Lethal Mutagenesis of RNA Viruses and Approved Drugs with Antiviral Mutagenic Activity
et al., Viruses, doi:10.3390/v14040841, Apr 2022
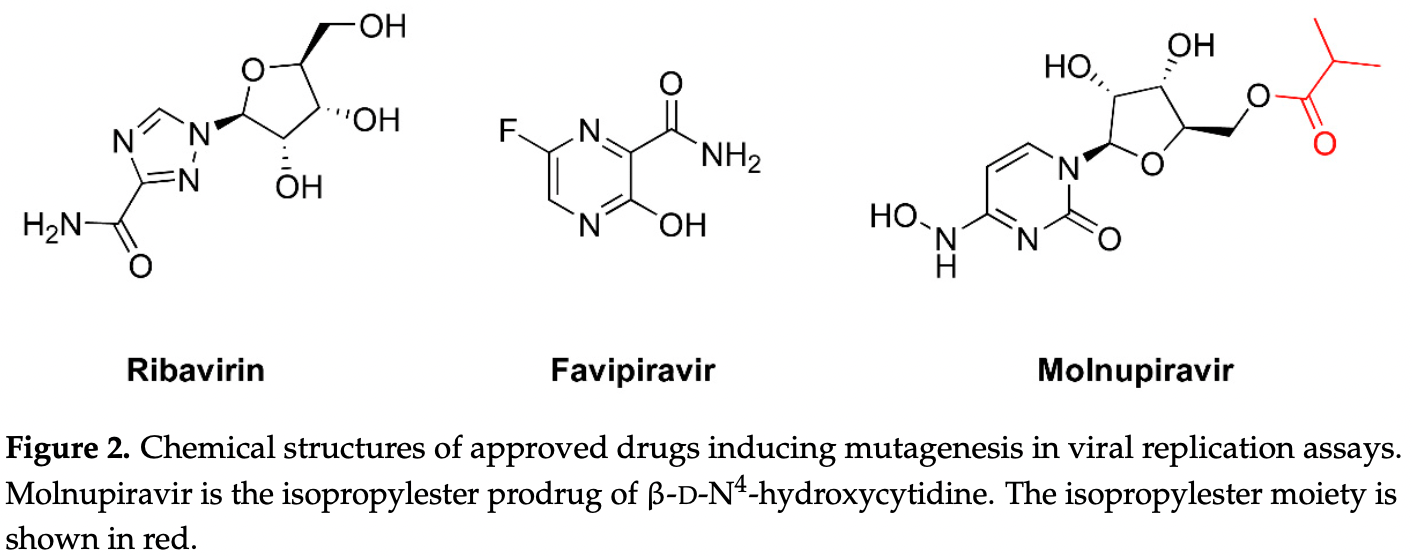
Review of lethal mutagenesis for RNA viruses, as used by molnupiravir, favipiravir, and ribavirin. Authors note the potential for permanently modifying the genomes of patients while causing teratogenicity or embryotoxicity, and the potential of creating novel virus variants with increased pathogenicity and transmissibility.
Authors recommend a registry of patients for long-term monitoring of potential adverse effects, including genetic, carcinogenic, teratogenic, and embryotoxic damage.
Review covers molnupiravir and favipiravir.
1.
Shen et al., Carboxylesterase Factors Influencing the Therapeutic Activity of Common Antiviral Medications Used for SARS-CoV-2 Infection, Pharmaceutics, doi:10.3390/pharmaceutics17070832.
2.
Saha et al., Inhaled Dry Powder of Antiviral Agents: A Promising Approach to Treating Respiratory Viral Pathogens, Viruses, doi:10.3390/v17020252.
3.
Lopez et al., SARS-CoV-2 Resistance to Small Molecule Inhibitors, Current Clinical Microbiology Reports, doi:10.1007/s40588-024-00229-6.
4.
Bacigalupo et al., Unveiling patenting strategies of therapeutics and vaccines: evergreening in the context of COVID-19 pandemic, Frontiers in Medicine, doi:10.3389/fmed.2023.1287542.
5.
Hadj Hassine et al., Lethal Mutagenesis of RNA Viruses and Approved Drugs with Antiviral Mutagenic Activity, Viruses, doi:10.3390/v14040841.
Hadj Hassine et al., 18 Apr 2022, peer-reviewed, 3 authors.
Contact: lmenendez@cbm.csic.es (corresponding author), hadj_hassine_ekbell@yahoo.fr, benmhadhebmanel@yahoo.fr.
Lethal Mutagenesis of RNA Viruses and Approved Drugs with Antiviral Mutagenic Activity
Viruses, doi:10.3390/v14040841
In RNA viruses, a small increase in their mutation rates can be sufficient to exceed their threshold of viability. Lethal mutagenesis is a therapeutic strategy based on the use of mutagens, driving viral populations to extinction. Extinction catastrophe can be experimentally induced by promutagenic nucleosides in cell culture models. The loss of HIV infectivity has been observed after passage in 5-hydroxydeoxycytidine or 5,6-dihydro-5-aza-2 -deoxycytidine while producing a two-fold increase in the viral mutation frequency. Among approved nucleoside analogs, experiments with polioviruses and other RNA viruses suggested that ribavirin can be mutagenic, although its mechanism of action is not clear. Favipiravir and molnupiravir exert an antiviral effect through lethal mutagenesis. Both drugs are broad-spectrum antiviral agents active against RNA viruses. Favipiravir incorporates into viral RNA, affecting the G→A and C→U transition rates. Molnupiravir (a prodrug of β-D-N 4 -hydroxycytidine) has been recently approved for the treatment of SARS-CoV-2 infection. Its triphosphate derivative can be incorporated into viral RNA and extended by the coronavirus RNA polymerase. Incorrect base pairing and inefficient extension by the polymerase promote mutagenesis by increasing the G→A and C→U transition frequencies. Despite having remarkable antiviral action and resilience to drug resistance, carcinogenic risks and genotoxicity are important concerns limiting their extended use in antiviral therapy.
Conflicts of Interest: The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
Abdelnabi, Foo, De Jonghe, Maes, Weynand et al., Molnupiravir inhibits replication of the emerging SARS-CoV-2 variants of concern in a hamster infection model, J. Infect. Dis, doi:10.1093/infdis/jiab361
Abdelnabi, Foo, Kaptein, Zhang, Do et al., The combined treatment of molnupiravir and favipiravir results in a potentiation of antiviral efficacy in a SARS-CoV-2 hamster infection model, EBioMedicine, doi:10.1016/j.ebiom.2021.103595
Agostini, Pruijssers, Chappell, Gribble, Lu et al., Small-molecule antiviral β-D-N 4 -hydroxycytidine inhibits a proofreading-intact coronavirus with a high genetic barrier to resistance, J. Virol, doi:10.1128/JVI.01348-19
Agudo, Ferrer-Orta, Arias, De La Higuera, Perales et al., A multi-step process of viral adaptation to a mutagenic nucleoside analogue by modulation of transition types leads to extinction-escape, PLoS Pathog, doi:10.1371/journal.ppat.1001072
Airaksinen, Pariente, Menéndez-Arias, Domingo, Curing of foot-and-mouth disease virus from persistently infected cells by ribavirin involves enhanced mutagenesis, Virology, doi:10.1016/S0042-6822(03)00144-2
Anderson, Daifuku, Loeb, Viral error catastrophe by mutagenic nucleosides, Annu. Rev. Microbiol, doi:10.1146/annurev.micro.58.030603.123649
Arias, Thorne, Goodfellow, Favipiravir elicits antiviral mutagenesis during virus replication in vivo, Elife, doi:10.7554/eLife.03679
Arnold, Cameron, Poliovirus RNA-dependent RNA polymerase (3Dpol) is sufficient for template switching in vitro, J. Biol. Chem, doi:10.1074/jbc.274.5.2706
Baranovich, Wong, Armstrong, Marjuki, Webby et al., T-705 (favipiravir) induces lethal mutagenesis in influenza A H1N1 viruses in vitro, J. Virol, doi:10.1128/JVI.02346-12
Barrioluengo, Alvarez, Barbieri, Menéndez-Arias, Thermostable HIV-1 group O reverse transcriptase variants with the same fidelity as murine leukaemia virus reverse transcriptase, Biochem. J, doi:10.1042/BJ20101852
Bassi, Sempere, Meyn, Polacek, Arias, Extinction of Zika virus and Usutu virus by lethal mutagenesis reveals different patterns of sensitivity to three mutagenic drugs, Antimicrob. Agents Chemother, doi:10.1128/AAC.00380-18
Beach, Rawson, Kim, Patterson, Mansky, Novel inhibitors of human immunodeficiency virus type 2 infectivity, J. Gen. Virol, doi:10.1099/vir.0.069864-0
Bernal, Gomes Da Silva, Musungaie, Kovalchuk, Gonzalez et al., Molnupiravir for oral treatment of COVID-19 in nonhospitalized patients, N. Engl. J. Med, doi:10.1056/NEJMoa2116044
Borrego, De Ávila, Domingo, Brun, Lethal mutagenesis of Rift Valley fever virus induced by favipiravir, Antimicrob. Agents Chemother, doi:10.1128/AAC.00669-19
Cao, Wang, Xi, Zhang, Wang et al., PIG-A gene mutation as a genotoxicity biomarker in human population studies: An investigation in lead-exposed workers, Environ. Mol. Mutagen
Cases-González, Gutiérrez-Rivas, Menéndez-Arias, Coupling ribose selection to fidelity of DNA synthesis: The role of Tyr-115 of human immunodeficiency virus type 1 reverse transcriptase, J. Biol. Chem, doi:10.1074/jbc.M910361199
Chung, Sun, Parker, Arterburn, Bartolucci et al., Ribavirin reveals a lethal threshold of allowable mutation frequency for Hantaan virus, J. Virol, doi:10.1128/JVI.00874-07
Clouser, Patterson, Mansky, Exploiting drug repositioning for discovery of a novel HIV combination therapy, J. Virol, doi:10.1128/JVI.01006-10
Coffin, HIV population dynamics in vivo: Implications for genetic variation, pathogenesis, and therapy, Science, doi:10.1126/science.7824947
Combe, Sanjuán, Variation in RNA virus mutation rates across host cells, PLoS Pathog, doi:10.1371/journal.ppat.1003855
Contreras, Hiasa, He, Terella, Schmidt et al., Viral RNA mutations are region specific and increased by ribavirin in a full-length hepatitis C virus replication system, J. Virol, doi:10.1128/JVI.76.17.8505-8517.2002
Cox, Wolf, Plemper, Therapeutically administered ribonucleoside analogue MK-4482/EIDD-2801 blocks SARS-CoV-2 transmission in ferrets, Nat. Microbiol, doi:10.1038/s41564-020-00835-2
Crotty, Maag, Arnold, Zhong, Lau et al., The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen, Nat. Med, doi:10.1038/82191
Cuevas, González-Candelas, Moya, Sanjuán, Effect of ribavirin on the mutation rate and spectrum of hepatitis C virus in vivo, J. Virol, doi:10.1128/JVI.00201-09
Dapp, Clouser, Patterson, Mansky, 5-Azacytidine can induce lethal mutagenesis in human immunodeficiency virus type 1, J. Virol, doi:10.1128/JVI.01406-09
Day, Smee, Julander, Yamshchikov, Sidwell et al., Error-prone replication of West Nile virus caused by ribavirin, Antiviral. Res, doi:10.1016/j.antiviral.2005.04.002
De Avila, Moreno, Perales, Domingo, Favipiravir can evoke lethal mutagenesis and extinction of foot-and-mouth disease virus, Virus Res, doi:10.1016/j.virusres.2017.03.014
De Ávila, Gallego, Soria, Gregori, Quer et al., Lethal mutagenesis of hepatitis C virus induced by favipiravir, PLoS ONE, doi:10.1371/journal.pone.0164691
Delang, Abdelnabi, Neyts, Favipiravir as a potential countermeasure against neglected and emerging RNA viruses, Antiviral. Res, doi:10.1016/j.antiviral.2018.03.003
Delang, Segura Guerrero, Tas, Quérat, Pastorino et al., Mutations in the Chikungunya virus non-structural proteins cause resistance to favipiravir (T-705), a broadspectrum antiviral, J. Antimicrob. Chemother, doi:10.1093/jac/dku209
Dietz, Schelhorn, Fitting, Mihm, Susser et al., Deep sequencing reveals mutagenic effects of ribavirin during monotherapy of hepatitis C virus genotype 1-infected patients, J. Virol, doi:10.1128/JVI.02778-12
Domingo, García-Crespo, Perales, Historical perspective on the discovery of the quasispecies concept, Annu. Rev. Virol, doi:10.1146/annurev-virology-091919-105900
Drake, Charlesworth, Charlesworth, Crow, Rates of spontaneous mutation, Genetics
Drake, Holland, Mutation rates among RNA viruses, doi:10.1073/pnas.96.24.13910
Díaz-Martínez, Brichette-Mieg, Pineño-Ramos, Domínguez-Huerta, Grande-Pérez, Lethal mutagenesis of an RNA plant virus via lethal defection, Sci. Rep, doi:10.1038/s41598-018-19829-6
Eckerle, Becker, Halpin, Li, Venter et al., Infidelity of SARS-CoV nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing, PLoS Pathog, doi:10.1371/journal.ppat.1000896
Eckerle, Lu, Sperry, Choi, Denison, High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants, J. Virol, doi:10.1128/JVI.01296-07
Eigen, Error catastrophe and antiviral strategy, doi:10.1073/pnas.212514799
Eigen, From Strange Simplicity to Complex Familiarity: A Treatise on Matter, Information, Life and Thought
Elder, Lerner, Hasselkus-Light, Fontenot, Hunter et al., Distinct subsets of retroviruses encode dUTPase, J. Virol, doi:10.1128/jvi.66.3.1791-1794.1992
Eron, Jr, Holman, Cohen, Fang et al., A phase 2a clinical trial of molnupiravir in patients with COVID-19 shows accelerated SARS-CoV-2 RNA clearance and elimination of infectious virus, Sci. Transl. Med, doi:10.1126/scitranslmed.abl7430
Escribano-Romero, Jiménez De Oya, Domingo, Saiz, Extinction of West Nile virus by favipiravir through lethal mutagenesis, Antimicrob. Agents Chemother, doi:10.1128/AAC.01400-17
Espy, Nagle, Pfeffer, Garcia, Chitty et al., T-705 induces lethal mutagenesis in Ebola and Marburg populations in macaques, Antiviral. Res, doi:10.1016/j.antiviral.2019.06.001
Ferrer-Orta, Sierra, Agudo, De La Higuera, Arias et al., Structure of foot-and-mouth disease virus mutant polymerases with reduced sensitivity to ribavirin, J. Virol, doi:10.1128/JVI.02420-09
Fischer, None
Fontecave, Lepoivre, Elleingand, Gerez, Guittet, Resveratrol, a remarkable inhibitor of ribonucleotide reductase, FEBS Lett, doi:10.1016/S0014-5793(97)01572-X
Gallego, Gregori, Soria, García-Crespo, García-Álvarez et al., Resistance of high fitness hepatitis C virus to lethal mutagenesis, Virology, doi:10.1016/j.virol.2018.07.030
Gallego, Soria, Gregori, De Ávila, García-Crespo et al., Synergistic lethal mutagenesis of hepatitis C virus, Antimicrob. Agents Chemother, doi:10.1128/AAC.01653-19
Geraghty, Aliota, Bonnac, Broad-spectrum antiviral strategies and nucleoside analogues, Viruses
Gohara, Crotty, Arnold, Yoder, Andino et al., Poliovirus RNA-dependent RNA polymerase (3Dpol): Structural, biochemical, and biological analysis of conserved structural motifs a and b, J. Biol. Chem, doi:10.1074/jbc.M002671200
Goldhill, Te, Velthuis, Fletcher, Langat et al., The mechanism of resistance to favipiravir in influenza, doi:10.1073/pnas.1811345115
Goldhill, Yan, Frise, Zhou, Shelley et al., Favipiravir-resistant influenza A virus shows potential for transmission, PLoS Pathog, doi:10.1371/journal.ppat.1008937
Gong, Peersen, Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase, Proc. Natl. Acad. Sci, doi:10.1073/pnas.1007626107
Gordon, Tchesnokov, Schinazi, Götte, Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA template, J. Biol. Chem, doi:10.1016/j.jbc.2021.100770
Graci, Cameron, Mechanisms of action of ribavirin against distinct viruses, Rev. Med. Virol, doi:10.1002/rmv.483
Graci, Gnädig, Galarraga, Castro, Vignuzzi et al., Mutational robustness of an RNA virus influences sensitivity to lethal mutagenesis, J. Virol, doi:10.1128/JVI.05712-11
Graci, Harki, Korneeva, Edathil, Too et al., Lethal mutagenesis of poliovirus mediated by a mutagenic pyrimidine analogue, J. Virol, doi:10.1128/JVI.01028-07
Graci, Too, Smidansky, Edathil, Barr et al., Lethal mutagenesis of picornaviruses with N-6-modified purine nucleoside analogues, Antimicrob. Agents Chemother, doi:10.1128/AAC.01056-07
Grande-Pérez, Lázaro, Lowenstein, Domingo, Manrubia, Suppression of viral infectivity through lethal defection, Proc. Natl. Acad. Sci, doi:10.1073/pnas.0408871102
Guarino, Salguero, Kearsey, Cellular regulation of ribonucleotide reductase in eukaryotes, Semin. Cell Dev. Biol, doi:10.1016/j.semcdb.2014.03.030
Harki, Graci, Galarraga, Chain, Cameron et al., Synthesis and antiviral activity of 5-substituted cytidine analogues: Identification of a potent inhibitor of viral RNA-dependent RNA polymerases, J. Med. Chem, doi:10.1021/jm060872x
Harris, Brabant, Styrchak, Gall, Daifuku, KP-1212/1461, a nucleoside designed for the treatment of HIV by viral mutagenesis, Antivir. Res, doi:10.1016/j.antiviral.2005.03.004
Haruna, Spiegelman, Recognition of size and sequence by an RNA replicase, Proc. Natl. Acad. Sci, doi:10.1073/pnas.54.4.1189
Haruna, Spiegelman, Specific template requirments of RNA replicases
Hillen, Structure and function of SARS-CoV-2 polymerase, Curr. Opin. Virol, doi:10.1016/j.coviro.2021.03.010
Holland, Domingo, De La Torre, Steinhauer, Mutation frequencies at defined single codon sites in vesicular stomatitis virus and poliovirus can be increased only slightly by chemical mutagenesis, J. Virol, doi:10.1128/jvi.64.8.3960-3962.1990
Holland, Spindler, Horodyski, Grabau, Nichol et al., Rapid evolution of RNA genomes, Science, doi:10.1126/science.7041255
Jena, Role of different tautomers in the base-pairing abilities of some of the vital antiviral drugs used against COVID-19, Phys. Chem. Chem. Phys, doi:10.1039/D0CP05297C
Jin, Smith, Rajwanshi, Kim, Deval, The ambiguous base-pairing and high substrate efficiency of T-705 (favipiravir) ribofuranosyl 5'-triphosphate towards influenza A virus polymerase, PLoS ONE, doi:10.1371/journal.pone.0068347
Jordan, Stevens, Deval, Nucleosides for the treatment of respiratory RNA virus infections, Antivir. Chem. Chemother, doi:10.1177/2040206618764483
Julias, Kim, Arnold, Pathak, The antiretrovirus drug 3 -azido-3 -deoxythymidine increases the retrovirus mutation rate, J. Virol, doi:10.1128/jvi.71.6.4254-4263.1997
Kabinger, Stiller, Schmitzová, Dienemann, Kokic et al., Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis, Nat. Struct. Mol. Biol, doi:10.1038/s41594-021-00651-0
Kentsis, Topisirovic, Culjkovic, Shao, Borden, Ribavirin suppresses EIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap, Proc. Natl. Acad. Sci, doi:10.1073/pnas.0406927102
Köppe, Menéndez-Arias, Oroszlan, Expression and purification of the mouse mammary tumor virus gag-pro transframe protein p30 and characterization of its dUTPase activity, J. Virol, doi:10.1128/jvi.68.4.2313-2319.1994
Lacasse, Remington, North, The mutation frequency of feline immunodeficiency virus enhanced by 3 -azido-3 -deoxythymidine, J. Acquir. Immune Defic. Syndr. Hum. Retrovirol, doi:10.1097/00042560-199605010-00004
Lanford, Chavez, Guerra, Lau, Hong et al., Ribavirin induces error-prone replication of GB virus B in primary tamarin hepatocytes, J. Virol, doi:10.1128/JVI.75.17.8074-8081.2001
Lerner, Wagaman, Phillips, Prospero-García, Henriksen et al., Increased mutation frequency of feline immunodeficiency virus lacking a functional deoxyuridine-triphosphatase, Proc. Natl. Acad. Sci, doi:10.1073/pnas.92.16.7480
Li, Fedeles, Singh, Peng, Silvestre et al., Tautomerism provides a molecular explanation for the mutagenic properties of the anti-HIV nucleoside 5-aza-5,6-dihydro-2 -deoxycytidine, Proc. Natl. Acad. Sci, doi:10.1073/pnas.1405635111
Li, Wang, Lavrijsen, Lamers, De Vries et al., SARS-CoV-2 omicron variant is highly sensitive to molnupiravir, nirmatrelvir, and the combination, Cell Res, doi:10.1038/s41422-022-00618-w
Loeb, Essigmann, Kazazi, Zhang, Rose et al., Lethal mutagenesis of HIV with mutagenic nucleoside analogs, Proc. Natl. Acad. Sci, doi:10.1073/pnas.96.4.1492
Lu, Gong, A structural view of the RNA-dependent RNA polymerases from the Flavivirus genus, Virus Res, doi:10.1016/j.virusres.2017.01.020
Lutchman, Danehower, Song, Liang, Hoofnagle et al., Mutation rate of the hepatitis C virus NS5B in patients undergoing treatment with ribavirin monotherapy, Gastroenterology, doi:10.1053/j.gastro.2007.03.035
Ma, Frutos-Beltrán, Kang, Pannecouque, De Clercq et al., Medicinal chemistry strategies for discovering antivirals effective against drug-resistant viruses, Chem. Soc. Rev, doi:10.1039/D0CS01084G
Mangeat, Turelli, Caron, Friedli, Perrin et al., Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts, Nature, doi:10.1038/nature01709
Mansky, Bernard, 3 -Azido-3 -deoxythymidine (AZT) and AZT-resistant reverse transcriptase can increase the in vivo mutation rate of human immunodeficiency virus type 1, J. Virol, doi:10.1128/JVI.74.20.9532-9539.2000
Mansky, Le Rouzic, Benichou, Gajary, Influence of reverse transcriptase variants, drugs, and Vpr on human immunodeficiency virus type 1 mutant frequencies, J. Virol, doi:10.1128/JVI.77.3.2071-2080.2003
Mansky, The mutation rate of human immunodeficiency virus type 1 is influenced by the vpr gene, Virology, doi:10.1006/viro.1996.0436
Martín-Hernández, Domingo, Menéndez-Arias, Human immunodeficiency virus type 1 reverse transcriptase: Role of Tyr115 in deoxynucleotide binding and misinsertion fidelity of DNA synthesis, EMBO J, doi:10.1002/j.1460-2075.1996.tb00816.x
Masters, The molecular biology of coronaviruses, Adv. Virus Res, doi:10.1016/S0065-3527(06)66005-3
Mcdaniel, Patterson, Mansky, Distinct dual antiviral mechanism that enhances hepatitis B virus mutagenesis and reduces viral DNA synthesis, Antiviral. Res, doi:10.1016/j.antiviral.2019.104540
Mejer, Fahnøe, Galli, Ramirez, Weiland et al., Mutations identified in the hepatitis C virus (HCV) polymerase of patients with chronic HCV treated with ribavirin cause resistance and affect viral replication fidelity, Antimicrob. Agents Chemother, doi:10.1128/AAC.01417-20
Mejer, Galli, Ramirez, Fahnøe, Benfield et al., Ribavirin inhibition of cell-culture infectious hepatitis C genotype 1-3 viruses is strain-dependent, Virology, doi:10.1016/j.virol.2019.09.014
Menéndez-Arias, Decoding molnupiravir-induced mutagenesis in SARS-CoV-2, J. Biol. Chem, doi:10.1016/j.jbc.2021.100867
Menéndez-Arias, Delgado, Update and latest advances in antiretroviral therapy, Trends Pharmacol. Sci, doi:10.1016/j.tips.2021.10.004
Menéndez-Arias, Molecular basis of fidelity of DNA synthesis and nucleotide specificity of retroviral reverse transcriptases, Prog. Nucleic Acid Res. Mol. Biol, doi:10.1016/s0079-6603(02)71042-8
Menéndez-Arias, Mutation rates and intrinsic fidelity of retroviral reverse transcriptases, CrossRef, doi:10.3390/v1031137
Menéndez-Arias, Sebastián-Martín, Álvarez, Viral reverse transcriptases, Virus Res, doi:10.1016/j.virusres.2016.12.019
Menéndez-Arias, Targeting HIV: Antiretroviral therapy and development of drug resistance, Trends Pharmacol. Sci, doi:10.1016/S0165-6147(02)02054-0
Moreno, Tejero, De La Torre, Domingo, Martín, Mutagenesis-mediated virus extinction: Virus-dependent effect of viral load on sensitivity to lethal defection, PLoS ONE, doi:10.1371/journal.pone.0032550
Mullins, Heath, Hughes, Kicha, Styrchak et al., Mutation of HIV-1 genomes in a clinical population treated with the mutagenic nucleoside KP1461, PLoS ONE
Musiałek, Rybaczek, Hydroxyurea-The good, the bad and the ugly, Genes, doi:10.3390/genes12071096
Naydenova, Muir, Wu, Zhang, Coscia et al., Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP, Proc. Natl. Acad. Sci, doi:10.1073/pnas.2021946118
O'neil, Sun, Yu, Ron, Dougherty et al., Mutational analysis of HIV-1 long terminal repeats to explore the relative contribution of reverse transcriptase and RNA polymerase II to viral mutagenesis, J. Biol. Chem, doi:10.1074/jbc.M204774200
Ortega-Prieto, Sheldon, Grande-Pérez, Tejero, Gregori et al., Extinction of hepatitis C virus by ribavirin in hepatoma cells involves lethal mutagenesis, PLoS ONE, doi:10.1371/journal.pone.0071039
Padhi, Dandapat, Saudagar, Uversky, Tripathi, Interface-based design of the favipiravir-binding site in SARS-CoV-2 RNA-dependent RNA polymerase reveals mutations conferring resistance to chain termination, FEBS Lett, doi:10.1002/1873-3468.14182
Paeshuyse, Dallmeier, Neyts, Ribavirin for the treatment of chronic hepatitis C virus infection: A review of the proposed mechanisms of action, Curr. Opin. Virol, doi:10.1016/j.coviro.2011.10.030
Painter, Natchus, Cohen, Holman, Painter, Developing a direct acting, orally available antiviral agent in a pandemic: The evolution of molnupiravir as a potential treatment for COVID-19, Curr. Opin. Virol, doi:10.1016/j.coviro.2021.06.003
Pariente, Airaksinen, Domingo, Mutagenesis versus inhibition in the efficiency of extinction of foot-and-mouth disease virus, J. Virol, doi:10.1128/JVI.77.12.7131-7138.2003
Pathak, Temin, 5-Azacytidine and RNA secondary structure increase the retrovirus mutation rate, J. Virol, doi:10.1128/jvi.66.5.3093-3100.1992
Pauly, Lauring, Effective lethal mutagenesis of influenza virus by three nucleoside analogs, J. Virol
Peck, Lauring, Complexities of viral mutation rates, J. Virol, doi:10.1128/JVI.01031-17
Peersen, Picornaviral polymerase structure, function, and fidelity modulation, Virus Res, doi:10.1016/j.virusres.2017.01.026
Perales, Domingo, Antiviral strategies based on lethal mutagenesis and error threshold, Curr. Top. Microbiol. Immunol, doi:10.1007/82_2015_459
Pfeiffer, Kirkegaard, A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity, Proc. Natl. Acad. Sci, doi:10.1073/pnas.1232294100
Qiu, Patterson, Bonnac, Geraghty, Nucleobases and corresponding nucleosides display potent antiviral activities against dengue virus possibly through viral lethal mutagenesis, PLoS Negl. Trop. Dis, doi:10.1371/journal.pntd.0006421
Ramírez-Olivencia, Estébanez, Membrillo, Ybarra, Uso de ribavirina en virus distintos de la hepatitis C. una revisión de la evidencia, Enferm. Infecc. Microbiol. Clin. (Engl, doi:10.1016/j.eimc.2018.05.008
Rawson, Daly, Xie, Clouser, Landman et al., 5-Azacytidine enhances the mutagenesis of HIV-1 by reduction to 5-aza-2 -deoxycytidine, Antimicrob. Agents Chemother, doi:10.1128/AAC.03084-15
Rawson, Heineman, Beach, Martin, Schnettler et al., 5,6-Dihydro-5-aza-2 -deoxycytidine potentiates the anti-HIV-1 activity of ribonucleotide reductase inhibitors, Bioorg. Med. Chem, doi:10.1016/j.bmc.2013.08.023
Rawson, Landman, Reilly, Bonnac, Patterson et al., Lack of mutational hot spots during decitabine-mediated HIV-1 mutagenesis, Antimicrob. Agents Chemother, doi:10.1128/AAC.01644-15
Rawson, Roth, Xie, Daly, Clouser et al., Synergistic reduction of HIV-1 infectivity by 5-azacytidine and inhibitors of ribonucleotide reductase, Bioorg. Med. Chem, doi:10.1016/j.bmc.2016.03.052
Robson, Khan, Le, Paris, Demirbag et al., Coronavirus RNA proofreading: Molecular basis and therapeutic targeting, Mol. Cell, doi:10.1016/j.molcel.2020.07.027
Rosenke, Hansen, Schwarz, Feldmann, Haddock et al., Orally delivered MK-4482 inhibits SARS-CoV-2 replication in the Syrian hamster model, Nat. Commun, doi:10.1038/s41467-021-22580-8
Roth, Mcdaniel, Daly, Talledge, Greggs et al., Distinct antiretroviral mechanisms elicited by a viral mutagen, J. Mol. Biol, doi:10.1016/j.jmb.2021.167111
Ruiz-Jarabo, Ly, Domingo, De La Torre, Lethal mutagenesis of the prototypic arenavirus lymphocytic choriomeningitis virus (LCMV), Virology, doi:10.1016/S0042-6822(02)00046-6
Sangawa, Komeno, Nishikawa, Yoshida, Takahashi et al., Mechanism of action of T-705 ribosyl triphosphate against influenza virus RNA polymerase, Antimicrob. Agents Chemother, doi:10.1128/AAC.00649-13
Sanjuán, Domingo-Calap, Mechanisms of viral mutation, Cell. Mol. Life Sci, doi:10.1007/s00018-016-2299-6
Sanjuán, Nebot, Chirico, Mansky, Belshaw, Viral mutation rates, J. Virol, doi:10.1128/JVI.00694-10
Schultz, Johnson, Ayyanathan, Miller, Whig et al., Pyrimidine inhibitors synergize with nucleoside analogues to block SARS-CoV-2, Nature
Seier, Zilberberg, Zeiger, Lovett, Azidothymidine and other chain terminators are mutagenic for template-switchgenerated genetic mutations, Proc. Natl. Acad. Sci, doi:10.1073/pnas.1116160109
Severson, Schmaljohn, Javadian, Jonsson, Ribavirin causes error catastrophe during Hantaan virus replication, J. Virol, doi:10.1128/JVI.77.1.481-488.2003
Shah, Curr, Hamburgh, Parniak, Mitsuya et al., Differential influence of nucleoside analog-resistance mutations K65R and L74V on the overall mutation rate and error specificity of human immunodeficiency virus type 1 reverse transcriptase, J. Biol. Chem, doi:10.1016/S0021-9258(19)61477-8
Shannon, Selisko, Le, Huchting, Touret et al., Rapid incorporation of favipiravir by the fast and permissive viral RNA polymerase complex results in SARS-CoV-2 lethal mutagenesis, Nat. Commun, doi:10.1038/s41467-020-18463-z
Sheahan, Sims, Zhou, Graham, Pruijssers et al., None
Sheehy, Gaddis, Choi, Malim, Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein, Nature, doi:10.1038/nature00939
Sidwell, Huffman, Khare Lois, Allen, Witkowski Roland et al., Broad-spectrum antiviral activity of virazole: 1-β-D-ribofuranosyl-1,2,4-triazole-3-carboxamide, Science, doi:10.1126/science.177.4050.705
Sierra, Airaksinen, González-López, Agudo, Arias et al., Foot-and-mouth disease virus mutant with decreased sensitivity to ribavirin: Implications for error catastrophe, J. Virol, doi:10.1128/JVI.01606-06
Stevens, An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice, Sci. Transl. Med, doi:10.1126/scitranslmed.abb5883
Sánchez-Jiménez, Olivares, De Ávila Lucas, Toledano, Gutiérrez-Rivas et al., Mutagen-mediated enhancement of HIV-1 replication in persistently infected cells, Virology, doi:10.1016/j.virol.2011.12.016
Takaori-Kondo, Shindo, HIV-1 Vif: A guardian of the virus that opens up a new era in the research field of restriction factors, Front Microbiol, doi:10.3389/fmicb.2013.00034
Tapia, Fernàndez, Parera, Gómez-Mariano, Clotet et al., Combination of a mutagenic agent with a reverse transcriptase inhibitor results in systematic inhibition of HIV-1 infection, Virology, doi:10.1016/j.virol.2005.05.008
Todt, Walter, Brown, Steinmann, Mutagenic effects of ribavirin on hepatitis E virus-Viral extinction versus selection of fitness-enhancing mutations, Viruses, doi:10.3390/v8100283
Toots, Yoon, Cox, Hart, Sticher et al., Characterization of orally efficacious influenza drug with high resistance barrier in ferrets and human airway epithelia, Sci. Transl. Med, doi:10.1126/scitranslmed.aax5866
Urakova, Kuznetsova, Crossman, Sokratian, Guthrie et al., β-D-N 4 -hydroxycytidine is a potent anti-alphavirus compound that induces a high level of mutations in the viral genome, J. Virol, doi:10.1128/JVI.01965-17
Vangeel, Chiu, De Jonghe, Maes, Slechten et al., molnupiravir and nirmatrelvir remain active against SARS-CoV-2 omicron and other variants of concern, Antiviral. Res, doi:10.1016/j.antiviral.2022.105252
Vignuzzi, Stone, Andino, Ribavirin and lethal mutagenesis of poliovirus: Molecular mechanisms, resistance and biological implications, Virus Res, doi:10.1016/j.virusres.2004.11.007
Vivet-Boudou, Isel, El Safadi, Smyth, Laumond et al., Evaluation of anti-HIV-1 mutagenic nucleoside analogues, J. Biol. Chem, doi:10.1074/jbc.M114.616383
Wang, Li, Yuan, Gao, Lan et al., In vitro assessment of combinations of enterovirus inhibitors against enterovirus 71, Antimicrob. Agents Chemother, doi:10.1128/AAC.01073-16
Waters, Warren, Hughes, Lewis, Zhang, Human genetic risk of treatment with antiviral nucleoside analog drugs that induce lethal mutagenesis: The special case of molnupiravir, Environ. Mol. Mutagen, doi:10.1002/em.22471
Weissmann, Billeter, Goodman, Hindley, Weber, Structure and function of phage RNA, Annu. Rev. Biochem, doi:10.1146/annurev.bi.42.070173.001511
Young, Lindsay, Lee, Liu, He et al., Identification of a ribavirin-resistant NS5B mutation of hepatitis C virus during ribavirin monotherapy, Hepatology, doi:10.1002/hep.1840380413
Zhao, Guo, Yi, Li, Ma et al., A cell-based assay to discover inhibitors of SARS-CoV-2 RNA dependent RNA polymerase, Antiviral. Res, doi:10.1016/j.antiviral.2021.105078
Zhou, Hill, Sarkar, Tse, Woodburn et al., β-D-N 4 -hydroxycytidine inhibits SARS-CoV-2 through lethal mutagenesis but is also mutagenic to mammalian cells, J. Infect. Dis, doi:10.1093/infdis/jiab247
Zhou, Liu, Baroudy, Malcolm, Reyes, The effect of ribavirin and IMPDH inhibitors on hepatitis C virus subgenomic replicon RNA, Virology, doi:10.1016/S0042-6822(03)00152-1
Álvarez, Sebastián-Martín, García-Marquina, Menéndez-Arias, Fidelity of classwide-resistant HIV-2 reverse transcriptase and differential contribution of K65R to the accuracy of HIV-1 and HIV-2 reverse transcriptases, Sci. Rep, doi:10.1038/srep44834
DOI record:
{
"DOI": "10.3390/v14040841",
"ISSN": [
"1999-4915"
],
"URL": "http://dx.doi.org/10.3390/v14040841",
"abstract": "<jats:p>In RNA viruses, a small increase in their mutation rates can be sufficient to exceed their threshold of viability. Lethal mutagenesis is a therapeutic strategy based on the use of mutagens, driving viral populations to extinction. Extinction catastrophe can be experimentally induced by promutagenic nucleosides in cell culture models. The loss of HIV infectivity has been observed after passage in 5-hydroxydeoxycytidine or 5,6-dihydro-5-aza-2′-deoxycytidine while producing a two-fold increase in the viral mutation frequency. Among approved nucleoside analogs, experiments with polioviruses and other RNA viruses suggested that ribavirin can be mutagenic, although its mechanism of action is not clear. Favipiravir and molnupiravir exert an antiviral effect through lethal mutagenesis. Both drugs are broad-spectrum antiviral agents active against RNA viruses. Favipiravir incorporates into viral RNA, affecting the G→A and C→U transition rates. Molnupiravir (a prodrug of β-d-N4-hydroxycytidine) has been recently approved for the treatment of SARS-CoV-2 infection. Its triphosphate derivative can be incorporated into viral RNA and extended by the coronavirus RNA polymerase. Incorrect base pairing and inefficient extension by the polymerase promote mutagenesis by increasing the G→A and C→U transition frequencies. Despite having remarkable antiviral action and resilience to drug resistance, carcinogenic risks and genotoxicity are important concerns limiting their extended use in antiviral therapy.</jats:p>",
"alternative-id": [
"v14040841"
],
"author": [
{
"ORCID": "http://orcid.org/0000-0003-1992-7976",
"affiliation": [],
"authenticated-orcid": false,
"family": "Hadj Hassine",
"given": "Ikbel",
"sequence": "first"
},
{
"ORCID": "http://orcid.org/0000-0003-0145-7240",
"affiliation": [],
"authenticated-orcid": false,
"family": "Ben M’hadheb",
"given": "Manel",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-1251-6640",
"affiliation": [],
"authenticated-orcid": false,
"family": "Menéndez-Arias",
"given": "Luis",
"sequence": "additional"
}
],
"container-title": "Viruses",
"container-title-short": "Viruses",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2022,
4,
20
]
],
"date-time": "2022-04-20T06:04:22Z",
"timestamp": 1650434662000
},
"deposited": {
"date-parts": [
[
2022,
4,
20
]
],
"date-time": "2022-04-20T06:12:44Z",
"timestamp": 1650435164000
},
"funder": [
{
"award": [
"PID2019-104176RB-I00/AEI/10.13039/501100011033"
],
"name": "Ministerio de Ciencia e Innovación"
}
],
"indexed": {
"date-parts": [
[
2022,
4,
21
]
],
"date-time": "2022-04-21T21:49:24Z",
"timestamp": 1650577764252
},
"is-referenced-by-count": 0,
"issue": "4",
"issued": {
"date-parts": [
[
2022,
4,
18
]
]
},
"journal-issue": {
"issue": "4",
"published-online": {
"date-parts": [
[
2022,
4
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2022,
4,
18
]
],
"date-time": "2022-04-18T00:00:00Z",
"timestamp": 1650240000000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/1999-4915/14/4/841/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "841",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2022,
4,
18
]
]
},
"published-online": {
"date-parts": [
[
2022,
4,
18
]
]
},
"publisher": "MDPI AG",
"reference": [
{
"DOI": "10.1128/JVI.00694-10",
"doi-asserted-by": "publisher",
"key": "ref1"
},
{
"DOI": "10.1146/annurev-virology-091919-105900",
"doi-asserted-by": "publisher",
"key": "ref2"
},
{
"DOI": "10.1073/pnas.212514799",
"doi-asserted-by": "publisher",
"key": "ref3"
},
{
"author": "Eigen",
"key": "ref4",
"series-title": "From Strange Simplicity to Complex Familiarity: A Treatise on Matter, Information, Life and Thought",
"year": "2013"
},
{
"DOI": "10.1146/annurev.bi.42.070173.001511",
"doi-asserted-by": "publisher",
"key": "ref5"
},
{
"DOI": "10.1073/pnas.54.4.1189",
"doi-asserted-by": "publisher",
"key": "ref6"
},
{
"DOI": "10.1073/pnas.54.2.579",
"doi-asserted-by": "publisher",
"key": "ref7"
},
{
"DOI": "10.1093/genetics/148.4.1667",
"doi-asserted-by": "publisher",
"key": "ref8"
},
{
"DOI": "10.1073/pnas.96.24.13910",
"doi-asserted-by": "publisher",
"key": "ref9"
},
{
"DOI": "10.1007/82_2015_459",
"doi-asserted-by": "publisher",
"key": "ref10"
},
{
"DOI": "10.1128/jvi.64.8.3960-3962.1990",
"doi-asserted-by": "publisher",
"key": "ref11"
},
{
"DOI": "10.1146/annurev.micro.58.030603.123649",
"doi-asserted-by": "publisher",
"key": "ref12"
},
{
"DOI": "10.1073/pnas.96.4.1492",
"doi-asserted-by": "publisher",
"key": "ref13"
},
{
"DOI": "10.1007/s00018-016-2299-6",
"doi-asserted-by": "publisher",
"key": "ref14"
},
{
"DOI": "10.1371/journal.ppat.1003855",
"doi-asserted-by": "publisher",
"key": "ref15"
},
{
"DOI": "10.1128/JVI.01031-17",
"doi-asserted-by": "publisher",
"key": "ref16"
},
{
"DOI": "10.1126/science.7824947",
"doi-asserted-by": "publisher",
"key": "ref17"
},
{
"DOI": "10.1016/j.virusres.2016.12.019",
"doi-asserted-by": "publisher",
"key": "ref18"
},
{
"DOI": "10.3390/v1031137",
"doi-asserted-by": "publisher",
"key": "ref19"
},
{
"DOI": "10.1074/jbc.M204774200",
"doi-asserted-by": "publisher",
"key": "ref20"
},
{
"DOI": "10.1016/s0079-6603(02)71042-8",
"doi-asserted-by": "publisher",
"key": "ref21"
},
{
"DOI": "10.1128/jvi.66.3.1791-1794.1992",
"doi-asserted-by": "publisher",
"key": "ref22"
},
{
"DOI": "10.1128/jvi.68.4.2313-2319.1994",
"doi-asserted-by": "publisher",
"key": "ref23"
},
{
"DOI": "10.1073/pnas.92.16.7480",
"doi-asserted-by": "publisher",
"key": "ref24"
},
{
"DOI": "10.1006/viro.1996.0436",
"doi-asserted-by": "publisher",
"key": "ref25"
},
{
"DOI": "10.1038/nature00939",
"doi-asserted-by": "publisher",
"key": "ref26"
},
{
"DOI": "10.1038/nature01709",
"doi-asserted-by": "publisher",
"key": "ref27"
},
{
"DOI": "10.3389/fmicb.2013.00034",
"doi-asserted-by": "publisher",
"key": "ref28"
},
{
"DOI": "10.1016/S0021-9258(19)61477-8",
"doi-asserted-by": "publisher",
"key": "ref29"
},
{
"DOI": "10.1042/BJ20101852",
"doi-asserted-by": "publisher",
"key": "ref30"
},
{
"DOI": "10.1002/j.1460-2075.1996.tb00816.x",
"doi-asserted-by": "publisher",
"key": "ref31"
},
{
"DOI": "10.1074/jbc.M910361199",
"doi-asserted-by": "publisher",
"key": "ref32"
},
{
"DOI": "10.1128/JVI.77.3.2071-2080.2003",
"doi-asserted-by": "publisher",
"key": "ref33"
},
{
"DOI": "10.1016/j.virusres.2017.01.026",
"doi-asserted-by": "publisher",
"key": "ref34"
},
{
"DOI": "10.1016/j.virusres.2017.01.020",
"doi-asserted-by": "publisher",
"key": "ref35"
},
{
"DOI": "10.1016/j.coviro.2021.03.010",
"doi-asserted-by": "publisher",
"key": "ref36"
},
{
"DOI": "10.1016/j.molcel.2020.07.027",
"doi-asserted-by": "publisher",
"key": "ref37"
},
{
"DOI": "10.1016/S0065-3527(06)66005-3",
"doi-asserted-by": "publisher",
"key": "ref38"
},
{
"DOI": "10.1128/JVI.01296-07",
"doi-asserted-by": "publisher",
"key": "ref39"
},
{
"DOI": "10.1371/journal.ppat.1000896",
"doi-asserted-by": "publisher",
"key": "ref40"
},
{
"DOI": "10.1128/jvi.66.5.3093-3100.1992",
"doi-asserted-by": "publisher",
"key": "ref41"
},
{
"DOI": "10.1097/00042560-199605010-00004",
"doi-asserted-by": "publisher",
"key": "ref42"
},
{
"DOI": "10.1128/jvi.71.6.4254-4263.1997",
"doi-asserted-by": "publisher",
"key": "ref43"
},
{
"DOI": "10.1128/JVI.74.20.9532-9539.2000",
"doi-asserted-by": "publisher",
"key": "ref44"
},
{
"DOI": "10.1128/JVI.01406-09",
"doi-asserted-by": "publisher",
"key": "ref45"
},
{
"DOI": "10.1016/j.jmb.2021.167111",
"doi-asserted-by": "publisher",
"key": "ref46"
},
{
"DOI": "10.1128/AAC.03084-15",
"doi-asserted-by": "publisher",
"key": "ref47"
},
{
"DOI": "10.1016/j.virol.2011.12.016",
"doi-asserted-by": "publisher",
"key": "ref48"
},
{
"DOI": "10.1074/jbc.M114.616383",
"doi-asserted-by": "publisher",
"key": "ref49"
},
{
"DOI": "10.1128/AAC.01644-15",
"doi-asserted-by": "publisher",
"key": "ref50"
},
{
"DOI": "10.1016/S0165-6147(02)02054-0",
"doi-asserted-by": "publisher",
"key": "ref51"
},
{
"DOI": "10.1016/j.tips.2021.10.004",
"doi-asserted-by": "publisher",
"key": "ref52"
},
{
"DOI": "10.1016/j.virol.2005.05.008",
"doi-asserted-by": "publisher",
"key": "ref53"
},
{
"DOI": "10.1073/pnas.1116160109",
"doi-asserted-by": "publisher",
"key": "ref54"
},
{
"DOI": "10.1016/j.antiviral.2005.03.004",
"doi-asserted-by": "publisher",
"key": "ref55"
},
{
"DOI": "10.1073/pnas.1405635111",
"doi-asserted-by": "publisher",
"key": "ref56"
},
{
"DOI": "10.1371/journal.pone.0015135",
"doi-asserted-by": "publisher",
"key": "ref57"
},
{
"DOI": "10.1016/S0014-5793(97)01572-X",
"doi-asserted-by": "publisher",
"key": "ref58"
},
{
"DOI": "10.1016/j.bmc.2013.08.023",
"doi-asserted-by": "publisher",
"key": "ref59"
},
{
"DOI": "10.1016/j.semcdb.2014.03.030",
"doi-asserted-by": "publisher",
"key": "ref60"
},
{
"DOI": "10.3390/genes12071096",
"doi-asserted-by": "publisher",
"key": "ref61"
},
{
"DOI": "10.1128/JVI.01006-10",
"doi-asserted-by": "publisher",
"key": "ref62"
},
{
"DOI": "10.1016/j.bmc.2016.03.052",
"doi-asserted-by": "publisher",
"key": "ref63"
},
{
"DOI": "10.1099/vir.0.069864-0",
"doi-asserted-by": "publisher",
"key": "ref64"
},
{
"DOI": "10.1038/srep44834",
"doi-asserted-by": "publisher",
"key": "ref65"
},
{
"DOI": "10.1016/j.antiviral.2019.104540",
"doi-asserted-by": "publisher",
"key": "ref66"
},
{
"DOI": "10.1126/science.7041255",
"doi-asserted-by": "publisher",
"key": "ref67"
},
{
"DOI": "10.1093/infdis/jiab247",
"doi-asserted-by": "publisher",
"key": "ref68"
},
{
"DOI": "10.1038/82191",
"doi-asserted-by": "publisher",
"key": "ref69"
},
{
"DOI": "10.1021/jm060872x",
"doi-asserted-by": "publisher",
"key": "ref70"
},
{
"DOI": "10.1128/JVI.01028-07",
"doi-asserted-by": "publisher",
"key": "ref71"
},
{
"DOI": "10.1128/AAC.01056-07",
"doi-asserted-by": "publisher",
"key": "ref72"
},
{
"DOI": "10.1128/JVI.05712-11",
"doi-asserted-by": "publisher",
"key": "ref73"
},
{
"DOI": "10.1371/journal.pone.0032550",
"doi-asserted-by": "publisher",
"key": "ref74"
},
{
"DOI": "10.1016/S0042-6822(03)00144-2",
"doi-asserted-by": "publisher",
"key": "ref75"
},
{
"DOI": "10.1128/JVI.77.12.7131-7138.2003",
"doi-asserted-by": "publisher",
"key": "ref76"
},
{
"DOI": "10.1128/JVI.01606-06",
"doi-asserted-by": "publisher",
"key": "ref77"
},
{
"DOI": "10.1371/journal.ppat.1001072",
"doi-asserted-by": "publisher",
"key": "ref78"
},
{
"DOI": "10.1016/j.virusres.2017.03.014",
"doi-asserted-by": "publisher",
"key": "ref79"
},
{
"DOI": "10.7554/eLife.03679",
"doi-asserted-by": "publisher",
"key": "ref80"
},
{
"DOI": "10.1371/journal.pntd.0006421",
"doi-asserted-by": "publisher",
"key": "ref81"
},
{
"DOI": "10.1128/AAC.00380-18",
"doi-asserted-by": "publisher",
"key": "ref82"
},
{
"DOI": "10.1016/j.antiviral.2005.04.002",
"doi-asserted-by": "publisher",
"key": "ref83"
},
{
"DOI": "10.1128/AAC.01400-17",
"doi-asserted-by": "publisher",
"key": "ref84"
},
{
"DOI": "10.1128/JVI.75.17.8074-8081.2001",
"doi-asserted-by": "publisher",
"key": "ref85"
},
{
"DOI": "10.1128/JVI.76.17.8505-8517.2002",
"doi-asserted-by": "publisher",
"key": "ref86"
},
{
"DOI": "10.1016/S0042-6822(03)00152-1",
"doi-asserted-by": "publisher",
"key": "ref87"
},
{
"DOI": "10.1128/JVI.00201-09",
"doi-asserted-by": "publisher",
"key": "ref88"
},
{
"DOI": "10.1371/journal.pone.0071039",
"doi-asserted-by": "publisher",
"key": "ref89"
},
{
"DOI": "10.1128/JVI.02778-12",
"doi-asserted-by": "publisher",
"key": "ref90"
},
{
"DOI": "10.1128/AAC.01653-19",
"doi-asserted-by": "publisher",
"key": "ref91"
},
{
"DOI": "10.1371/journal.pone.0164691",
"doi-asserted-by": "publisher",
"key": "ref92"
},
{
"DOI": "10.3390/v8100283",
"doi-asserted-by": "publisher",
"key": "ref93"
},
{
"DOI": "10.1128/JVI.01965-17",
"doi-asserted-by": "publisher",
"key": "ref94"
},
{
"DOI": "10.1038/s41467-020-18463-z",
"doi-asserted-by": "publisher",
"key": "ref95"
},
{
"DOI": "10.1128/JVI.01348-19",
"doi-asserted-by": "publisher",
"key": "ref96"
},
{
"DOI": "10.1038/s41598-018-19829-6",
"doi-asserted-by": "publisher",
"key": "ref97"
},
{
"DOI": "10.1128/JVI.03483-14",
"doi-asserted-by": "publisher",
"key": "ref98"
},
{
"DOI": "10.1126/scitranslmed.aax5866",
"doi-asserted-by": "publisher",
"key": "ref99"
},
{
"DOI": "10.1073/pnas.0408871102",
"doi-asserted-by": "publisher",
"key": "ref100"
},
{
"DOI": "10.1128/JVI.77.1.481-488.2003",
"doi-asserted-by": "publisher",
"key": "ref101"
},
{
"DOI": "10.1128/JVI.00874-07",
"doi-asserted-by": "publisher",
"key": "ref102"
},
{
"DOI": "10.1128/AAC.00669-19",
"doi-asserted-by": "publisher",
"key": "ref103"
},
{
"DOI": "10.1016/S0042-6822(02)00046-6",
"doi-asserted-by": "publisher",
"key": "ref104"
},
{
"DOI": "10.1016/j.antiviral.2019.06.001",
"doi-asserted-by": "publisher",
"key": "ref105"
},
{
"DOI": "10.1016/j.virusres.2004.11.007",
"doi-asserted-by": "publisher",
"key": "ref106"
},
{
"DOI": "10.1016/j.eimc.2018.05.008",
"doi-asserted-by": "publisher",
"key": "ref107"
},
{
"DOI": "10.1002/rmv.483",
"doi-asserted-by": "publisher",
"key": "ref108"
},
{
"DOI": "10.1016/j.coviro.2011.10.030",
"doi-asserted-by": "publisher",
"key": "ref109"
},
{
"DOI": "10.1073/pnas.0406927102",
"doi-asserted-by": "publisher",
"key": "ref110"
},
{
"DOI": "10.1126/science.177.4050.705",
"doi-asserted-by": "publisher",
"key": "ref111"
},
{
"DOI": "10.3390/v13040667",
"doi-asserted-by": "publisher",
"key": "ref112"
},
{
"DOI": "10.1074/jbc.274.5.2706",
"doi-asserted-by": "publisher",
"key": "ref113"
},
{
"DOI": "10.1074/jbc.M002671200",
"doi-asserted-by": "publisher",
"key": "ref114"
},
{
"DOI": "10.1073/pnas.1232294100",
"doi-asserted-by": "publisher",
"key": "ref115"
},
{
"DOI": "10.1128/JVI.02420-09",
"doi-asserted-by": "publisher",
"key": "ref116"
},
{
"DOI": "10.1002/hep.1840380413",
"doi-asserted-by": "publisher",
"key": "ref117"
},
{
"DOI": "10.1053/j.gastro.2007.03.035",
"doi-asserted-by": "publisher",
"key": "ref118"
},
{
"DOI": "10.1016/j.virol.2019.09.014",
"doi-asserted-by": "publisher",
"key": "ref119"
},
{
"DOI": "10.1128/AAC.01417-20",
"doi-asserted-by": "publisher",
"key": "ref120"
},
{
"DOI": "10.1073/pnas.1007626107",
"doi-asserted-by": "publisher",
"key": "ref121"
},
{
"DOI": "10.1016/j.virol.2018.07.030",
"doi-asserted-by": "publisher",
"key": "ref122"
},
{
"DOI": "10.1016/j.antiviral.2018.03.003",
"doi-asserted-by": "publisher",
"key": "ref123"
},
{
"DOI": "10.1177/2040206618764483",
"doi-asserted-by": "publisher",
"key": "ref124"
},
{
"DOI": "10.1128/AAC.00649-13",
"doi-asserted-by": "publisher",
"key": "ref125"
},
{
"DOI": "10.1371/journal.pone.0068347",
"doi-asserted-by": "publisher",
"key": "ref126"
},
{
"DOI": "10.1128/JVI.02346-12",
"doi-asserted-by": "publisher",
"key": "ref127"
},
{
"DOI": "10.1073/pnas.1811345115",
"doi-asserted-by": "publisher",
"key": "ref128"
},
{
"DOI": "10.1371/journal.ppat.1008937",
"doi-asserted-by": "publisher",
"key": "ref129"
},
{
"DOI": "10.1093/jac/dku209",
"doi-asserted-by": "publisher",
"key": "ref130"
},
{
"DOI": "10.1128/AAC.01073-16",
"doi-asserted-by": "publisher",
"key": "ref131"
},
{
"DOI": "10.1073/pnas.2021946118",
"doi-asserted-by": "publisher",
"key": "ref132"
},
{
"DOI": "10.1002/1873-3468.14182",
"doi-asserted-by": "publisher",
"key": "ref133"
},
{
"DOI": "10.1016/j.antiviral.2021.105078",
"doi-asserted-by": "publisher",
"key": "ref134"
},
{
"DOI": "10.1126/scitranslmed.abb5883",
"doi-asserted-by": "publisher",
"key": "ref135"
},
{
"DOI": "10.1016/j.jbc.2021.100770",
"doi-asserted-by": "publisher",
"key": "ref136"
},
{
"DOI": "10.1038/s41594-021-00651-0",
"doi-asserted-by": "publisher",
"key": "ref137"
},
{
"DOI": "10.1039/D0CP05297C",
"doi-asserted-by": "publisher",
"key": "ref138"
},
{
"DOI": "10.1038/s41467-021-22580-8",
"doi-asserted-by": "publisher",
"key": "ref139"
},
{
"DOI": "10.1038/s41564-020-00835-2",
"doi-asserted-by": "publisher",
"key": "ref140"
},
{
"DOI": "10.1056/NEJMoa2116044",
"doi-asserted-by": "publisher",
"key": "ref141"
},
{
"DOI": "10.1126/scitranslmed.abl7430",
"doi-asserted-by": "publisher",
"key": "ref142"
},
{
"DOI": "10.1093/infdis/jiab361",
"doi-asserted-by": "publisher",
"key": "ref143"
},
{
"DOI": "10.1038/s41422-022-00618-w",
"doi-asserted-by": "publisher",
"key": "ref144"
},
{
"DOI": "10.1016/j.antiviral.2022.105252",
"doi-asserted-by": "publisher",
"key": "ref145"
},
{
"DOI": "10.1016/j.jbc.2021.100867",
"doi-asserted-by": "publisher",
"key": "ref146"
},
{
"DOI": "10.1016/j.ebiom.2021.103595",
"doi-asserted-by": "publisher",
"key": "ref147"
},
{
"DOI": "10.1038/s41586-022-04482-x",
"doi-asserted-by": "publisher",
"key": "ref148"
},
{
"DOI": "10.1016/j.coviro.2021.06.003",
"doi-asserted-by": "publisher",
"key": "ref149"
},
{
"DOI": "10.1002/em.22373",
"doi-asserted-by": "publisher",
"key": "ref150"
},
{
"DOI": "10.1002/em.22471",
"doi-asserted-by": "publisher",
"key": "ref151"
},
{
"DOI": "10.1039/D0CS01084G",
"doi-asserted-by": "publisher",
"key": "ref152"
}
],
"reference-count": 152,
"references-count": 152,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.mdpi.com/1999-4915/14/4/841"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"Virology",
"Infectious Diseases"
],
"subtitle": [],
"title": "Lethal Mutagenesis of RNA Viruses and Approved Drugs with Antiviral Mutagenic Activity",
"type": "journal-article",
"volume": "14"
}
hadjhassine