A pseudovirus-based method to dynamically mimic SARS-CoV-2-associated cell-to-cell fusion and transmission
et al., Acta Biochimica et Biophysica Sinica, doi:10.3724/abbs.2023129, Jul 2023
56th treatment shown to reduce risk in
August 2025, now with p = 0.0069 from 7 studies.
Lower risk for recovery.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
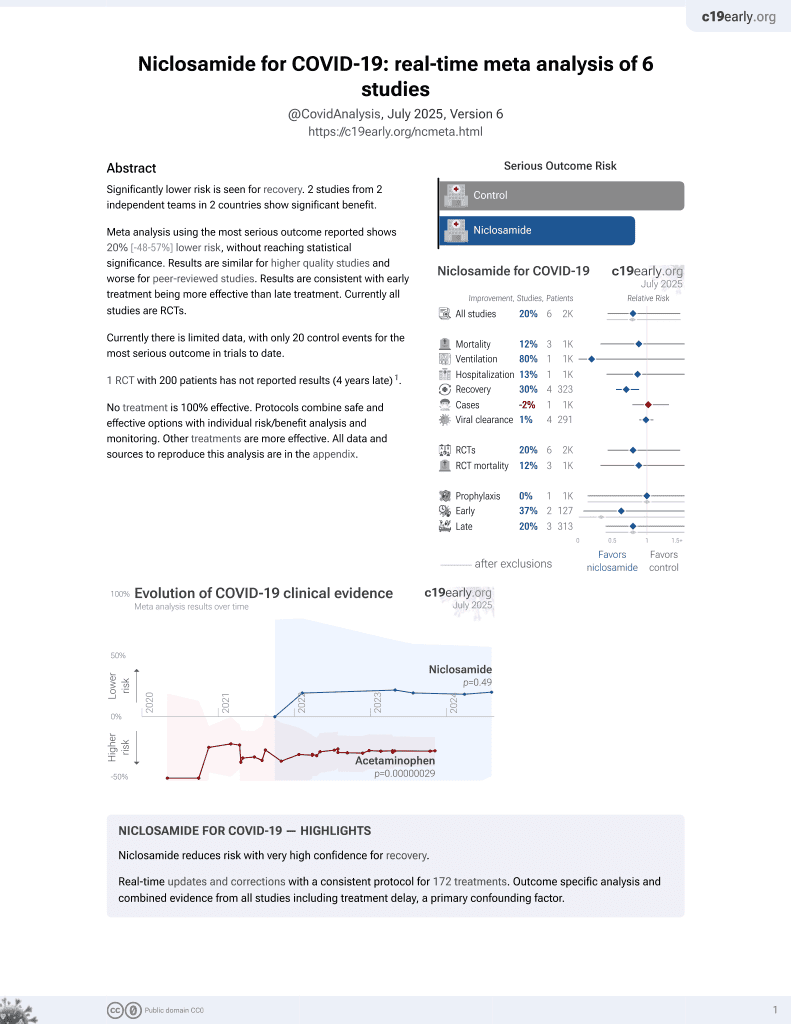
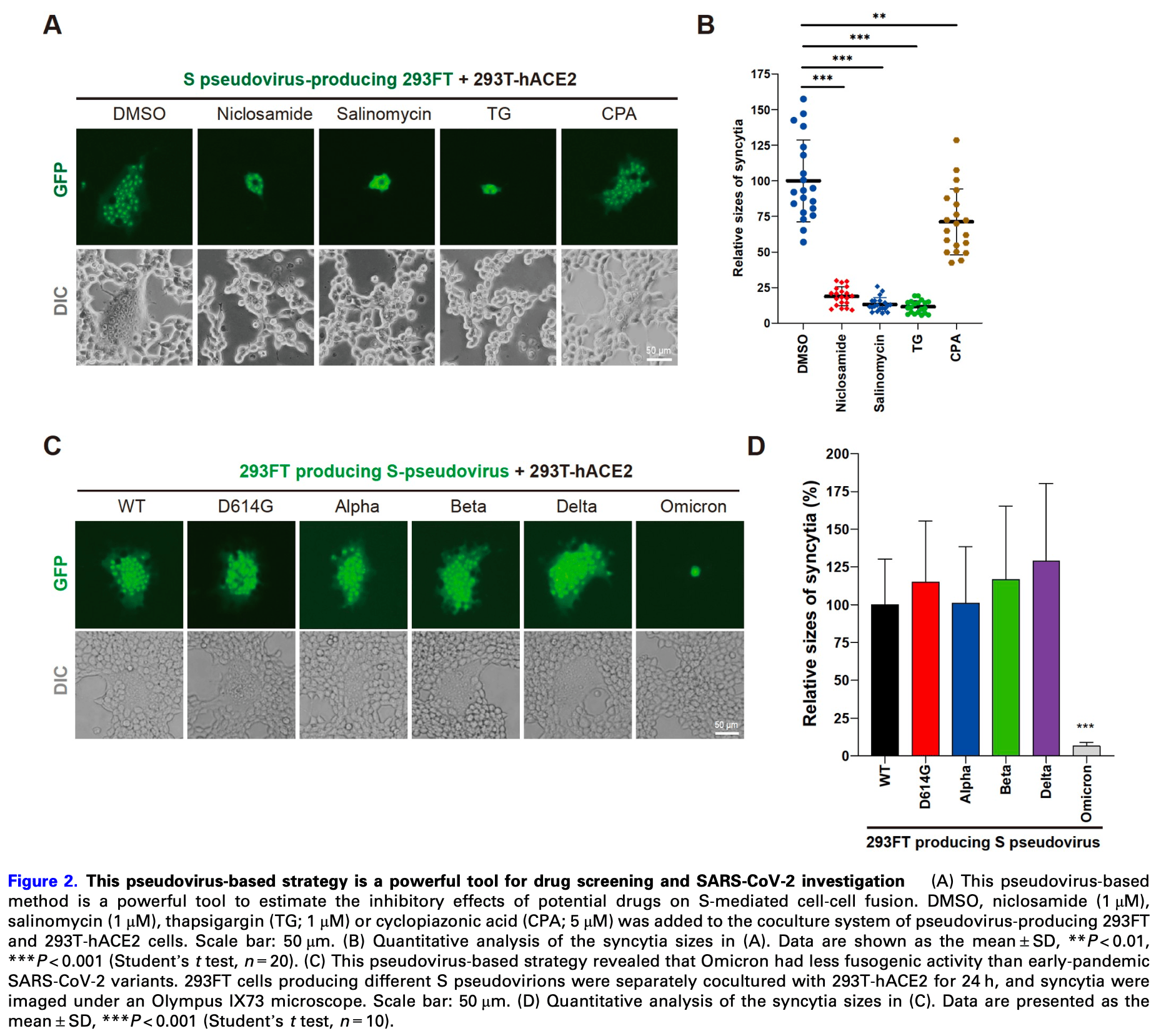
In vitro study showing that niclosamide dramatically blocked the formation of syncytia mediated by SARS-CoV-2 spike protein pseudovirus-producing 293FT cells when cocultured with hACE2-expressing 293T cells at 1μM concentration. Authors developed a novel pseudovirus-based method to dynamically investigate cell fusion and cell-to-cell transmission of SARS-CoV-2. The method was used to test the effects of several compounds, with niclosamide, salinomycin and thapsigargin significantly inhibiting syncytia formation, while cyclopiazonic acid showed a weaker effect.
9 preclinical studies support the efficacy of niclosamide for COVID-19:
In silico studies predict inhibition of SARS-CoV-2 with niclosamide or metabolites via binding to the spikeA,1, MproB,1, RNA-dependent RNA polymeraseC,1, PLproD,1, nucleocapsidE,1, and helicaseF,1 proteins.
Niclosamide inhibits endolysosomal acidification and suppresses
TLR3-mediated pro-inflammatory signaling in human small airway
epithelial cells stimulated with TLR3 agonists mimicking viral RNA2, modulates host lipid metabolism and reduces
infectious SARS-CoV-2 virion production in Vero E6 cells4, reduces CD147 protein levels and inhibits
SARS-CoV-2-induced upregulation of CD147 in A549-ACE2 cells, including
the highly glycosylated form of CD147 which has been implicated in
COVID-19 disease progression and post-COVID-19 cardiac complications5, blocked the formation of syncytia mediated by
SARS-CoV-2 spike protein pseudovirus-producing cells6, may reduce inflammation, NLRP3 formation, and
caspase-1 activity9, may inhibit viral uncoating, replication, and
assembly via disruption of pH gradients and reduced ATP production in
host cells8, may counter immune evasion by reversing E-, ORF7a-, and ORF8-mediated down-regulation of MHC-I, preserving CD8⁺ T-cell recognition10, and shows strong synergy when combined with
ivermectin7.
1.
Haque et al., Exploring potential therapeutic candidates against COVID-19: a molecular docking study, Discover Molecules, doi:10.1007/s44345-024-00005-5.
2.
Pejler et al., Blockade of endolysosomal acidification suppresses TLR3-mediated pro-inflammatory signaling in airway epithelial cells, Journal of Allergy and Clinical Immunology, doi:10.1016/j.jaci.2024.05.031.
3.
Walia et al., SARS-CoV-2 virulence factor ORF3a blocks lysosome function by modulating TBC1D5-dependent Rab7 GTPase cycle, Nature Communications, doi:10.1038/s41467-024-46417-2.
4.
Garrett et al., Niclosamide as a chemical probe for analyzing SARS-CoV-2 modulation of host cell lipid metabolism, Frontiers in Microbiology, doi:10.3389/fmicb.2023.1251065.
5.
Yang et al., Repurposing Niclosamide as a Novel Anti-SARS-CoV-2 Drug by Restricting Entry Protein CD147, Biomedicines, doi:10.3390/biomedicines11072019.
6.
Sheng et al., A pseudovirus-based method to dynamically mimic SARS-CoV-2-associated cell-to-cell fusion and transmission, Acta Biochimica et Biophysica Sinica, doi:10.3724/abbs.2023129.
7.
Jitobaom et al., Synergistic anti-SARS-CoV-2 activity of repurposed anti-parasitic drug combinations, BMC Pharmacology and Toxicology, doi:10.1186/s40360-022-00580-8.
8.
Needham, D., The pH Dependence of Niclosamide Solubility, Dissolution, and Morphology: Motivation for Potentially Universal Mucin-Penetrating Nasal and Throat Sprays for COVID19, its Variants and other Viral Infections, Pharmaceutical Research, doi:10.1007/s11095-021-03112-x.
a.
The trimeric spike (S) protein is a glycoprotein that mediates viral entry by binding to the host ACE2 receptor, is critical for SARS-CoV-2's ability to infect host cells, and is a target of neutralizing antibodies. Inhibition of the spike protein prevents viral attachment, halting infection at the earliest stage.
b.
The main protease or Mpro, also known as 3CLpro or nsp5, is a cysteine protease that cleaves viral polyproteins into functional units needed for replication. Inhibiting Mpro disrupts the SARS-CoV-2 lifecycle within the host cell, preventing the creation of new copies.
c.
RNA-dependent RNA polymerase (RdRp), also called nsp12, is the core enzyme of the viral replicase-transcriptase complex that copies the positive-sense viral RNA genome into negative-sense templates for progeny RNA synthesis. Inhibiting RdRp blocks viral genome replication and transcription.
d.
The papain-like protease (PLpro) has multiple functions including cleaving viral polyproteins and suppressing the host immune response by deubiquitination and deISGylation of host proteins. Inhibiting PLpro may block viral replication and help restore normal immune responses.
e.
The nucleocapsid (N) protein binds and encapsulates the viral genome by coating the viral RNA. N enables formation and release of infectious virions and plays additional roles in viral replication and pathogenesis. N is also an immunodominant antigen used in diagnostic assays.
f.
The helicase, or nsp13, protein unwinds the double-stranded viral RNA, a crucial step in replication and transcription. Inhibition may prevent viral genome replication and the creation of new virus components.
Sheng et al., 1 Jul 2023, peer-reviewed, 6 authors.
Contact: coryhu@sibcb.ac.cn, shengxiangpeng@ucas.ac.cn.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Abstract: Acta Biochim Biophys Sin 2023, 55(11): 1840–1843
https://doi.org/10.3724/abbs.2023129
Advance Access Publication Date: 6 July 2023
Lab Note
Lab Note
A pseudovirus-based method to dynamically mimic
SARS-CoV-2-associated cell-to-cell fusion and transmission
Xiangpeng Sheng1,2,3,†,*, Yi Yang4,†, Fang Zhu5,†, Fan Yang2,†, Honghua Wang1, and
Ronggui Hu1,2,*
1Key Laboratory of Systems Health Science of Zhejiang Province, School of Life Science, Hangzhou Institute for Advanced Study, University of
Chinese Academy of Sciences, Hangzhou 310024, China, 2State Key Laboratory of Molecular Biology, Shanghai Institute of Biochemistry and
Cell Biology, Center for Excellence in Molecular Cell Science, University of Chinese Academy of Sciences, Chinese Academy of Sciences,
Shanghai 200031, China, 3State Key Laboratory of Animal Disease Control, Harbin Veterinary Research Institute, Chinese Academy of
Agricultural Sciences, Harbin 150069, China, 4Department of Thoracic Surgery, Ruijin Hospital, Shanghai Jiaotong University School of
Medicine, Shanghai 200025, China, and 5School of Medicine, Guizhou University, Guiyang 550025, China
†These authors contributed equally to this work.
*Correspondence address. Tel: +86-17717541320; E-mail: coryhu@sibcb.ac.cn (R.H.) / Tel: +86-13701790678; E-mail: shengxiangpeng@ucas.ac.cn (X.S.)
Received 8 June 2023 Accepted 25 June 2023
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2),
responsible for the COVID-19 pandemic, has caused tremendous
global loss and continues to evolve to generate variants. Entry of
SARS-CoV-2 into target host cells is primarily mediated by spike (S),
which binds to the host receptor hACE2 and initiates virus-cell
membrane fusion [1]. Most COVID-19 patients show pneumocyte
syncytia in the lungs [2]. Cell fusion contributes to viral entry, cellto-cell transmission and tissue damage, and thus attracts much
attention. Because authentic SARS-CoV-2 live virions can only be
handled in biosafety level-3 (BSL-3) facilities, many researchers
have developed different assays to study cell fusion in BSL-1/2 by
directly expressing S and hACE2 on mammalian cells [2‒5]. Briefly,
in regular cell fusion assays, S-expressing cells and hACE2-positive
cells are cocultured at approximately a 1:1 ratio, which induces cellcell fusion and usually activates a fusion reporter. Although these
strategies are useful, they cannot efficiently simulate cell-cell fusion
and transmission in SARS-CoV-2 infection, in which virions from
one target cell are transmitted to neighboring cells, resulting in
syncytia. Here, we design a pseudovirus-based method to dynamically and highly mimic cell-to-cell fusion and virus transmission of
SARS-CoV-2.
First, we generated spike-pseudotyped virions (S pseudovirions)
in HEK293FT cells by co-transfecting three plasmids, including
psPAX2, pCDH-sfGFP, and a plasmid expressing SARS-CoV-2 S into
cells (Figure 1A), and collected the viral supernatant, which is
similar to a previous report [6]. S pseudovirus was found to
efficiently infect hACE2-positive 293T cells (293T-hACE2) but not
the control 293T or 293FT cell lines (Supplementary Figure S1A,B),
suggesting that S indeed envelops the pseudovirions. However,
fluorescence microscopy revealed that the infection of 293T-hACE2
cells by pseudovirus supernatant could not trigger cell-cell
membrane fusion events (Supplementary Figure S1C). Given that
the authentic SARS-CoV-2-infected host cells can continue to
generate live virions to..
DOI record:
{
"DOI": "10.3724/abbs.2023129",
"ISSN": [
"1672-9145"
],
"URL": "http://dx.doi.org/10.3724/abbs.2023129",
"author": [
{
"affiliation": [],
"family": "Sheng",
"given": "Xiangpeng",
"sequence": "first"
},
{
"affiliation": [],
"family": "Yang",
"given": "Yi",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Zhu",
"given": "Fang",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Yang",
"given": "Fan",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Wang",
"given": "Honghua",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Hu",
"given": "Ronggui",
"sequence": "additional"
}
],
"container-title": "Acta Biochimica et Biophysica Sinica",
"container-title-short": "ABBS",
"content-domain": {
"crossmark-restriction": false,
"domain": [
"engine.scichina.com"
]
},
"created": {
"date-parts": [
[
2023,
7,
3
]
],
"date-time": "2023-07-03T02:15:48Z",
"timestamp": 1688350548000
},
"deposited": {
"date-parts": [
[
2023,
7,
7
]
],
"date-time": "2023-07-07T01:44:54Z",
"timestamp": 1688694294000
},
"indexed": {
"date-parts": [
[
2023,
7,
7
]
],
"date-time": "2023-07-07T04:20:01Z",
"timestamp": 1688703601422
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2023,
7,
1
]
]
},
"language": "en",
"link": [
{
"URL": "https://engine.scichina.com/doi/pdf/C0B6CFF296134A179C6D9C2CC6DD07D6",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://engine.scichina.com/doi/10.3724/abbs.2023129",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://engine.scichina.com/doi/pdf/C0B6CFF296134A179C6D9C2CC6DD07D6",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "2026",
"original-title": [],
"prefix": "10.3724",
"published": {
"date-parts": [
[
2023,
7,
1
]
]
},
"published-online": {
"date-parts": [
[
2023,
7,
6
]
]
},
"published-print": {
"date-parts": [
[
2023,
7,
1
]
]
},
"publisher": "China Science Publishing & Media Ltd.",
"reference": [
{
"DOI": "10.1016/j.cell.2020.03.045",
"author": "Wang Q",
"doi-asserted-by": "publisher",
"first-page": "894",
"journal-title": "Cell",
"key": "CITATION1",
"unstructured": "Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell, 2020, 181: 894-904.",
"volume": "181",
"year": "2020"
},
{
"DOI": "10.1038/s41586-021-03491-6",
"author": "Braga L",
"doi-asserted-by": "publisher",
"first-page": "88",
"journal-title": "Nature",
"key": "CITATION2",
"unstructured": "Braga L, Ali H, Secco I, Chiavacci E, Neves G, Goldhill D, Penn R. Drugs that inhibit TMEM16 proteins block SARS-CoV-2 spike-induced syncytia. Nature, 2021, 594: 88-93.",
"volume": "594",
"year": "2021"
},
{
"DOI": "10.1371/journal.ppat.1009246",
"doi-asserted-by": "crossref",
"key": "CITATION3",
"unstructured": "Papa G, Mallery DL, Albecka A, Welch LG, Cattin-Ortolá J, Luptak J, Paul D, et al. Furin cleavage of SARS-CoV-2 Spike promotes but is not essential for infection and cell-cell fusion. PLoS Pathogens 2021, 17: e1009246."
},
{
"DOI": "10.15252/embj.2020106267",
"author": "Buchrieser J",
"doi-asserted-by": "publisher",
"first-page": "e106267",
"journal-title": "EMBO J",
"key": "CITATION4",
"unstructured": "Buchrieser J, Dufloo J, Hubert M, Monel B, Planas D, Rajah MM, Planchais C. Syncytia formation by SARS‐CoV‐2‐infected cells. EMBO J, 2020, 39:",
"volume": "39",
"year": "2020"
},
{
"DOI": "10.1073/pnas.2111400119",
"author": "Zeng C",
"doi-asserted-by": "publisher",
"first-page": "e2111400119",
"journal-title": "Proc Natl Acad Sci USA",
"key": "CITATION5",
"unstructured": "Zeng C, Evans JP, King T, Zheng YM, Oltz EM, Whelan SPJ, Saif LJ. SARS-CoV-2 spreads through cell-to-cell transmission. Proc Natl Acad Sci USA, 2022, 119:",
"volume": "119",
"year": "2022"
},
{
"DOI": "10.1038/s41467-020-15562-9",
"author": "Ou X",
"doi-asserted-by": "publisher",
"first-page": "1620",
"journal-title": "Nat Commun",
"key": "CITATION6",
"unstructured": "Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun, 2020, 11:",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1038/s41579-022-00841-7",
"author": "Carabelli AM",
"doi-asserted-by": "publisher",
"first-page": "162",
"journal-title": "Nat Rev Microbiol",
"key": "CITATION7",
"unstructured": "Carabelli AM, Peacock TP, Thorne LG, Harvey WT, Hughes J, de Silva TI, Peacock SJ. SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nat Rev Microbiol, 2023, 21: 162-177.",
"volume": "21",
"year": "2023"
},
{
"DOI": "10.1038/s41586-022-04442-5",
"author": "Shuai H",
"doi-asserted-by": "publisher",
"first-page": "693",
"journal-title": "Nature",
"key": "CITATION8",
"unstructured": "Shuai H, Chan JFW, Hu B, Chai Y, Yuen TTT, Yin F, Huang X. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature, 2022, 603: 693-699.",
"volume": "603",
"year": "2022"
},
{
"DOI": "10.1038/s41564-022-01143-7",
"author": "Willett BJ",
"doi-asserted-by": "publisher",
"first-page": "1161",
"journal-title": "Nat Microbiol",
"key": "CITATION9",
"unstructured": "Willett BJ, Grove J, MacLean OA, Wilkie C, De Lorenzo G, Furnon W, Cantoni D. SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway. Nat Microbiol, 2022, 7: 1161-1179.",
"volume": "7",
"year": "2022"
}
],
"reference-count": 9,
"references-count": 9,
"relation": {},
"resource": {
"primary": {
"URL": "https://engine.scichina.com/doi/10.3724/abbs.2023129"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"General Medicine",
"Biochemistry",
"Biophysics",
"Molecular Biology"
],
"subtitle": [],
"title": "A pseudovirus-based method to dynamically mimic SARS-CoV-2-associated cell-to-cell fusion and transmission",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1360/scp-crossmark-policy-page"
}