/bioconf/20248800007"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Exploring Therapeutic Potential of Nutraceutical Compounds from Propolis on MAPK1 Protein Using Bioinformatics Approaches as Anti-Coronavirus Disease 2019 (COVID-19)",
"type": "journal-article",
"volume": "88"
}