The Endolysosomal System: The Acid Test for SARS-CoV-2
et al., International Journal of Molecular Sciences, doi:10.3390/ijms23094576, Apr 2022
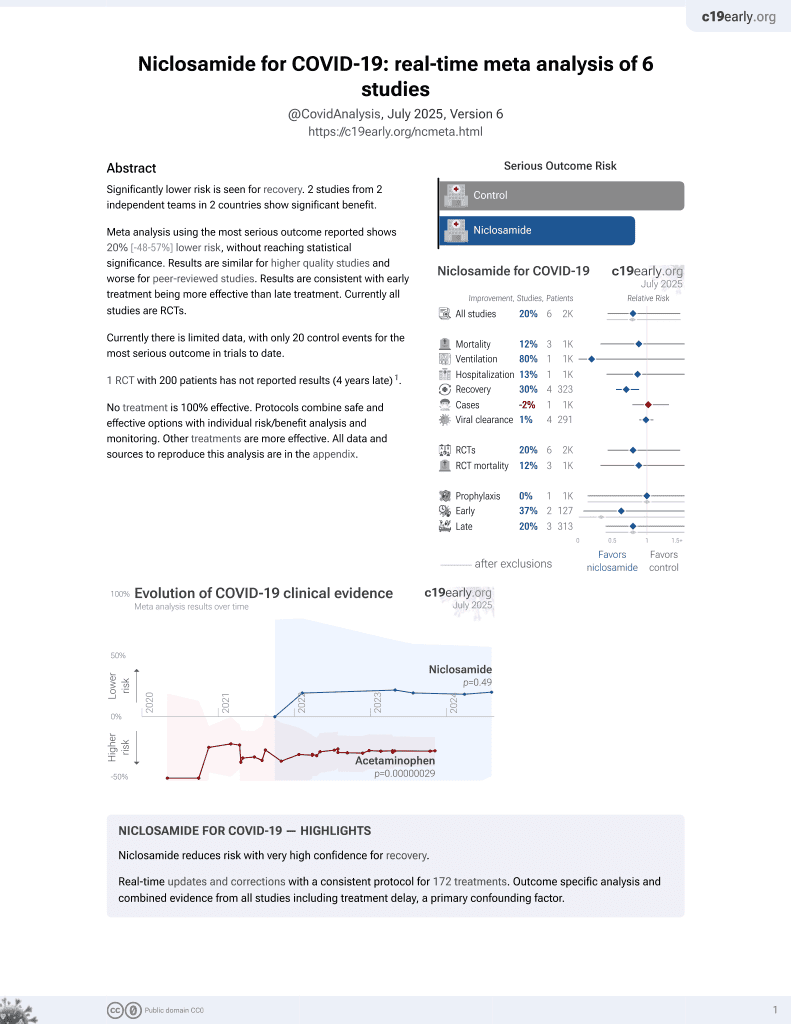
56th treatment shown to reduce risk in
August 2025, now with p = 0.0069 from 7 studies.
Lower risk for recovery.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
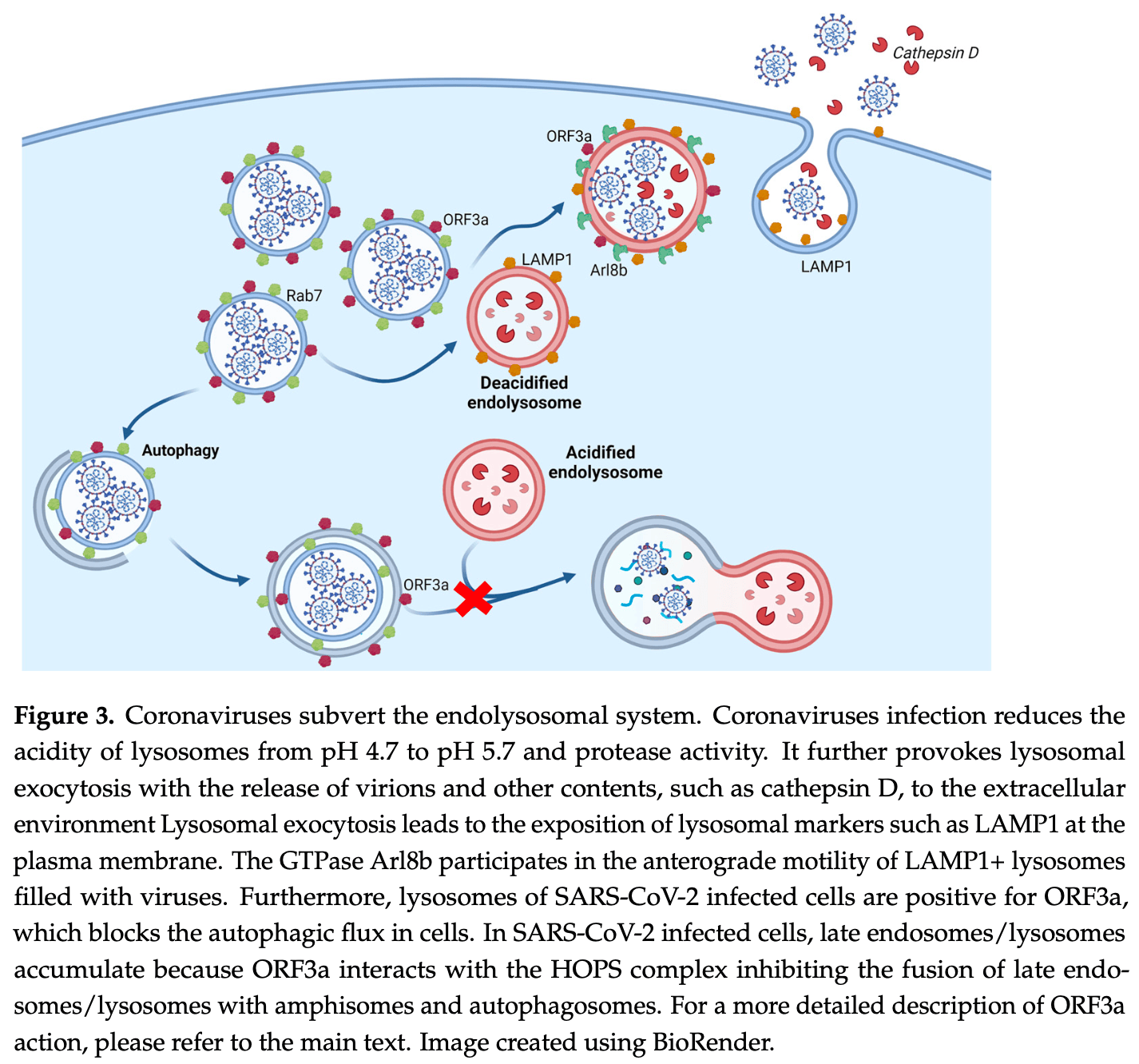
Review of the role of the endolysosomal system in SARS-CoV-2 infection. The endolysosomal system, which includes endosomes and lysosomes, is crucial for cellular homeostasis and host defense against pathogens. SARS-CoV-2 can enter cells by direct fusion with the plasma membrane or by endocytosis and subsequent fusion with endosomal membranes. The viral protein ORF3a plays a key role in subverting the endolysosomal system by blocking autophagy, inhibiting fusion of lysosomes with autophagosomes, and promoting lysosomal exocytosis for viral egress. ORF3a achieves this through interactions with host proteins like Vps39 and the HOPS complex. Genome-wide studies have identified multiple components of the endolysosomal system as potential therapeutic targets against COVID-19. Authors suggest that targeting the endolysosomal system could be a promising antiviral strategy, as it would not directly select for resistant viral variants.
1.
Mothae et al., SARS-CoV-2 host-pathogen interactome: insights into more players during pathogenesis, Virology, doi:10.1016/j.virol.2025.110607.
2.
Saha et al., Inhaled Dry Powder of Antiviral Agents: A Promising Approach to Treating Respiratory Viral Pathogens, Viruses, doi:10.3390/v17020252.
3.
Ali et al., SARS-CoV-2 Syncytium under the Radar: Molecular Insights of the Spike-Induced Syncytia and Potential Strategies to Limit SARS-CoV-2 Replication, Journal of Clinical Medicine, doi:10.3390/jcm12186079.
4.
Vibhute et al., Niclosamide: a potential treatment option for COVID-19, International Journal of Applied Pharmaceutics, doi:10.22159/ijap.2023v15i1.45850.
5.
Abed et al., Evaluation the plausibility of repurpose of levamisole and niclosamide in treatment of covid-19, Journal of Pharmaceutical Technolgy, doi:10.37662/jpt.2022.999481.
6.
Cesar-Silva et al., The Endolysosomal System: The Acid Test for SARS-CoV-2, International Journal of Molecular Sciences, doi:10.3390/ijms23094576.
Cesar-Silva et al., 20 Apr 2022, multiple countries, peer-reviewed, 4 authors.
Contact: calmeida@ioc.fiocruz.br (corresponding author), daniellasilva@aluno.fiocruz.br, filipe.dutra@ioc.fiocruz.br, ana.giannini@ufrj.br, calmeidaioc@gmail.com.
The Endolysosomal System: The Acid Test for SARS-CoV-2
International Journal of Molecular Sciences, doi:10.3390/ijms23094576
This review aims to describe and discuss the different functions of the endolysosomal system, from homeostasis to its vital role during viral infections. We will initially describe endolysosomal system's main functions, presenting recent data on how its compartments are essential for host defense to explore later how SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2) and other coronaviruses subvert these organelles for their benefit. It is clear that to succeed, pathogens' evolution favored the establishment of ways to avoid, escape, or manipulate lysosomal function. The unavoidable coexistence with such an unfriendly milieu imposed on viruses the establishment of a vast array of strategies to make the most out of the invaded cell's machinery to produce new viruses and maneuvers to escape the host's defense system.
Conflicts of Interest: The authors declare no conflict of interest.
References
Amraei, Yin, Napoleon, Suder, Berrigan et al., CD209L/L-SIGN and CD209/DC-SIGN Act as Receptors for SARS-CoV-2, ACS Cent. Sci, doi:10.1021/acscentsci.0c01537
Bach, Larance, James, Ramm, The Serine/Threonine Kinase ULK1 Is a Target of Multiple Phosphorylation Events, Biochem. J, doi:10.1042/BJ20101894
Backer, Sjöbring, Sonne, Weiss, Hostrup et al., A Randomized, Double-Blind, Placebo-Controlled Phase 1 Trial of Inhaled and Intranasal Niclosamide: A Broad Spectrum Antiviral Candidate for Treatment of COVID-19, Lancet Reg. Health Eur, doi:10.1016/j.lanepe.2021.100084
Balderhaar, Ungermann, CORVET and HOPS Tethering Complexes-Coordinators of Endosome and Lysosome Fusion, J. Cell Sci, doi:10.1242/jcs.107805
Bayati, Kumar, Francis, Mcpherson, SARS-CoV-2 Infects Cells after Viral Entry via Clathrin-Mediated Endocytosis, J. Biol. Chem, doi:10.1016/j.jbc.2021.100306
Boda, Lőrincz, Takáts, Csizmadia, Tóth et al., Drosophila Arl8 Is a General Positive Regulator of Lysosomal Fusion Events, Biochim. Biophys. Acta Mol. Cell Res, doi:10.1016/j.bbamcr.2018.12.011
Bollavaram, Leeman, Lee, Kulkarni, Upshaw et al., Multiple Sites on SARS-CoV-2 Spike Protein Are Susceptible to Proteolysis by Cathepsins B, Protein Sci, doi:10.1002/pro.4073
Bracquemond, Muriaux, Betacoronavirus Assembly: Clues and Perspectives for Elucidating SARS-CoV-2 Particle Formation and Egress, mBio, doi:10.1128/mBio.02371-21
Bright, Davis, Luzio, Endolysosomes Are the Principal Intracellular Sites of Acid Hydrolase Activity, Curr. Biol, doi:10.1016/j.cub.2016.06.046
Brunaugh, Seo, Warnken, Ding, Seo et al., Development and Evaluation of Inhalable Composite Niclosamide-Lysozyme Particles: A Broad-Spectrum, Patient-Adaptable Treatment for Coronavirus Infections and Sequalae, PLoS ONE, doi:10.1371/journal.pone.0246803
Buchrieser, Dufloo, Hubert, Monel, Planas et al., Syncytia Formation by SARS-CoV-2-infected Cells, EMBO J, doi:10.15252/embj.2020106267
Carlos, Ha, Yeh, Van Krieken, Tseng et al., The Chaperone GRP78 Is a Host Auxiliary Factor for SARS-CoV-2 and GRP78 Depleting Antibody Blocks Viral Entry and Infection, J. Biol. Chem, doi:10.1016/j.jbc.2021.100759
Carty, Guy, Bowie, Detection of Viral Infections by Innate Immunity, Biochem. Pharmacol, doi:10.1016/j.bcp.2020.114316
Castaño-Rodriguez, Honrubia, Gutiérrez-Álvarez, Dediego, Nieto-Torres et al., Role of Severe Acute Respiratory Syndrome Coronavirus Viroporins E, 3a, and 8a in Replication and Pathogenesis, MBio, doi:10.1128/mBio.02325-17
Catalano, O'driscoll, Inhibiting Extracellular Vesicles Formation and Release: A Review of EV Inhibitors, J. Extracell. Vesicles, doi:10.1080/20013078.2019.1703244
Chambers, Yu, Valdes, Arulanandam, SARS-CoV-2, Early Entry Events, J. Pathog, doi:10.1155/2020/9238696
Chaudhary, Gomez, Howes, Lo, Mcmahon et al., Endocytic Crosstalk: Cavins, Caveolins, and Caveolae Regulate Clathrin-Independent Endocytosis, PLoS Biol, doi:10.1371/journal.pbio.1001832
Chen, Yu, Autophagic Lysosome Reformation, Exp. Cell Res, doi:10.1016/j.yexcr.2012.09.004
Chen, Zheng, Sun, Ji, Li et al., ORF3a of SARS-CoV-2 Promotes Lysosomal Exocytosis-Mediated Viral Egress, Dev. Cell, doi:10.1016/j.devcel.2021.10.006
Cheng, Nichols, Caveolae: One Function or Many?, Trends Cell Biol, doi:10.1016/j.tcb.2015.10.010
Clausen, Sandoval, Spliid, Pihl, Perrett et al., SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2, Cell, doi:10.1016/j.cell.2020.09.033
Cohen, Lin, Machamer, Identification of a Golgi Complex-Targeting Signal in the Cytoplasmic Tail of the Severe Acute Respiratory Syndrome Coronavirus Envelope Protein, J. Virol, doi:10.1128/JVI.00060-11
Cottam, Whelband, Wileman, Coronavirus NSP6 Restricts Autophagosome Expansion, Autophagy, doi:10.4161/auto.29309
Da, Dias, Soares, Ferreira, Sacramento et al., Lipid Droplets Fuel SARS-CoV-2 Replication and Production of Inflammatory Mediators, PLoS Pathog, doi:10.1371/journal.ppat.1009127
Daly, Simonetti, Klein, Chen, Williamson et al., Neuropilin-1 Is a Host Factor for SARS-CoV-2 Infection, Science, doi:10.1126/science.abd3072
Damm, Pelkmans, Kartenbeck, Mezzacasa, Kurzchalia et al., Clathrin-and Caveolin-1-Independent Endocytosis: Entry of Simian Virus 40 into Cells Devoid of Caveolae, J. Cell Biol, doi:10.1083/jcb.200407113
Daniloski, Jordan, Wessels, Hoagland, Kasela et al., Identification of Required Host Factors for SARS-CoV-2 Infection in Human Cells, Cell, doi:10.1016/j.cell.2020.10.030
Datta, Kim, Lai, Mcgee, Johnson et al., Manumycin A Suppresses Exosome Biogenesis and Secretion via Targeted Inhibition of Ras/Raf/ERK1/2 Signaling and HnRNP H1 in Castration-Resistant Prostate Cancer Cells, Cancer Lett, doi:10.1016/j.canlet.2017.08.020
Datta, Miller, Halcrow, Khan, Colwell et al., SARS-CoV-2 S1 Protein Induces Endolysosome Dysfunction and Neuritic Dystrophy, Front. Cell. Neurosci, doi:10.3389/fncel.2021.777738
Dikic, Elazar, Mechanism and Medical Implications of Mammalian Autophagy, Nat. Rev. Mol. Cell Biol, doi:10.1038/s41580-018-0003-4
Dossou, Bosu, The Emerging Roles of MTORC1 in Macromanaging Autophagy, Cancers, doi:10.3390/cancers11101422
Ducatelle, Hoorens, Archives of Virology Significance of Lysosomes in the Morphogenesis of Coronaviruses, Arch. ViroIogy, doi:10.1007/BF01314299
Earnest, Hantak, Li, Mccray, Perlman et al., The Tetraspanin CD9 Facilitates MERS-Coronavirus Entry by Scaffolding Host Cell Receptors and Proteases, PLoS Pathog, doi:10.1371/journal.ppat.1006546
Egan, Chun, Vamos, Zou, Rong et al., Small Molecule Inhibition of the Autophagy Kinase ULK1 and Identification of ULK1 Substrates, Mol. Cell, doi:10.1016/j.molcel.2015.05.031
Ewers, Römer, Smith, Bacia, Dmitrieff et al., GM1 Structure Determines SV40-Induced Membrane Invagination and Infection, Nat. Cell Biol, doi:10.1038/ncb1999
Gagescu, Demaurex, Parton, Hunziker, Huber et al., The Recycling Endosome of Madin-Darby Canine Kidney Cells Is a Mildly Acidic Compartment Rich in Raft Components, Mol. Biol. Cell, doi:10.1091/mbc.11.8.2775
Ganley, Lam, Wang, Ding, Chen et al., ULK1•ATG13•FIP200 Complex Mediates MTOR Signaling and Is Essential for Autophagy, J. Biol. Chem, doi:10.1074/jbc.M900573200
Gassen, Papies, Bajaj, Emanuel, Dethloff et al., SARS-CoV-2-Mediated Dysregulation of Metabolism and Autophagy Uncovers Host-Targeting Antivirals, Nat. Commun, doi:10.1038/s41467-021-24007-w
Ghosh, Dellibovi-Ragheb, Kerviel, Pak, Qiu et al., β-Coronaviruses Use Lysosomes for Egress Instead of the Biosynthetic Secretory Pathway, Cell, doi:10.1016/j.cell.2020.10.039
Gordon, Jang, Bouhaddou, Xu, Obernier et al., A SARS-CoV-2 Protein Interaction Map Reveals Targets for Drug Repurposing, Nature, doi:10.1038/s41586-020-2286-9
Grasso, Renna, Vaccaro, Initial Steps in Mammalian Autophagosome Biogenesis, Front. Cell Dev. Biol, doi:10.3389/fcell.2018.00146
Hayer, Stoeber, Ritz, Engel, Meyer et al., Caveolin-1 Is Ubiquitinated and Targeted to Intralumenal Vesicles in Endolysosomes for Degradation, J. Cell Biol, doi:10.1083/jcb.201003086
Hill, Bastiani, Luetterforst, Kirkham, Kirkham et al., a Conserved Cytoplasmic Protein Required for Caveola Formation and Function, Cell, doi:10.1016/j.cell.2007.11.042
Hoffmann, Berking, Agerer, Buntru, Neske et al., Caveolin Limits Membrane Microdomain Mobility and Integrin-Mediated Uptake of Fibronectin-Binding Pathogens, J. Cell Sci, doi:10.1242/jcs.064006
Hoffmann, Kleine-Weber, Pöhlmann, A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells, Mol. Cell, doi:10.1016/j.molcel.2020.04.022
Hoffmann, Kleine-Weber, Schroeder, Krüger, Herrler et al., SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor, Cell, doi:10.1016/j.cell.2020.02.052
Hofmann, Munro, An N-Terminally Acetylated Arf-like GTPase Is Localised to Lysosomes and Affects Their Motility, J. Cell Sci, doi:10.1242/jcs.02958
Hosokawa, Sasaki, Iemura, Natsume, Hara et al., Atg101, a Novel Mammalian Autophagy Protein Interacting with Atg13, Autophagy, doi:10.4161/auto.5.7.9296
Hsu, Morohashi, Yoshimura, Manrique-Hoyos, Jung et al., Regulation of Exosome Secretion by Rab35 and Its GTPase-Activating Proteins TBC1D10A-C, J. Cell Biol, doi:10.1083/jcb.200911018
Huotari, Helenius, Maturation, None, EMBO J, doi:10.1038/emboj.2011.286
Hurley, Escrts, Are Everywhere, EMBO J, doi:10.15252/embj.201592484
Ichimura, Mori, Aschauer, Padmanabha Das, Padera et al., KIM-1/TIM-1 Is a Receptor for SARS-CoV-2 in Lung and Kidney, Medrxiv Prepr. Serv. Health Sci, doi:10.1101/2020.09.16.20190694
Inoue, Tanaka, Tanaka, Inoue, Morita et al., Clathrin-Dependent Entry of Severe Acute Respiratory Syndrome Coronavirus into Target Cells Expressing ACE2 with the Cytoplasmic Tail Deleted, J. Virol, doi:10.1128/JVI.00253-07
Itakura, Kishi-Itakura, Mizushima, The Hairpin-Type Tail-Anchored SNARE Syntaxin 17 Targets to Autophagosomes for Fusion with Endosomes/Lysosomes, Cell, doi:10.1016/j.cell.2012.11.001
Itakura, Mizushima, Atg14 and UVRAG: Mutually Exclusive Subunits of Mammalian Beclin 1-PI3K Complexes, Autophagy, doi:10.4161/auto.5.4.8062
Itakura, Mizushima, Characterization of Autophagosome Formation Site by a Hierarchical Analysis of Mammalian Atg Proteins, Autophagy, doi:10.4161/auto.6.6.12709
Iwata-Yoshikawa, Okamura, Shimizu, Hasegawa, Takeda et al., TMPRSS2 Contributes to Virus Spread and Immunopathology in the Airways of Murine Models after Coronavirus Infection, J. Virol, doi:10.1128/JVI.01815-18
Jackson, Farzan, Chen, Choe, Mechanisms of SARS-CoV-2 Entry into Cells, Nat. Rev. Mol. Cell Biol, doi:10.1038/s41580-021-00418-x
Jahn, Scheller, SNAREs-Engines for Membrane Fusion, Nat. Rev. Mol. Cell Biol, doi:10.1038/nrm2002
Jeon, Ko, Lee, Choi, Byun et al., Identification of Antiviral Drug Candidates against SARS-CoV-2 from FDA-Approved Drugs, Antimicrob. Agents Chemother, doi:10.1128/AAC.00819-20
Jiang, Nishimura, Sakamaki, Itakura, Hatta et al., The HOPS Complex Mediates Autophagosome-Lysosome Fusion through Interaction with Syntaxin 17, Mol. Biol. Cell, doi:10.1091/mbc.e13-08-0447
Johnson, Ostrowski, Jaumouillé, Grinstein, The Position of Lysosomes within the Cell Determines Their Luminal PH, J. Cell Biol, doi:10.1083/jcb.201507112
Jung, Jun, Ro, Kim, Otto et al., ULK-Atg13-FIP200 Complexes Mediate MTOR Signaling to the Autophagy Machinery, Mol. Biol. Cell, doi:10.1091/mbc.e08-12-1249
Kang, Chou, Rothlauf, Liu, Soh et al., Inhibition of PIKfyve Kinase Prevents Infection by Zaire Ebolavirus and SARS-CoV-2, Proc. Natl. Acad. Sci, doi:10.1073/pnas.2007837117
Kanzawa, Nishigaki, Hayashi, Ishii, Furukawa et al., Augmentation of Chemokine Production by Severe Acute Respiratory Syndrome Coronavirus 3a/X1 and 7a/X4 Proteins through NF-KB Activation, FEBS Lett, doi:10.1016/j.febslet.2006.11.046
Kawase, Shirato, Van Der Hoek, Taguchi, Matsuyama, Simultaneous Treatment of Human Bronchial Epithelial Cells with Serine and Cysteine Protease Inhibitors Prevents Severe Acute Respiratory Syndrome Coronavirus Entry, J. Virol, doi:10.1128/JVI.00094-12
Khatter, Sindhwani, Sharma, Arf-like GTPase Arl8: Moving from the Periphery to the Center of Lysosomal Biology, Cell. Logist, doi:10.1080/21592799.2015.1086501
Klumperman, Hille, Veenendaal, Oorschot, Stoorvogel et al., Differences in the Endosomal Distributions of the Two Mannose 6-Phosphate Receptors, J. Cell Biol, doi:10.1083/jcb.121.5.997
Koch, Uckeley, Doldan, Stanifer, Boulant et al., TMPRSS2 Expression Dictates the Entry Route Used by SARS-CoV-2 to Infect Host Cells, EMBO J, doi:10.15252/embj.2021107821
Korolchuk, Saiki, Lichtenberg, Siddiqi, Roberts et al., Lysosomal Positioning Coordinates Cellular Nutrient Responses, Nat. Cell Biol, doi:10.1038/ncb2204
Lajoie, Kojic, Nim, Li, Dennis et al., Caveolin-1 Regulation of Dynamin-Dependent, Raft-Mediated Endocytosis of Cholera Toxin-B Sub-Unit Occurs Independently of Caveolae, J. Cell. Mol. Med, doi:10.1111/j.1582-4934.2009.00732.x
Lajoie, Nabi, Lipid Rafts, Caveolae, and Their Endocytosis
Lakpa, Khan, Afghah, Chen, Geiger, Lysosomal Stress Response (LSR): Physiological Importance and Pathological Relevance, J. Neuroimmune Pharmacol, doi:10.1007/s11481-021-09990-7
Le, Guay, Altschuler, Nabi, Caveolin-1 Is a Negative Regulator of Caveolae-Mediated Endocytosis to the Endoplasmic Reticulum, J. Biol. Chem, doi:10.1074/jbc.M111240200
Letko, Marzi, Munster, Functional Assessment of Cell Entry and Receptor Usage for SARS-CoV-2 and Other Lineage B Betacoronaviruses, Nat. Microbiol, doi:10.1038/s41564-020-0688-y
Li, Li, Felder, Periasamy, Jose, Rab4 and Rab11 Coordinately Regulate the Recycling of Angiotensin II Type I Receptor as Demonstrated by Fluorescence Resonance Energy Transfer Microscopy, J. Biomed. Opt, doi:10.1117/1.2943286
Li, Li, Yamate, Li, Ikuta, Lipid Rafts Play an Important Role in the Early Stage of Severe Acute Respiratory Syndrome-Coronavirus Life Cycle, Microbes Infect, doi:10.1016/j.micinf.2006.10.015
Li, Zhu, Fan, Zhang, Peng et al., Dependence of SARS-CoV-2 Infection on Cholesterol-Rich Lipid Raft and Endosomal Acidification, Comput. Struct. Biotechnol. J, doi:10.1016/j.csbj.2021.04.001
Liang, Feng, Ku, Dotan, Canaani et al., Autophagic and Tumour Suppressor Activity of a Novel Beclin1-Binding Protein UVRAG, Nat. Cell Biol, doi:10.1038/ncb1426
Liang, Lee, Inn, Gack, Li et al., Beclin1-Binding UVRAG Targets the Class C Vps Complex to Coordinate Autophagosome Maturation and Endocytic Trafficking, Nat. Cell Biol, doi:10.1038/ncb1740
Liebl, Difato, Horníková, Mannová, Štokrová et al., Mouse Polyomavirus Enters Early Endosomes, Requires Their Acidic PH for Productive Infection, and Meets Transferrin Cargo in Rab11-Positive Endosomes, J. Virol, doi:10.1128/JVI.80.9.4610-4622.2006
Lin, Hurley, Structure and Function of the ULK1 Complex in Autophagy, Curr. Opin. Cell Biol, doi:10.1016/j.ceb.2016.02.010
Liu, Chopra, Li, Bouwman, Tompkins et al., Heparan Sulfate Proteoglycans as Attachment Factor for SARS-CoV-2, ACS Cent. Sci, doi:10.1021/acscentsci.1c00010
Liu, Lear, Larsen, Lin, Cao et al., Modulation of Lysosomal Function as a Therapeutic Approach for Coronaviral Infections, Res. Sq, doi:10.21203/rs.3.rs-419305/v1
Luzio, Hackmann, Dieckmann, Griffiths, The Biogenesis of Lysosomes and Lysosome-Related Organelles, Cold Spring Harb. Perspect. Biol, doi:10.1101/cshperspect.a016840
Mayor, Parton, Donaldson, Clathrin-Independent Pathways of Endocytosis, Cold Spring Harb. Perspect. Biol, doi:10.1101/cshperspect.a016758
Meier, Boucke, Hammer, Keller, Stidwill et al., Adenovirus Triggers Macropinocytosis and Endosomal Leakage Together with Its Clathrin-Mediated Uptake, J. Cell Biol, doi:10.1083/jcb.200112067
Mercer, Gubas, Tooze, A Molecular Perspective of Mammalian Autophagosome Biogenesis, J. Biol. Chem, doi:10.1074/jbc.R117.810366
Mercer, Schelhaas, Helenius, Virus Entry by Endocytosis, Annu. Rev. Biochem, doi:10.1146/annurev-biochem-060208-104626
Miao, Zhao, Li, Ji, Chen et al., ORF3a of the COVID-19 Virus SARS-CoV-2 Blocks HOPS Complex-Mediated Assembly of the SNARE Complex Required for Autolysosome Formation, Dev. Cell, doi:10.1016/j.devcel.2020.12.010
Mille, Whittaker, Host Cell Entry of Middle East Respiratory Syndrome Coronavirus after Two-Step, Furin-Mediated Activation of the Spike Protein, Proc. Natl. Acad. Sci, doi:10.1073/pnas.1407087111
Mingo, Simmons, Shoemaker, Nelson, Schornberg et al., Ebola Virus and Severe Acute Respiratory Syndrome Coronavirus Display Late Cell Entry Kinetics: Evidence That Transport to NPC1 + Endolysosomes Is a Rate-Defining Step, J. Virol, doi:10.1128/JVI.03398-14
Ostrowski, Carmo, Krumeich, Fanget, Raposo et al., Rab27a and Rab27b Control Different Steps of the Exosome Secretion Pathway, Nat. Cell Biol, doi:10.1038/ncb2000
Ou, Liu, Lei, Li, Mi et al., Characterization of Spike Glycoprotein of SARS-CoV-2 on Virus Entry and Its Immune Cross-Reactivity with SARS-CoV, Nat. Commun, doi:10.1038/s41467-020-15562-9
Papa, Mallery, Albecka, Welch, Cattin-Ortolá et al., Furin Cleavage of SARS-CoV-2 Spike Promotes but Is Not Essential for Infection and Cell-Cell Fusion, PLoS Pathog, doi:10.1371/journal.ppat.1009246
Park, Li, Barlan, Fehr, Perlman et al., Proteolytic Processing of Middle East Respiratory Syndrome Coronavirus Spikes Expands Virus Tropism, Proc. Natl. Acad. Sci, doi:10.1073/pnas.1608147113
Parton, Caveolae, Structure, Function, and Relationship to Disease, Annu. Rev. Cell Dev. Biol, doi:10.1146/annurev-cellbio-100617-062737
Parton, Howes, Revisiting Caveolin Trafficking: The End of the Caveosome, J. Cell Biol, doi:10.1083/jcb.201009093
Parton, Joggerst, Simons, Regulated Internalization of Caveolae, J. Cell Biol, doi:10.1083/jcb.127.5.1199
Pelkmans, Bürli, Zerial, Helenius, Caveolin-Stabilized Membrane Domains as Multifunctional Transport and Sorting Devices in Endocytic Membrane Traffic, Cell, doi:10.1016/j.cell.2004.09.003
Pelkmans, Kartenbeck, Helenius, Caveolar Endocytosis of Simian Virus 40 Reveals a New Two-Step Vesicular-Transport Pathway to the ER, Nat. Cell Biol, doi:10.1038/35074539
Pelkmans, Zerial, Kinase-Regulated Quantal Assemblies and Kiss-and-Run Recycling of Caveolae, Nature, doi:10.1038/nature03866
Perrier, Bonnin, Desmarets, Danneels, Goffard et al., The C-Terminal Domain of the MERS CoronavirusMprotein Contains a Trans-Golgi Network Localization Signal, J. Biol. Chem, doi:10.1074/jbc.RA119.008964
Pol, Lu, Pons, Peiró, Enrich, Epidermal Growth Factor-Mediated Caveolin Recruitment to Early Endosomes and MAPK Activation. Role of Cholesterol and Actin Cytoskeleton, J. Biol. Chem, doi:10.1074/jbc.M001131200
Ponsford, Ryan, Raimondi, Cocucci, Wycislo et al., Live Imaging of Intra-Lysosome PH in Cell Lines and Primary Neuronal Culture Using a Novel Genetically Encoded Biosensor, Autophagy, doi:10.1080/15548627.2020.1771858
Pu, Guardia, Keren-Kaplan, Bonifacino, Mechanisms and Functions of Lysosome Positioning, J. Cell Sci, doi:10.1242/jcs.196287
Pu, Schindler, Jia, Jarnik, Backlund et al., a Multisubunit Complex That Regulates Lysosome Positioning, Dev. Cell, doi:10.1016/j.devcel.2015.02.011
Puertollano, MTOR and Lysosome Regulation, Prime Rep, doi:10.12703/P6-52
Qu, Wang, Zhu, Wang, Wang et al., ORF3a-Mediated Incomplete Autophagy Facilitates Severe Acute Respiratory Syndrome Coronavirus-2 Replication, Front. Cell Dev. Biol, doi:10.3389/fcell.2021.716208
Rao, Huynh, Proux-Gillardeaux, Galli, Andrews, Identification of SNAREs Involved in Synaptotagmin VII-Regulated Lysosomal Exocytosis, J. Biol. Chem, doi:10.1074/jbc.M400798200
Ripa, Andreu, López-Guerrero, Bello-Morales, Membrane Rafts: Portals for Viral Entry, Front. Microbiol, doi:10.3389/fmicb.2021.631274
Rosa-Ferreira, Munro, Arl8 and SKIP Act Together to Link Lysosomes to Kinesin-1, Dev. Cell, doi:10.1016/j.devcel.2011.10.007
Russell, Tian, Yuan, Park, Chang et al., ULK1 Induces Autophagy by Phosphorylating Beclin-1 and Activating VPS34 Lipid Kinase, Nat. Cell Biol, doi:10.1038/ncb2757
Saftig, Klumperman, Lysosome Biogenesis and Lysosomal Membrane Proteins: Trafficking Meets Function, Nat. Rev. Mol. Cell Biol, doi:10.1038/nrm2745
Sanders, Jumper, Ackerman, Bracha, Donlic et al., SARS-CoV-2 Requires Cholesterol for Viral Entry and Pathological Syncytia Formation, eLife, doi:10.7554/eLife.65962
Sandvig, Kavaliauskiene, Skotland, Clathrin-Independent Endocytosis: An Increasing Degree of Complexity, Histochem. Cell Biol, doi:10.1007/s00418-018-1678-5
Sardiello, Ballabio, Lysosomal Enhancement: A CLEAR Answer to Cellular Degradative Needs, Cell Cycle, doi:10.4161/cc.8.24.10263
Savina, Vidal, Colombo, The Exosome Pathway in K562 Cells Is Regulated by Rab11, J. Cell Sci, doi:10.1242/jcs.115.12.2505
Saxton, Sabatini, Mtor, Signaling in Growth, Metabolism, and Disease, Cell, doi:10.1016/j.cell.2017.02.004
Schweizer, Haugk, Mckiernan, Gulati, Cheng et al., A Phase I Study of Niclosamide in Combination with Enzalutamide in Men with Castration-Resistant Prostate Cancer, PLoS ONE, doi:10.1371/journal.pone.0198389
Settembre, Di Malta, Polito, Arencibia, Vetrini et al., TFEB Links Autophagy to Lysosomal Biogenesis, Science, doi:10.1126/science.1204592
Seyfoori, Barough, Mokarram, Ahmadi, Mehrbod et al., Emerging Advances of Nanotechnology in Drug and Vaccine Delivery against Viral Associated Respiratory Infectious Diseases (VARID), Int. J. Mol. Sci, doi:10.3390/ijms22136937
Shang, Wang, AMPK and MTOR Coordinate the Regulation of Ulk1 and Mammalian Autophagy Initiation, Autophagy, doi:10.4161/auto.7.8.15860
Shang, Ye, Shi, Wan, Luo et al., Structural Basis of Receptor Recognition by SARS-CoV-2, Nature, doi:10.1038/s41586-020-2179-y
Sherman, Emmer, ACE2 Protein Expression within Isogenic Cell Lines Is Heterogeneous and Associated with Distinct Transcriptomes, Sci. Rep, doi:10.1038/s41598-021-95308-9
Shirato, Kawase, Matsuyama, Wild-Type Human Coronaviruses Prefer Cell-Surface TMPRSS2 to Endosomal Cathepsins for Cell Entry, Virology, doi:10.1016/j.virol.2017.11.012
Siu, Yuen, Castano-Rodriguez, Ye, Yeung et al., Severe Acute Respiratory Syndrome Coronavirus ORF3a Protein Activates the NLRP3 Inflammasome by Promoting TRAF3-Dependent Ubiquitination of ASC, FASEB J, doi:10.1096/fj.201802418R
Snijder, Decroly, Ziebuhr, The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing, Adv. Virus Res, doi:10.1016/bs.aivir.2016.08.008
Solinger, Spang, Tethering Complexes in the Endocytic Pathway: CORVET and HOPS, FEBS J, doi:10.1111/febs.12151
Stukalov, Girault, Grass, Karayel, Bergant et al., Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV, Nature, doi:10.1038/s41586-021-03493-4
Takáts, Pircs, Nagy, Varga, Kárpáti et al., Interaction of the HOPS Complex with Syntaxin 17 Mediates Autophagosome Clearance in Drosophila, Mol. Biol. Cell, doi:10.1091/mbc.e13-08-0449
Tan, Lim, Hong, Understanding the Accessory Viral Proteins Unique to the Severe Acute Respiratory Syndrome (SARS) Coronavirus, Antivir. Res, doi:10.1016/j.antiviral.2006.05.010
Tancini, Buratta, Delo, Sagini, Chiaradia et al., Lysosomal Exocytosis: The Extracellular Role of an Intracellular Organelle, Membranes, doi:10.3390/membranes10120406
Thomsen, Roepstorff, Stahlhut, Van Deurs, Caveolae Are Highly Immobile Plasma Membrane Microdomains, Which Are Not Involved in Constitutive Endocytic Trafficking, Mol. Biol. Cell, doi:10.1091/mbc.01-06-0317
Tooze, Tooze, Fuller, Sorting of Progeny Coronavirus from Condensed Secretory Protein at the Exit from the Trans-Golgi Network at AtT20 Cells, J. Cell Biol, doi:10.1083/jcb.105.3.1215
Trajkovic, Hsu, Chiantia, Rajendran, Wenzel et al., Ceramide Triggers Budding of Exosome Vesicles into Multivesicular Endosomes, Science, doi:10.1126/science.1153124
Tran, Carpentier, Sawano, Gorden, Orci, Ligands Internalized through Coated or Noncoated Invaginations Follow a Common Intracellular Pathway, Proc. Natl. Acad. Sci, doi:10.1073/pnas.84.22.7957
Tripathi, Chowdhury, Trudel, Tee, Slack et al., Reactive Nitrogen Species Regulate Autophagy through ATM-AMPK-TSC2-Mediated Suppression of MTORC1, Proc. Natl. Acad. Sci, doi:10.1073/pnas.1307736110
V'kovski, Kratzel, Steiner, Stalder, Thiel, Coronavirus Biology and Replication: Implications for SARS-CoV-2, Nat. Rev. Microbiol, doi:10.1038/s41579-020-00468-6
Van Der Kant, Jonker, Wijdeven, Bakker, Janssen et al., Characterization of the Mammalian CORVET and HOPS Complexes and Their Modular Restructuring for Endosome Specificity, J. Biol. Chem, doi:10.1074/jbc.M115.688440
Walls, Park, Tortorici, Wall, Mcguire et al., Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein, Cell, doi:10.1016/j.cell.2020.02.058
Wandinger-Ness, Zerial, Rab Proteins and the Compartmentalization of the Endosomal System, Cold Spring Harb. Perspect. Biol, doi:10.1101/cshperspect.a022616
Wang, Chen, Zhou, Lian, Zhang et al., SARS-CoV-2 Invades Host Cells via a Novel Route: CD147-Spike Protein, bioRxiv, doi:10.1101/2020.03.14.988345
Wang, Li, Hui, Tiwari, Zhang et al., Cholesterol 25-Hydroxylase Inhibits SARS -CoV-2 and Other Coronaviruses by Depleting Membrane Cholesterol, EMBO J, doi:10.15252/embj.2020106057
Wang, Qiu, Hou, Deng, Xu et al., AXL Is a Candidate Receptor for SARS-CoV-2 That Promotes Infection of Pulmonary and Bronchial Epithelial Cells, Cell Res, doi:10.1038/s41422-020-00460-y
Wang, Simoneau, Kulsuptrakul, Bouhaddou, Travisano et al., Genetic Screens Identify Host Factors for SARS-CoV-2 and Common Cold Coronaviruses, Cell, doi:10.1016/j.cell.2020.12.004
Wang, Yang, Liu, Guo, Zhang et al., SARS Coronavirus Entry into Host Cells through a Novel Clathrin-and Caveolae-Independent Endocytic Pathway, Cell Res, doi:10.1038/cr.2008.15
Ward, Martinez, Vaccaro, Zhou, Tang et al., From Sorting Endosomes to Exocytosis: Association of Rab4 and Rab11 GTPases with the Fc Receptor, FcRn, during Recycling, Mol. Biol. Cell, doi:10.1091/mbc.e04-08-0735
Weiss, Touret, Baronti, Gilles, Hoen et al., Niclosamide Shows Strong Antiviral Activity in a Human Airway Model of SARS-CoV-2 Infection and a Conserved Potency against the Alpha (B.1.1.7), Beta (B.1.351) and Delta Variant (B.1.617.2, PLoS ONE, doi:10.1371/journal.pone.0260958
Wolff, Limpens, Zevenhoven-Dobbe, Laugks, Zheng et al., A Molecular Pore Spans the Double Membrane of the Coronavirus Replication Organelle, Science, doi:10.1126/science.abd3629
Wrapp, Wang, Corbett, Goldsmith, Hsieh et al., Cryo-EM Structure of the 2019-NCoV Spike in the Prefusion Conformation, Science, doi:10.1126/science.abb2507
Yamauchi, Helenius, Virus Entry at a Glance, J. Cell Sci, doi:10.1242/jcs.119685
Yuan, Song, The Emerging Role of Rab5 in Membrane Receptor Trafficking and Signaling Pathways, Biochem. Res. Int, doi:10.1155/2020/4186308
Yuan, Yan, Cao, Ye, Liang et al., SARS-CoV-2 Exploits Host DGAT and ADRP for Efficient Replication, Cell Discov, doi:10.1038/s41421-021-00338-2
Yue, Nabar, Shi, Kamenyeva, Xiao et al., SARS-Coronavirus Open Reading Frame-3a Drives Multimodal Necrotic Cell Death, Cell Death Dis, doi:10.1038/s41419-018-0917-y
Zang, Case, Yutuc, Ma, Shen et al., Cholesterol 25-Hydroxylase Suppresses SARS-CoV-2 Replication by Blocking Membrane Fusion, Proc. Natl. Acad. Sci, doi:10.1073/pnas.2012197117
Zhang, Sun, Pei, Mao, Zhao et al., The SARS-CoV-2 Protein ORF3a Inhibits Fusion of Autophagosomes with Lysosomes, Cell Discov, doi:10.1038/s41421-021-00268-z
Zhao, Zhang, Autophagosome Maturation: An Epic Journey from the ER to Lysosomes, J. Cell Biol, doi:10.1083/jcb.201810099
Zhou, Vedantham, Lu, Agudelo, Carrion et al., Protease Inhibitors Targeting Coronavirus and Filovirus Entry, Antivir. Res, doi:10.1016/j.antiviral.2015.01.011
Zhou, Yang, Wang, Hu, Zhang et al., A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin, Nature, doi:10.1038/s41586-020-2012-7
Zhu, Feng, Hu, Wang, Yu et al., A Genome-wide CRISPR Screen Identifies Host Factors That Regulate SARS-CoV-2 Entry, Nat. Commun, doi:10.1038/s41467-021-21213-4
DOI record:
{
"DOI": "10.3390/ijms23094576",
"ISSN": [
"1422-0067"
],
"URL": "http://dx.doi.org/10.3390/ijms23094576",
"abstract": "<jats:p>This review aims to describe and discuss the different functions of the endolysosomal system, from homeostasis to its vital role during viral infections. We will initially describe endolysosomal system’s main functions, presenting recent data on how its compartments are essential for host defense to explore later how SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2) and other coronaviruses subvert these organelles for their benefit. It is clear that to succeed, pathogens’ evolution favored the establishment of ways to avoid, escape, or manipulate lysosomal function. The unavoidable coexistence with such an unfriendly milieu imposed on viruses the establishment of a vast array of strategies to make the most out of the invaded cell’s machinery to produce new viruses and maneuvers to escape the host’s defense system.</jats:p>",
"alternative-id": [
"ijms23094576"
],
"author": [
{
"affiliation": [],
"family": "Cesar-Silva",
"given": "Daniella",
"sequence": "first"
},
{
"ORCID": "http://orcid.org/0000-0003-1917-2876",
"affiliation": [],
"authenticated-orcid": false,
"family": "Pereira-Dutra",
"given": "Filipe S.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Moraes Giannini",
"given": "Ana Lucia Moraes",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-9931-6627",
"affiliation": [],
"authenticated-orcid": false,
"family": "Jacques G. de Almeida",
"given": "Cecília Jacques G.",
"sequence": "additional"
}
],
"container-title": "International Journal of Molecular Sciences",
"container-title-short": "IJMS",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2022,
4,
21
]
],
"date-time": "2022-04-21T05:55:51Z",
"timestamp": 1650520551000
},
"deposited": {
"date-parts": [
[
2022,
4,
21
]
],
"date-time": "2022-04-21T07:00:21Z",
"timestamp": 1650524421000
},
"funder": [
{
"award": [
"VPPCB-005-FIO-20-2-19",
"E-26/010.001244/2016"
],
"name": "Inova Program/FIOCRUZ"
},
{
"DOI": "10.13039/501100003593",
"award": [
"401700/2020-8"
],
"doi-asserted-by": "publisher",
"name": "Conselho Nacional de Desenvolvimento Científico e Tecnológico"
}
],
"indexed": {
"date-parts": [
[
2024,
2,
22
]
],
"date-time": "2024-02-22T16:01:37Z",
"timestamp": 1708617697402
},
"is-referenced-by-count": 5,
"issue": "9",
"issued": {
"date-parts": [
[
2022,
4,
20
]
]
},
"journal-issue": {
"issue": "9",
"published-online": {
"date-parts": [
[
2022,
5
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2022,
4,
20
]
],
"date-time": "2022-04-20T00:00:00Z",
"timestamp": 1650412800000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/1422-0067/23/9/4576/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "4576",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2022,
4,
20
]
]
},
"published-online": {
"date-parts": [
[
2022,
4,
20
]
]
},
"publisher": "MDPI AG",
"reference": [
{
"DOI": "10.1016/j.cub.2016.06.046",
"doi-asserted-by": "publisher",
"key": "ref1"
},
{
"DOI": "10.1242/jcs.196287",
"doi-asserted-by": "publisher",
"key": "ref2"
},
{
"DOI": "10.1101/cshperspect.a016840",
"doi-asserted-by": "publisher",
"key": "ref3"
},
{
"DOI": "10.1007/s00418-018-1678-5",
"doi-asserted-by": "publisher",
"key": "ref4"
},
{
"DOI": "10.1091/mbc.e04-08-0735",
"doi-asserted-by": "publisher",
"key": "ref5"
},
{
"DOI": "10.1038/emboj.2011.286",
"doi-asserted-by": "publisher",
"key": "ref6"
},
{
"DOI": "10.1155/2020/4186308",
"doi-asserted-by": "publisher",
"key": "ref7"
},
{
"DOI": "10.1101/cshperspect.a022616",
"doi-asserted-by": "publisher",
"key": "ref8"
},
{
"DOI": "10.1117/1.2943286",
"doi-asserted-by": "publisher",
"key": "ref9"
},
{
"DOI": "10.1038/nrm2745",
"doi-asserted-by": "publisher",
"key": "ref10"
},
{
"DOI": "10.1038/nrm2002",
"doi-asserted-by": "publisher",
"key": "ref11"
},
{
"DOI": "10.1242/jcs.107805",
"doi-asserted-by": "publisher",
"key": "ref12"
},
{
"DOI": "10.1111/febs.12151",
"doi-asserted-by": "publisher",
"key": "ref13"
},
{
"DOI": "10.1074/jbc.M115.688440",
"doi-asserted-by": "publisher",
"key": "ref14"
},
{
"DOI": "10.1083/jcb.121.5.997",
"doi-asserted-by": "publisher",
"key": "ref15"
},
{
"DOI": "10.15252/embj.201592484",
"doi-asserted-by": "publisher",
"key": "ref16"
},
{
"DOI": "10.1080/20013078.2019.1703244",
"doi-asserted-by": "publisher",
"key": "ref17"
},
{
"DOI": "10.1126/science.1153124",
"doi-asserted-by": "publisher",
"key": "ref18"
},
{
"DOI": "10.1016/j.canlet.2017.08.020",
"doi-asserted-by": "publisher",
"key": "ref19"
},
{
"DOI": "10.3390/membranes10120406",
"doi-asserted-by": "publisher",
"key": "ref20"
},
{
"DOI": "10.1242/jcs.02958",
"doi-asserted-by": "publisher",
"key": "ref21"
},
{
"DOI": "10.1016/j.devcel.2011.10.007",
"doi-asserted-by": "publisher",
"key": "ref22"
},
{
"DOI": "10.1080/21592799.2015.1086501",
"doi-asserted-by": "publisher",
"key": "ref23"
},
{
"DOI": "10.1016/j.devcel.2015.02.011",
"doi-asserted-by": "publisher",
"key": "ref24"
},
{
"DOI": "10.1242/jcs.115.12.2505",
"doi-asserted-by": "publisher",
"key": "ref25"
},
{
"DOI": "10.1038/ncb2000",
"doi-asserted-by": "publisher",
"key": "ref26"
},
{
"DOI": "10.1083/jcb.200911018",
"doi-asserted-by": "publisher",
"key": "ref27"
},
{
"DOI": "10.1074/jbc.M400798200",
"doi-asserted-by": "publisher",
"key": "ref28"
},
{
"DOI": "10.1038/ncb2204",
"doi-asserted-by": "publisher",
"key": "ref29"
},
{
"DOI": "10.12703/P6-52",
"doi-asserted-by": "publisher",
"key": "ref30"
},
{
"DOI": "10.1016/j.cell.2017.02.004",
"doi-asserted-by": "publisher",
"key": "ref31"
},
{
"DOI": "10.3390/cancers11101422",
"doi-asserted-by": "publisher",
"key": "ref32"
},
{
"DOI": "10.1038/s41580-018-0003-4",
"doi-asserted-by": "publisher",
"key": "ref33"
},
{
"DOI": "10.1074/jbc.M900573200",
"doi-asserted-by": "publisher",
"key": "ref34"
},
{
"DOI": "10.4161/auto.5.7.9296",
"doi-asserted-by": "publisher",
"key": "ref35"
},
{
"DOI": "10.4161/auto.7.8.15860",
"doi-asserted-by": "publisher",
"key": "ref36"
},
{
"DOI": "10.1091/mbc.e08-12-1249",
"doi-asserted-by": "publisher",
"key": "ref37"
},
{
"DOI": "10.1042/BJ20101894",
"doi-asserted-by": "publisher",
"key": "ref38"
},
{
"DOI": "10.1016/j.molcel.2015.05.031",
"doi-asserted-by": "publisher",
"key": "ref39"
},
{
"DOI": "10.1016/j.ceb.2016.02.010",
"doi-asserted-by": "publisher",
"key": "ref40"
},
{
"DOI": "10.1038/ncb2757",
"doi-asserted-by": "publisher",
"key": "ref41"
},
{
"DOI": "10.4161/auto.5.4.8062",
"doi-asserted-by": "publisher",
"key": "ref42"
},
{
"DOI": "10.4161/auto.6.6.12709",
"doi-asserted-by": "publisher",
"key": "ref43"
},
{
"DOI": "10.1038/ncb1740",
"doi-asserted-by": "publisher",
"key": "ref44"
},
{
"DOI": "10.1038/ncb1426",
"doi-asserted-by": "publisher",
"key": "ref45"
},
{
"DOI": "10.1074/jbc.R117.810366",
"doi-asserted-by": "publisher",
"key": "ref46"
},
{
"DOI": "10.1083/jcb.201810099",
"doi-asserted-by": "publisher",
"key": "ref47"
},
{
"DOI": "10.3389/fcell.2018.00146",
"doi-asserted-by": "publisher",
"key": "ref48"
},
{
"DOI": "10.1073/pnas.1307736110",
"doi-asserted-by": "publisher",
"key": "ref49"
},
{
"DOI": "10.4161/cc.8.24.10263",
"doi-asserted-by": "publisher",
"key": "ref50"
},
{
"DOI": "10.1126/science.1204592",
"doi-asserted-by": "publisher",
"key": "ref51"
},
{
"DOI": "10.1016/j.bbamcr.2018.12.011",
"doi-asserted-by": "publisher",
"key": "ref52"
},
{
"DOI": "10.1016/j.cell.2012.11.001",
"doi-asserted-by": "publisher",
"key": "ref53"
},
{
"DOI": "10.1091/mbc.e13-08-0447",
"doi-asserted-by": "publisher",
"key": "ref54"
},
{
"DOI": "10.1091/mbc.e13-08-0449",
"doi-asserted-by": "publisher",
"key": "ref55"
},
{
"DOI": "10.1016/j.yexcr.2012.09.004",
"doi-asserted-by": "publisher",
"key": "ref56"
},
{
"DOI": "10.1007/s11481-021-09990-7",
"doi-asserted-by": "publisher",
"key": "ref57"
},
{
"DOI": "10.1083/jcb.201507112",
"doi-asserted-by": "publisher",
"key": "ref58"
},
{
"DOI": "10.1080/15548627.2020.1771858",
"doi-asserted-by": "publisher",
"key": "ref59"
},
{
"DOI": "10.1242/jcs.119685",
"doi-asserted-by": "publisher",
"key": "ref60"
},
{
"DOI": "10.1016/j.bcp.2020.114316",
"doi-asserted-by": "publisher",
"key": "ref61"
},
{
"DOI": "10.3389/fmicb.2021.631274",
"doi-asserted-by": "publisher",
"key": "ref62"
},
{
"DOI": "10.1101/cshperspect.a016758",
"doi-asserted-by": "publisher",
"key": "ref63"
},
{
"DOI": "10.1038/35074539",
"doi-asserted-by": "publisher",
"key": "ref64"
},
{
"DOI": "10.1083/jcb.200407113",
"doi-asserted-by": "publisher",
"key": "ref65"
},
{
"DOI": "10.1038/ncb1999",
"doi-asserted-by": "publisher",
"key": "ref66"
},
{
"DOI": "10.1016/j.tcb.2015.10.010",
"doi-asserted-by": "publisher",
"key": "ref67"
},
{
"DOI": "10.1146/annurev-cellbio-100617-062737",
"doi-asserted-by": "publisher",
"key": "ref68"
},
{
"DOI": "10.1091/mbc.01-06-0317",
"doi-asserted-by": "publisher",
"key": "ref69"
},
{
"DOI": "10.1074/jbc.M111240200",
"doi-asserted-by": "publisher",
"key": "ref70"
},
{
"DOI": "10.1242/jcs.064006",
"doi-asserted-by": "publisher",
"key": "ref71"
},
{
"DOI": "10.1111/j.1582-4934.2009.00732.x",
"doi-asserted-by": "publisher",
"key": "ref72"
},
{
"DOI": "10.1038/nature03866",
"doi-asserted-by": "publisher",
"key": "ref73"
},
{
"author": "Lajoie",
"key": "ref74",
"series-title": "Lipid Rafts, Caveolae, and Their Endocytosis",
"volume": "Volume 282",
"year": "2010"
},
{
"DOI": "10.1083/jcb.127.5.1199",
"doi-asserted-by": "publisher",
"key": "ref75"
},
{
"DOI": "10.1074/jbc.M001131200",
"doi-asserted-by": "publisher",
"key": "ref76"
},
{
"DOI": "10.1073/pnas.84.22.7957",
"doi-asserted-by": "publisher",
"key": "ref77"
},
{
"DOI": "10.1016/j.cell.2004.09.003",
"doi-asserted-by": "publisher",
"key": "ref78"
},
{
"DOI": "10.1091/mbc.11.8.2775",
"doi-asserted-by": "publisher",
"key": "ref79"
},
{
"DOI": "10.1128/JVI.80.9.4610-4622.2006",
"doi-asserted-by": "publisher",
"key": "ref80"
},
{
"DOI": "10.1016/j.cell.2007.11.042",
"doi-asserted-by": "publisher",
"key": "ref81"
},
{
"DOI": "10.1083/jcb.201003086",
"doi-asserted-by": "publisher",
"key": "ref82"
},
{
"DOI": "10.1083/jcb.201009093",
"doi-asserted-by": "publisher",
"key": "ref83"
},
{
"DOI": "10.1146/annurev-biochem-060208-104626",
"doi-asserted-by": "publisher",
"key": "ref84"
},
{
"DOI": "10.1371/journal.pbio.1001832",
"doi-asserted-by": "publisher",
"key": "ref85"
},
{
"DOI": "10.1083/jcb.200112067",
"doi-asserted-by": "publisher",
"key": "ref86"
},
{
"DOI": "10.1038/s41586-020-2012-7",
"doi-asserted-by": "publisher",
"key": "ref87"
},
{
"DOI": "10.1038/s41564-020-0688-y",
"doi-asserted-by": "publisher",
"key": "ref88"
},
{
"DOI": "10.1126/science.abd3072",
"doi-asserted-by": "publisher",
"key": "ref89"
},
{
"DOI": "10.1101/2020.03.14.988345",
"doi-asserted-by": "publisher",
"key": "ref90"
},
{
"DOI": "10.1021/acscentsci.1c00010",
"doi-asserted-by": "publisher",
"key": "ref91"
},
{
"DOI": "10.1016/j.cell.2020.09.033",
"doi-asserted-by": "publisher",
"key": "ref92"
},
{
"DOI": "10.1021/acscentsci.0c01537",
"doi-asserted-by": "publisher",
"key": "ref93"
},
{
"DOI": "10.1016/j.jbc.2021.100759",
"doi-asserted-by": "publisher",
"key": "ref94"
},
{
"DOI": "10.1038/s41422-020-00460-y",
"doi-asserted-by": "publisher",
"key": "ref95"
},
{
"DOI": "10.1101/2020.09.16.20190694",
"doi-asserted-by": "publisher",
"key": "ref96"
},
{
"DOI": "10.1016/j.molcel.2020.04.022",
"doi-asserted-by": "publisher",
"key": "ref97"
},
{
"DOI": "10.1016/j.cell.2020.02.052",
"doi-asserted-by": "publisher",
"key": "ref98"
},
{
"DOI": "10.1038/s41467-021-21213-4",
"doi-asserted-by": "publisher",
"key": "ref99"
},
{
"DOI": "10.1371/journal.ppat.1009246",
"doi-asserted-by": "publisher",
"key": "ref100"
},
{
"DOI": "10.1038/s41586-020-2179-y",
"doi-asserted-by": "publisher",
"key": "ref101"
},
{
"DOI": "10.1016/j.cell.2020.02.058",
"doi-asserted-by": "publisher",
"key": "ref102"
},
{
"DOI": "10.1126/science.abb2507",
"doi-asserted-by": "publisher",
"key": "ref103"
},
{
"DOI": "10.15252/embj.2021107821",
"doi-asserted-by": "publisher",
"key": "ref104"
},
{
"DOI": "10.1128/JVI.00094-12",
"doi-asserted-by": "publisher",
"key": "ref105"
},
{
"DOI": "10.1016/j.virol.2017.11.012",
"doi-asserted-by": "publisher",
"key": "ref106"
},
{
"DOI": "10.1038/s41467-020-15562-9",
"doi-asserted-by": "publisher",
"key": "ref107"
},
{
"DOI": "10.1002/pro.4073",
"doi-asserted-by": "publisher",
"key": "ref108"
},
{
"DOI": "10.1073/pnas.1407087111",
"doi-asserted-by": "publisher",
"key": "ref109"
},
{
"DOI": "10.1073/pnas.1608147113",
"doi-asserted-by": "publisher",
"key": "ref110"
},
{
"DOI": "10.1016/j.antiviral.2015.01.011",
"doi-asserted-by": "publisher",
"key": "ref111"
},
{
"DOI": "10.1128/JVI.01815-18",
"doi-asserted-by": "publisher",
"key": "ref112"
},
{
"DOI": "10.1128/JVI.03398-14",
"doi-asserted-by": "publisher",
"key": "ref113"
},
{
"DOI": "10.1371/journal.ppat.1006546",
"doi-asserted-by": "publisher",
"key": "ref114"
},
{
"DOI": "10.1155/2020/9238696",
"doi-asserted-by": "publisher",
"key": "ref115"
},
{
"DOI": "10.1038/s41598-021-95308-9",
"doi-asserted-by": "publisher",
"key": "ref116"
},
{
"DOI": "10.1016/j.jbc.2021.100306",
"doi-asserted-by": "publisher",
"key": "ref117"
},
{
"DOI": "10.1016/j.csbj.2021.04.001",
"doi-asserted-by": "publisher",
"key": "ref118"
},
{
"DOI": "10.1016/j.micinf.2006.10.015",
"doi-asserted-by": "publisher",
"key": "ref119"
},
{
"DOI": "10.15252/embj.2020106057",
"doi-asserted-by": "publisher",
"key": "ref120"
},
{
"DOI": "10.1073/pnas.2012197117",
"doi-asserted-by": "publisher",
"key": "ref121"
},
{
"DOI": "10.7554/eLife.65962",
"doi-asserted-by": "publisher",
"key": "ref122"
},
{
"DOI": "10.1073/pnas.2007837117",
"doi-asserted-by": "publisher",
"key": "ref123"
},
{
"DOI": "10.1016/j.cell.2020.12.004",
"doi-asserted-by": "publisher",
"key": "ref124"
},
{
"DOI": "10.1038/cr.2008.15",
"doi-asserted-by": "publisher",
"key": "ref125"
},
{
"DOI": "10.1128/JVI.00253-07",
"doi-asserted-by": "publisher",
"key": "ref126"
},
{
"DOI": "10.15252/embj.2020106267",
"doi-asserted-by": "publisher",
"key": "ref127"
},
{
"DOI": "10.1038/s41580-021-00418-x",
"doi-asserted-by": "publisher",
"key": "ref128"
},
{
"DOI": "10.1016/bs.aivir.2016.08.008",
"doi-asserted-by": "publisher",
"key": "ref129"
},
{
"DOI": "10.1126/science.abd3629",
"doi-asserted-by": "publisher",
"key": "ref130"
},
{
"DOI": "10.1083/jcb.105.3.1215",
"doi-asserted-by": "publisher",
"key": "ref131"
},
{
"DOI": "10.1128/JVI.00060-11",
"doi-asserted-by": "publisher",
"key": "ref132"
},
{
"DOI": "10.1074/jbc.RA119.008964",
"doi-asserted-by": "publisher",
"key": "ref133"
},
{
"DOI": "10.1016/j.cell.2020.10.039",
"doi-asserted-by": "publisher",
"key": "ref134"
},
{
"DOI": "10.1038/s41579-020-00468-6",
"doi-asserted-by": "publisher",
"key": "ref135"
},
{
"DOI": "10.1128/mBio.02371-21",
"doi-asserted-by": "publisher",
"key": "ref136"
},
{
"DOI": "10.1007/BF01314299",
"doi-asserted-by": "publisher",
"key": "ref137"
},
{
"DOI": "10.1038/s41419-018-0917-y",
"doi-asserted-by": "publisher",
"key": "ref138"
},
{
"DOI": "10.1016/j.devcel.2021.10.006",
"doi-asserted-by": "publisher",
"key": "ref139"
},
{
"DOI": "10.1016/j.devcel.2020.12.010",
"doi-asserted-by": "publisher",
"key": "ref140"
},
{
"DOI": "10.3389/fcell.2021.716208",
"doi-asserted-by": "publisher",
"key": "ref141"
},
{
"DOI": "10.1038/s41421-021-00268-z",
"doi-asserted-by": "publisher",
"key": "ref142"
},
{
"DOI": "10.4161/auto.29309",
"doi-asserted-by": "publisher",
"key": "ref143"
},
{
"DOI": "10.1016/j.antiviral.2006.05.010",
"doi-asserted-by": "publisher",
"key": "ref144"
},
{
"DOI": "10.1096/fj.201802418R",
"doi-asserted-by": "publisher",
"key": "ref145"
},
{
"DOI": "10.1016/j.febslet.2006.11.046",
"doi-asserted-by": "publisher",
"key": "ref146"
},
{
"DOI": "10.1128/mBio.02325-17",
"doi-asserted-by": "publisher",
"key": "ref147"
},
{
"DOI": "10.1038/s41467-021-24007-w",
"doi-asserted-by": "publisher",
"key": "ref148"
},
{
"DOI": "10.21203/rs.3.rs-419305/v1",
"doi-asserted-by": "publisher",
"key": "ref149"
},
{
"DOI": "10.3389/fncel.2021.777738",
"doi-asserted-by": "publisher",
"key": "ref150"
},
{
"DOI": "10.1016/j.cell.2020.10.030",
"doi-asserted-by": "publisher",
"key": "ref151"
},
{
"DOI": "10.1371/journal.ppat.1009127",
"doi-asserted-by": "publisher",
"key": "ref152"
},
{
"DOI": "10.1038/s41421-021-00338-2",
"doi-asserted-by": "publisher",
"key": "ref153"
},
{
"DOI": "10.1038/s41586-020-2286-9",
"doi-asserted-by": "publisher",
"key": "ref154"
},
{
"DOI": "10.1038/s41586-021-03493-4",
"doi-asserted-by": "publisher",
"key": "ref155"
},
{
"DOI": "10.3390/ijms22136937",
"doi-asserted-by": "publisher",
"key": "ref156"
},
{
"DOI": "10.1128/AAC.00819-20",
"doi-asserted-by": "publisher",
"key": "ref157"
},
{
"DOI": "10.1371/journal.pone.0260958",
"doi-asserted-by": "publisher",
"key": "ref158"
},
{
"DOI": "10.1371/journal.pone.0198389",
"doi-asserted-by": "publisher",
"key": "ref159"
},
{
"DOI": "10.1371/journal.pone.0246803",
"doi-asserted-by": "publisher",
"key": "ref160"
},
{
"DOI": "10.1016/j.lanepe.2021.100084",
"doi-asserted-by": "publisher",
"key": "ref161"
}
],
"reference-count": 161,
"references-count": 161,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.mdpi.com/1422-0067/23/9/4576"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"Inorganic Chemistry",
"Organic Chemistry",
"Physical and Theoretical Chemistry",
"Computer Science Applications",
"Spectroscopy",
"Molecular Biology",
"General Medicine",
"Catalysis"
],
"subtitle": [],
"title": "The Endolysosomal System: The Acid Test for SARS-CoV-2",
"type": "journal-article",
"volume": "23"
}