Bromhexine inhibits SARS-CoV-2 Omicron and variant pseudovirus infection via ACE2-targeted mechanisms
et al., Frontiers in Pharmacology, doi:10.3389/fphar.2025.1745277, Jan 2026
In vitro study showing that bromhexine inhibits SARS-CoV-2 Omicron and variant pseudovirus infection via ACE2-targeted mechanisms. Authors found that the antiviral mechanism involves destabilization of the SARS-CoV-2 spike-ACE2 interface, preventing viral entry through direct disruption of virus-host receptor interaction rather than solely through TMPRSS2 inhibition. The consistent effectiveness across multiple variants suggests bromhexine's potential as a broad-spectrum entry inhibitor for current and emerging SARS-CoV-2 variants.
Bromhexine efficacy may vary depending on the degree of TMPRSS-dependent fusion for different variants1,2.
6 preclinical studies support the efficacy of bromhexine for COVID-19:
In silico studies predict inhibition of SARS-CoV-2 with bromhexine or metabolites via binding to the spikeA,3, MproB,3, RNA-dependent RNA polymeraseC,3, and TMPRSS2D,4 proteins.
In vitro studies demonstrate inhibition of the TMPRSS2D,6 and acid sphingomyelinaseE,7 proteins.
Bromhexine is a mucolytic agent that helps thin mucus
secretions in the respiratory tract and has been shown to have antiviral
properties against respiratory viruses.
Bromhexine inhibits the expression of TMPRSS2 which plays an important role in SARS-CoV-2 cell entry and replication4,6,8 , may prevent SARS-CoV-2 infection by disrupting spike-ACE2 binding through direct interaction with the ACE2 receptor5, and bromhexine metabolite ambroxol inhibits SARS-CoV-2 via inhibition of acid sphingomyelinase in epithelial cells7.
1.
Willett et al., The hyper-transmissible SARS-CoV-2 Omicron variant exhibits significant antigenic change, vaccine escape and a switch in cell entry mechanism, medRxiv, doi:10.1101/2022.01.03.21268111.
2.
Peacock et al., The SARS-CoV-2 variant, Omicron, shows rapid replication in human primary nasal epithelial cultures and efficiently uses the endosomal route of entry, bioRxiv, doi:10.1101/2021.12.31.474653.
3.
Haque et al., Exploring potential therapeutic candidates against COVID-19: a molecular docking study, Discover Molecules, doi:10.1007/s44345-024-00005-5.
4.
Sgrignani et al., Computational Identification of a Putative Allosteric Binding Pocket in TMPRSS2, Frontiers in Molecular Biosciences, doi:10.3389/fmolb.2021.666626.
5.
Zúñiga et al., Bromhexine inhibits SARS-CoV-2 Omicron and variant pseudovirus infection via ACE2-targeted mechanisms, Frontiers in Pharmacology, doi:10.3389/fphar.2025.1745277.
6.
Martins et al., In Vitro Inhibition of SARS-CoV-2 Infection by Bromhexine hydrochloride, bioRxiv, doi:10.1101/2022.12.23.521817.
a.
The trimeric spike (S) protein is a glycoprotein that mediates viral entry by binding to the host ACE2 receptor, is critical for SARS-CoV-2's ability to infect host cells, and is a target of neutralizing antibodies. Inhibition of the spike protein prevents viral attachment, halting infection at the earliest stage.
b.
The main protease or Mpro, also known as 3CLpro or nsp5, is a cysteine protease that cleaves viral polyproteins into functional units needed for replication. Inhibiting Mpro disrupts the SARS-CoV-2 lifecycle within the host cell, preventing the creation of new copies.
c.
RNA-dependent RNA polymerase (RdRp), also called nsp12, is the core enzyme of the viral replicase-transcriptase complex that copies the positive-sense viral RNA genome into negative-sense templates for progeny RNA synthesis. Inhibiting RdRp blocks viral genome replication and transcription.
d.
Transmembrane protease serine 2 (TMPRSS2) is a host cell protease that primes the spike protein, facilitating cellular entry. TMPRSS2 activity helps enable cleavage of the spike protein required for membrane fusion and virus entry. Inhibition may especially protect respiratory epithelial cells, buy may have physiological effects.
e.
Acid sphingomyelinase (ASM) is a lysosomal enzyme that hydrolyzes sphingomyelin into ceramide and phosphorylcholine. ASM activity is upregulated by SARS-CoV-2 infection, leading to ceramide-enriched membrane domains that facilitate viral entry and replication. Inhibiting ASM may disrupt viral entry and reduce infection severity while potentially restoring membrane stability and immune homeostasis.
Zúñiga et al., 12 Jan 2026, Chile, peer-reviewed, 9 authors.
Contact: lzuniga@utalca.cl.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Bromhexine inhibits SARS-CoV-2 Omicron and variant pseudovirus infection via ACE2-targeted mechanisms
Frontiers in Pharmacology, doi:10.3389/fphar.2025.1745277
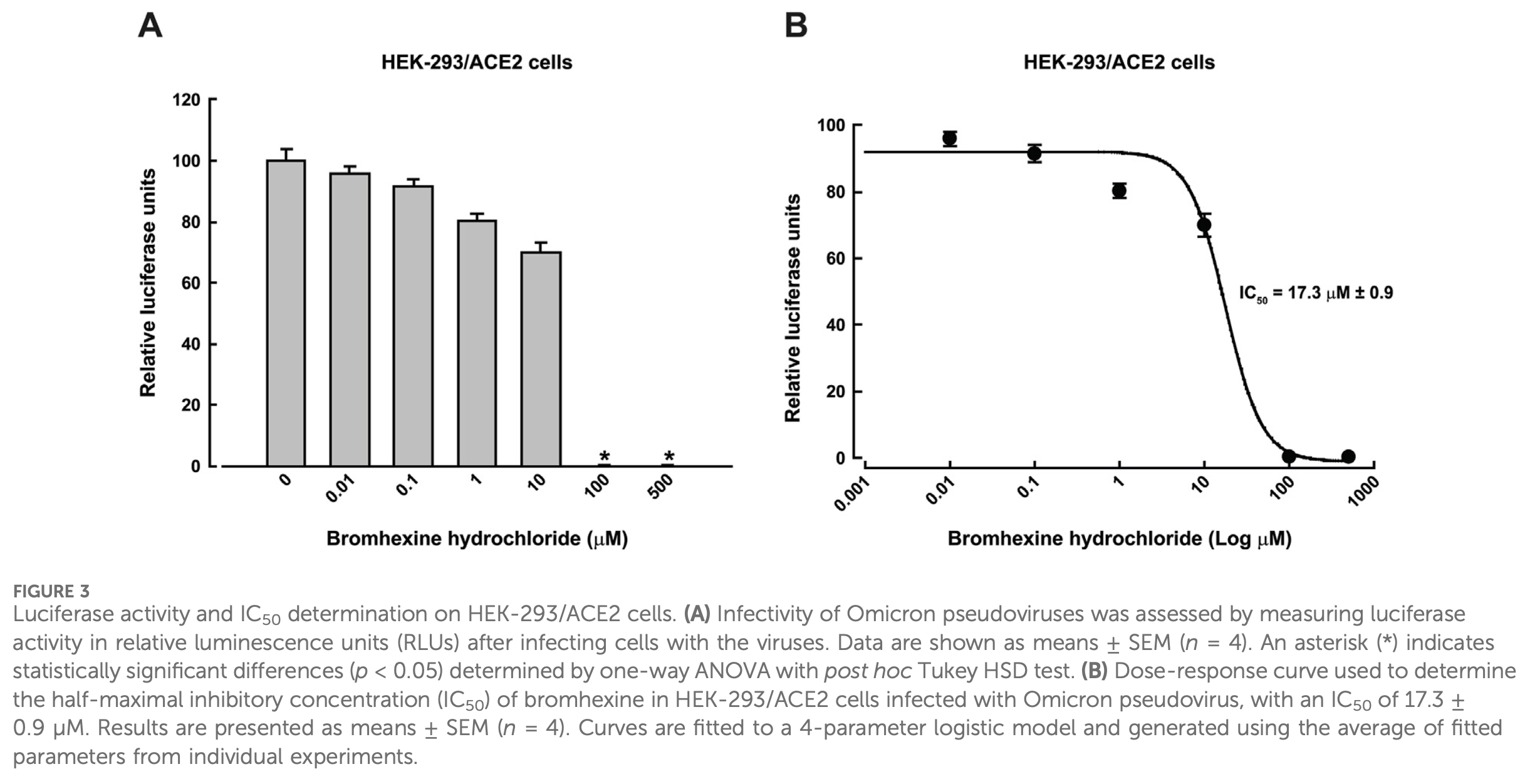
Background: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes a highly infectious disease characterized by fever, acute respiratory illness, and pneumonia, known as coronavirus disease 2019 (COVID-19). SARS-CoV-2 infects host cells through the interaction of its spike glycoprotein (S protein) with human angiotensin-converting enzyme 2 (hACE2). Structural studies have shown that hACE2 interacts exclusively with the receptor-binding domain (RBD) of the spike. A high binding affinity between spike and hACE2 has been linked to increased viral infection. Disrupting this interaction can reduce viral infectivity. Methods: This study aimed to assess infection using Omicron variant pseudovirus in a stable HEK-293 cell line expressing hACE2 (HEK-293/ACE2), treated with bromhexine hydrochloride. First, immunofluorescence and Western blot confirmed the presence of hACE2 in the stable line. Then, bromhexine concentrations for treatment were determined by cytotoxicity assays. Next, infection was evaluated using Omicron pseudoviruses carrying GFP and luciferase reporter genes. Infection levels were measured through fluorescence or luciferase activity. Results: Bromhexine reduced infection with an IC 50 of 17.3 ± 0.9 µM. About 40% inhibition was also observed against Alpha, Beta, and Delta variants at 40 µM. Computational docking followed by molecular dynamics simulations showed that bromhexine binds to the extracellular domain of hACE2, with recurrent contacts near Phe40, Phe390, and Asn394. Conclusion: Consistent with this model, our findings support an entry-inhibition mechanism whereby bromhexine destabilizes the SARS-CoV-2 spike-ACE2 interface, preventing viral entry. Overall, these results suggest bromhexine as a potential repurposing candidate and support its inclusion in therapeutic strategies aimed at both current and emerging SARS-CoV-2 variants.
Ethics statement Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
Conflict of interest The author(s) declared that this work was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement The author(s) declared that generative AI was not used in the creation of this manuscript. Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material The Supplementary Material for this article..
References
Alegría-Arcos, Barbosa, Sepúlveda, Combariza, González et al., Network pharmacology reveals multitarget mechanism of action of drugs to be repurposed for COVID-19, Front. Pharmacol, doi:10.3389/fphar.2022.952192
Andersen, Rambaut, Lipkin, Holmes, Garry, The proximal origin of SARS-CoV-2, Nat. Med, doi:10.1038/s41591-020-0820-9
Bosch, Van Der Zee, De Haan, Rottier, The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex, J. Virol, doi:10.1128/jvi.77.16.8801-8811.2003
Cai, Zhang, Xiao, Peng, Sterling et al., Distinct conformational states of SARS-CoV-2 spike protein, Science, doi:10.1126/science.abd4251
Chan, Kok, Zhu, Chu, To et al., Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting wuhan, Emerg. Microb. Infect, doi:10.1080/22221751.2020.1719902
Chen, Liu, Guo, Emerging coronaviruses: genome structure, replication, and pathogenesis, J. Med. Virol, doi:10.1002/jmv.25681
Dasmahapatra, Kumar, Das, Subramanian, Murali et al., In-silico molecular modelling, MM/GBSA binding free energy and molecular dynamics simulation study of novel pyrido fused imidazo[4,5-c]quinolines as potential anti-tumor agents, Front. Chem, doi:10.3389/fchem.2022.991369
Dilipkumar, Karthik, Dk, Gowramma, Lakshmanan, Insilico screening and molecular dynamics simulation of quinazolinone derivatives as PARP1 and STAT3 dual inhibitors: a novel DML approaches, J. Biomol. Struct. Dyn, doi:10.1080/07391102.2023.2259476
Donoghue, Hsieh, Baronas, Godbout, Gosselin et al., A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9, Circ. Res, doi:10.1161/01.RES.87.5.e1
Dutta, Acharya, Cryo-electron microscopy in the study of virus entry and infection, Front. Mol. Biosci, doi:10.3389/fmolb.2024.1429180
Fehr, Perlman, Coronaviruses: an overview of their replication and pathogenesis, Methods Mol. Biol, doi:10.1007/978-1-4939-2438-7_1
Gil, Ginex, Maestro, Nozal, Barrado-Gil et al., COVID-19: drug targets and potential treatments, J. Med. Chem, doi:10.1021/acs.jmedchem.0c00606
Gorbalenya, Baker, Baric, De Groot, Drosten et al., The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2, Nat. Microbiol, doi:10.1038/s41564-020-0695-z
Hardenbrook, Zhang, A structural view of the SARS-CoV-2 virus and its assembly, Curr. Opin. Virol, doi:10.1016/j.coviro.2021.11.011
Hoffmann, Kleine-Weber, Pöhlmann, A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells, Mol. Cell, doi:10.1016/j.molcel.2020.04.022
Hoffmann, Kleine-Weber, Schroeder, Krüger, Herrler et al., SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor, Cell, doi:10.1016/j.cell.2020.02.052
Jackson, Farzan, Chen, Choe, Mechanisms of SARS-CoV-2 entry into cells, Nat. Rev. Mol. Cell Biol, doi:10.1038/s41580-021-00418-x
Kehinde, Egbejimi, Kaur, Onyenaka, Adebusuyi et al., Inhibitory mechanism of ambroxol and bromhexine hydrochlorides as potent blockers of molecular interaction between SARS-CoV-2 spike protein and human angiotensinconverting Enzyme-2, J. Mol. Graph. Model, doi:10.1016/j.jmgm.2022.108201
Li, Moore, Vasilieva, Sui, Wong et al., Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus, Nature, doi:10.1038/nature02145
Lu, Wu, Ghoreishi, Chen, Wang et al., OPLS4: improving force field accuracy on challenging regimes of chemical space, J. Chem. Theory Comp, doi:10.1021/acs.jctc.1c00302
Lu, Zhao, Li, Niu, Yang et al., Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding, Lancet, doi:10.1016/S0140-6736(20)30251-8
Lucas, Heinlein, Kim, Hernandez, Malik et al., The androgen-regulated protease TMPRSS2 activates a proteolytic cascade involving components of the tumor microenvironment and promotes prostate cancer metastasis, Cancer Discov, doi:10.1158/2159-8290.CD-13-1010
Pirolli, Righino, Camponeschi, Ria, Di Sante et al., Virtual screening and molecular dynamics simulations provide insight into repurposing drugs against SARS-CoV-2 variants spike protein/ACE2 interface, Sci. Rep, doi:10.1038/s41598-023-28716-8
Sastry, Adzhigirey, Day, Annabhimoju, Sherman, Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments, J. Comput. Aided Mol. Des, doi:10.1007/s10822-013-9644-8
Shang, Wan, Luo, Ye, Geng et al., Cell entry mechanisms of SARS-CoV-2, Proc. Natl. Acad. Sci. U. S. A, doi:10.1073/pnas.2003138117
Shen, Mao, Wu, Tanaka, Zhang, TMPRSS2: a potential target for treatment of influenza virus and coronavirus infections, Biochimie, doi:10.1016/j.biochi.2017.07.016
Stadler, Masignani, Eickmann, Becker, Abrignani et al., SARS -beginning to understand a new virus, Nat. Rev. Microbiol, doi:10.1038/nrmicro775
Sándor, Kiss, Keserű, Virtual fragment docking by glide: a validation study on 190 protein-fragment complexes, J. Chem. Inf. Model, doi:10.1021/ci1000407
Tikellis, Bernardi, Burns, Angiotensin-converting enzyme 2 is a key modulator of the renin-angiotensin system in cardiovascular and renal disease, Curr. Opin. Nephrol. Hypertens, doi:10.1097/MNH.0b013e328341164a
Walls, Park, Tortorici, Wall, Mcguire et al., Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein, Cell, doi:10.1016/j.cell.2020.02.058
Wang, Horby, Hayden, Gao, A novel coronavirus outbreak of global health concern, Pharmacology frontiersin
Wang, Wang, Luo, Huang, Xiao et al., A comprehensive investigation of the mRNA and protein level of ACE2, the putative receptor of SARS-CoV-2, in human tissues and blood cells, Int. J. Med. Sci, doi:10.7150/ijms.46695
Wang, Zhang, Wu, Niu, Song et al., Structural and functional basis of SARS-CoV-2 entry by using human ACE2, Cell, doi:10.1016/j.cell.2020.03.045
Wang, Zhao, Gao, Gao, Wang et al., SARS-CoV-2: structure, biology, and structure-based therapeutics development, Front. Cell. Infect. Microbiol, doi:10.3389/fcimb.2020.587269
Wrapp, Wang, Corbett, Goldsmith, Hsieh et al., Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation, Science, doi:10.1126/science.abb2507
Wu, Peng, Huang, Ding, Wang et al., Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China, Cell Host Microbe, doi:10.1016/j.chom.2020.02.001
Xiao, Lu, Zhang, Johnson, Mckay et al., A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent, Nat. Struct. Mol. Biol, doi:10.1038/s41594-020-00549-3
Yan, Zhang, Li, Xia, Guo et al., Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2, Science, doi:10.1126/science.abb2762
Yang, Rao, Structural biology of SARS-CoV-2 and implications for therapeutic development, Nat. Rev. Microbiol, doi:10.1038/s41579-021-00630-8
Zhou, Tsybovsky, Gorman, Rapp, Cerutti et al., Cryo-EM structures of SARS-CoV-2 spike without and with ACE2 reveal a pHdependent switch to mediate endosomal positioning of receptor-binding domains, Cell Host Microbe, doi:10.1016/j.chom.2020.11.004
Zhou, Yang, Huang, Jiang, Du, Advances in MERS-CoV vaccines and therapeutics based on the receptor-binding domain, Viruses, doi:10.3390/v11010060
Zhou, Yang, Wang, Hu, Zhang et al., A pneumonia outbreak associated with a new coronavirus of probable bat origin, Nature, doi:10.1038/s41586-020-2012-7
Zhu, Zhang, Wang, Li, Yang et al., A novel coronavirus from patients with pneumonia in China, N. Engl. J. Med, doi:10.1056/NEJMoa2001017
DOI record:
{
"DOI": "10.3389/fphar.2025.1745277",
"ISSN": [
"1663-9812"
],
"URL": "http://dx.doi.org/10.3389/fphar.2025.1745277",
"abstract": "<jats:sec>\n <jats:title>Background</jats:title>\n <jats:p>Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) causes a highly infectious disease characterized by fever, acute respiratory illness, and pneumonia, known as coronavirus disease 2019 (COVID‐19). SARS‐CoV‐2 infects host cells through the interaction of its spike glycoprotein (S protein) with human angiotensin‐converting enzyme 2 (hACE2). Structural studies have shown that hACE2 interacts exclusively with the receptor‐binding domain (RBD) of the spike. A high binding affinity between spike and hACE2 has been linked to increased viral infection. Disrupting this interaction can reduce viral infectivity.</jats:p>\n </jats:sec>\n <jats:sec>\n <jats:title>Methods</jats:title>\n <jats:p>This study aimed to assess infection using Omicron variant pseudovirus in a stable HEK‐293 cell line expressing hACE2 (HEK‐293/ACE2), treated with bromhexine hydrochloride. First, immunofluorescence and Western blot confirmed the presence of hACE2 in the stable line. Then, bromhexine concentrations for treatment were determined by cytotoxicity assays. Next, infection was evaluated using Omicron pseudoviruses carrying GFP and luciferase reporter genes. Infection levels were measured through fluorescence or luciferase activity.</jats:p>\n </jats:sec>\n <jats:sec>\n <jats:title>Results</jats:title>\n <jats:p>\n Bromhexine reduced infection with an IC\n <jats:sub>50</jats:sub>\n of 17.3 ± 0.9 μM. About 40% inhibition was also observed against Alpha, Beta, and Delta variants at 40 μM. Computational docking followed by molecular dynamics simulations showed that bromhexine binds to the extracellular domain of hACE2, with recurrent contacts near Phe40, Phe390, and Asn394.\n </jats:p>\n </jats:sec>\n <jats:sec>\n <jats:title>Conclusion</jats:title>\n <jats:p>Consistent with this model, our findings support an entry‐inhibition mechanism whereby bromhexine destabilizes the SARS‐CoV‐2 spike–ACE2 interface, preventing viral entry. Overall, these results suggest bromhexine as a potential repurposing candidate and support its inclusion in therapeutic strategies aimed at both current and emerging SARS‐CoV‐2 variants.</jats:p>\n </jats:sec>",
"alternative-id": [
"10.3389/fphar.2025.1745277"
],
"article-number": "1745277",
"author": [
{
"affiliation": [],
"family": "Zúñiga",
"given": "Rafael",
"sequence": "first"
},
{
"affiliation": [],
"family": "Venturini",
"given": "Whitney",
"sequence": "additional"
},
{
"affiliation": [],
"family": "González",
"given": "Natalia",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Valenzuela-Hormazábal",
"given": "Paulina",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Sánchez-Aros",
"given": "Laura",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Ramírez",
"given": "David",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Cayo",
"given": "Angel",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Vilos",
"given": "Cristian",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Zúñiga",
"given": "Leandro",
"sequence": "additional"
}
],
"container-title": "Frontiers in Pharmacology",
"container-title-short": "Front. Pharmacol.",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"frontiersin.org"
]
},
"created": {
"date-parts": [
[
2026,
1,
12
]
],
"date-time": "2026-01-12T08:12:31Z",
"timestamp": 1768205551000
},
"deposited": {
"date-parts": [
[
2026,
1,
12
]
],
"date-time": "2026-01-12T08:12:34Z",
"timestamp": 1768205554000
},
"indexed": {
"date-parts": [
[
2026,
1,
12
]
],
"date-time": "2026-01-12T11:23:43Z",
"timestamp": 1768217023691,
"version": "3.49.0"
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2026,
1,
12
]
]
},
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2026,
1,
12
]
],
"date-time": "2026-01-12T00:00:00Z",
"timestamp": 1768176000000
}
}
],
"link": [
{
"URL": "https://www.frontiersin.org/articles/10.3389/fphar.2025.1745277/full",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1965",
"original-title": [],
"prefix": "10.3389",
"published": {
"date-parts": [
[
2026,
1,
12
]
]
},
"published-online": {
"date-parts": [
[
2026,
1,
12
]
]
},
"publisher": "Frontiers Media SA",
"reference": [
{
"DOI": "10.3389/fphar.2022.952192",
"article-title": "Network pharmacology reveals multitarget mechanism of action of drugs to be repurposed for COVID-19",
"author": "Alegría-Arcos",
"doi-asserted-by": "publisher",
"first-page": "952192",
"journal-title": "Front. Pharmacol.",
"key": "B1",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.1038/s41591-020-0820-9",
"article-title": "The proximal origin of SARS-CoV-2",
"author": "Andersen",
"doi-asserted-by": "publisher",
"first-page": "450",
"journal-title": "Nat. Med.",
"key": "B2",
"volume": "26",
"year": "2020"
},
{
"DOI": "10.1128/jvi.77.16.8801-8811.2003",
"article-title": "The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex",
"author": "Bosch",
"doi-asserted-by": "publisher",
"first-page": "8801",
"journal-title": "J. Virol.",
"key": "B3",
"volume": "77",
"year": "2003"
},
{
"DOI": "10.1126/science.abd4251",
"article-title": "Distinct conformational states of SARS-CoV-2 spike protein",
"author": "Cai",
"doi-asserted-by": "publisher",
"first-page": "1586",
"journal-title": "Science",
"key": "B4",
"volume": "369",
"year": "2020"
},
{
"DOI": "10.1080/22221751.2020.1719902",
"article-title": "Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting wuhan",
"author": "Chan",
"doi-asserted-by": "publisher",
"first-page": "221",
"journal-title": "Emerg. Microb. Infect.",
"key": "B5",
"volume": "9",
"year": "2020"
},
{
"DOI": "10.1002/jmv.25681",
"article-title": "Emerging coronaviruses: genome structure, replication, and pathogenesis",
"author": "Chen",
"doi-asserted-by": "publisher",
"first-page": "418",
"journal-title": "J. Med. Virol.",
"key": "B6",
"volume": "92",
"year": "2020"
},
{
"DOI": "10.3389/fchem.2022.991369",
"article-title": "In-silico molecular modelling, MM/GBSA binding free energy and molecular dynamics simulation study of novel pyrido fused imidazo[4,5-c]quinolines as potential anti-tumor agents",
"author": "Dasmahapatra",
"doi-asserted-by": "publisher",
"first-page": "991369",
"journal-title": "Front. Chem.",
"key": "B7",
"volume": "10",
"year": "2022"
},
{
"DOI": "10.1080/07391102.2023.2259476",
"article-title": "In-silico screening and molecular dynamics simulation of quinazolinone derivatives as PARP1 and STAT3 dual inhibitors: a novel DML approaches",
"author": "Dilipkumar",
"doi-asserted-by": "publisher",
"first-page": "10824",
"journal-title": "J. Biomol. Struct. Dyn.",
"key": "B8",
"volume": "42",
"year": "2024"
},
{
"DOI": "10.1161/01.RES.87.5.e1",
"article-title": "A novel angiotensin-converting enzyme–related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9",
"author": "Donoghue",
"doi-asserted-by": "publisher",
"first-page": "e1",
"journal-title": "Circ. Res.",
"key": "B9",
"volume": "87",
"year": "2000"
},
{
"DOI": "10.3389/fmolb.2024.1429180",
"article-title": "Cryo-electron microscopy in the study of virus entry and infection",
"author": "Dutta",
"doi-asserted-by": "publisher",
"first-page": "1429180",
"journal-title": "Front. Mol. Biosci.",
"key": "B10",
"volume": "11",
"year": "2024"
},
{
"DOI": "10.1007/978-1-4939-2438-7_1",
"article-title": "Coronaviruses: an overview of their replication and pathogenesis",
"author": "Fehr",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Methods Mol. Biol.",
"key": "B11",
"volume": "1282",
"year": "2015"
},
{
"DOI": "10.1021/acs.jmedchem.0c00606",
"article-title": "COVID-19: drug targets and potential treatments",
"author": "Gil",
"doi-asserted-by": "publisher",
"first-page": "12359",
"journal-title": "J. Med. Chem.",
"key": "B12",
"volume": "63",
"year": "2020"
},
{
"DOI": "10.1038/s41564-020-0695-z",
"article-title": "The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2",
"author": "Gorbalenya",
"doi-asserted-by": "publisher",
"first-page": "536",
"journal-title": "Nat. Microbiol.",
"key": "B13",
"volume": "5",
"year": "2020"
},
{
"DOI": "10.1016/j.coviro.2021.11.011",
"article-title": "A structural view of the SARS-CoV-2 virus and its assembly",
"author": "Hardenbrook",
"doi-asserted-by": "publisher",
"first-page": "123",
"journal-title": "Curr. Opin. Virol.",
"key": "B14",
"volume": "52",
"year": "2022"
},
{
"DOI": "10.1016/j.molcel.2020.04.022",
"article-title": "A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells",
"author": "Hoffmann",
"doi-asserted-by": "publisher",
"first-page": "779",
"journal-title": "Mol. Cell",
"key": "B15",
"volume": "78",
"year": ""
},
{
"DOI": "10.1016/j.cell.2020.02.052",
"article-title": "SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor",
"author": "Hoffmann",
"doi-asserted-by": "publisher",
"first-page": "271",
"journal-title": "Cell",
"key": "B16",
"volume": "181",
"year": ""
},
{
"DOI": "10.1038/s41580-021-00418-x",
"article-title": "Mechanisms of SARS-CoV-2 entry into cells",
"author": "Jackson",
"doi-asserted-by": "publisher",
"first-page": "3",
"journal-title": "Nat. Rev. Mol. Cell Biol.",
"key": "B17",
"volume": "23",
"year": "2022"
},
{
"DOI": "10.1016/j.jmgm.2022.108201",
"article-title": "Inhibitory mechanism of ambroxol and bromhexine hydrochlorides as potent blockers of molecular interaction between SARS-CoV-2 spike protein and human angiotensin-converting Enzyme-2",
"author": "Kehinde",
"doi-asserted-by": "publisher",
"first-page": "108201",
"journal-title": "J. Mol. Graph. Model.",
"key": "B18",
"volume": "114",
"year": "2022"
},
{
"DOI": "10.1038/nature02145",
"article-title": "Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus",
"author": "Li",
"doi-asserted-by": "publisher",
"first-page": "450",
"journal-title": "Nature",
"key": "B19",
"volume": "426",
"year": "2003"
},
{
"DOI": "10.1016/S0140-6736(20)30251-8",
"article-title": "Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding",
"author": "Lu",
"doi-asserted-by": "publisher",
"first-page": "565",
"journal-title": "Lancet",
"key": "B20",
"volume": "395",
"year": "2020"
},
{
"DOI": "10.1021/acs.jctc.1c00302",
"article-title": "OPLS4: improving force field accuracy on challenging regimes of chemical space",
"author": "Lu",
"doi-asserted-by": "publisher",
"first-page": "4291",
"journal-title": "J. Chem. Theory Comp.",
"key": "B21",
"volume": "17",
"year": "2021"
},
{
"DOI": "10.1158/2159-8290.CD-13-1010",
"article-title": "The androgen-regulated protease TMPRSS2 activates a proteolytic cascade involving components of the tumor microenvironment and promotes prostate cancer metastasis",
"author": "Lucas",
"doi-asserted-by": "publisher",
"first-page": "1310",
"journal-title": "Cancer Discov.",
"key": "B22",
"volume": "4",
"year": "2014"
},
{
"DOI": "10.1038/s41598-023-28716-8",
"article-title": "Virtual screening and molecular dynamics simulations provide insight into repurposing drugs against SARS-CoV-2 variants spike protein/ACE2 interface",
"author": "Pirolli",
"doi-asserted-by": "publisher",
"first-page": "1494",
"journal-title": "Sci. Rep.",
"key": "B23",
"volume": "13",
"year": "2023"
},
{
"DOI": "10.1021/ci1000407",
"article-title": "Virtual fragment docking by glide: a validation study on 190 protein−fragment complexes",
"author": "Sándor",
"doi-asserted-by": "publisher",
"first-page": "1165",
"journal-title": "J. Chem. Inf. Model.",
"key": "B24",
"volume": "50",
"year": "2010"
},
{
"DOI": "10.1007/s10822-013-9644-8",
"article-title": "Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments",
"author": "Sastry",
"doi-asserted-by": "publisher",
"first-page": "221",
"journal-title": "J. Comput. Aided Mol. Des.",
"key": "B25",
"volume": "27",
"year": "2013"
},
{
"DOI": "10.1073/pnas.2003138117",
"article-title": "Cell entry mechanisms of SARS-CoV-2",
"author": "Shang",
"doi-asserted-by": "publisher",
"first-page": "11727",
"journal-title": "Proc. Natl. Acad. Sci. U. S. A.",
"key": "B26",
"volume": "117",
"year": "2020"
},
{
"DOI": "10.1016/j.biochi.2017.07.016",
"article-title": "TMPRSS2: a potential target for treatment of influenza virus and coronavirus infections",
"author": "Shen",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Biochimie",
"key": "B27",
"volume": "142",
"year": "2017"
},
{
"DOI": "10.1038/nrmicro775",
"article-title": "SARS — beginning to understand a new virus",
"author": "Stadler",
"doi-asserted-by": "publisher",
"first-page": "209",
"journal-title": "Nat. Rev. Microbiol.",
"key": "B28",
"volume": "1",
"year": "2003"
},
{
"DOI": "10.1097/MNH.0b013e328341164a",
"article-title": "Angiotensin-converting enzyme 2 is a key modulator of the renin-angiotensin system in cardiovascular and renal disease",
"author": "Tikellis",
"doi-asserted-by": "publisher",
"first-page": "62",
"journal-title": "Curr. Opin. Nephrol. Hypertens.",
"key": "B29",
"volume": "20",
"year": "2011"
},
{
"DOI": "10.1016/j.cell.2020.02.058",
"article-title": "Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein",
"author": "Walls",
"doi-asserted-by": "publisher",
"first-page": "281",
"journal-title": "Cell",
"key": "B30",
"volume": "181",
"year": "2020"
},
{
"DOI": "10.1016/S0140-6736(20)30185-9",
"article-title": "A novel coronavirus outbreak of global health concern",
"author": "Wang",
"doi-asserted-by": "publisher",
"first-page": "470",
"journal-title": "Lancet",
"key": "B31",
"volume": "395",
"year": ""
},
{
"DOI": "10.3389/fcimb.2020.587269",
"article-title": "SARS-CoV-2: structure, biology, and structure-based therapeutics development",
"author": "Wang",
"doi-asserted-by": "publisher",
"first-page": "587269",
"journal-title": "Front. Cell. Infect. Microbiol.",
"key": "B32",
"volume": "10",
"year": ""
},
{
"DOI": "10.1016/j.cell.2020.03.045",
"article-title": "Structural and functional basis of SARS-CoV-2 entry by using human ACE2",
"author": "Wang",
"doi-asserted-by": "publisher",
"first-page": "894",
"journal-title": "Cell",
"key": "B33",
"volume": "181",
"year": ""
},
{
"DOI": "10.7150/ijms.46695",
"article-title": "A comprehensive investigation of the mRNA and protein level of ACE2, the putative receptor of SARS-CoV-2, in human tissues and blood cells",
"author": "Wang",
"doi-asserted-by": "publisher",
"first-page": "1522",
"journal-title": "Int. J. Med. Sci.",
"key": "B34",
"volume": "17",
"year": ""
},
{
"DOI": "10.1126/science.abb2507",
"article-title": "Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation",
"author": "Wrapp",
"doi-asserted-by": "publisher",
"first-page": "1260",
"journal-title": "Science",
"key": "B35",
"volume": "367",
"year": "2020"
},
{
"DOI": "10.1016/j.chom.2020.02.001",
"article-title": "Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China",
"author": "Wu",
"doi-asserted-by": "publisher",
"first-page": "325",
"journal-title": "Cell Host Microbe",
"key": "B36",
"volume": "27",
"year": "2020"
},
{
"DOI": "10.1038/s41594-020-00549-3",
"article-title": "A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent",
"author": "Xiao",
"doi-asserted-by": "publisher",
"first-page": "202",
"journal-title": "Nat. Struct. Mol. Biol.",
"key": "B37",
"volume": "28",
"year": "2021"
},
{
"DOI": "10.1126/science.abb2762",
"article-title": "Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2",
"author": "Yan",
"doi-asserted-by": "publisher",
"first-page": "1444",
"journal-title": "Science",
"key": "B38",
"volume": "367",
"year": "2020"
},
{
"DOI": "10.1038/s41579-021-00630-8",
"article-title": "Structural biology of SARS-CoV-2 and implications for therapeutic development",
"author": "Yang",
"doi-asserted-by": "publisher",
"first-page": "685",
"journal-title": "Nat. Rev. Microbiol.",
"key": "B39",
"volume": "19",
"year": "2021"
},
{
"DOI": "10.3390/v11010060",
"article-title": "Advances in MERS-CoV vaccines and therapeutics based on the receptor-binding domain",
"author": "Zhou",
"doi-asserted-by": "publisher",
"first-page": "60",
"journal-title": "Viruses",
"key": "B40",
"volume": "11",
"year": "2019"
},
{
"DOI": "10.1038/s41586-020-2012-7",
"article-title": "A pneumonia outbreak associated with a new coronavirus of probable bat origin",
"author": "Zhou",
"doi-asserted-by": "publisher",
"first-page": "270",
"journal-title": "Nature",
"key": "B41",
"volume": "579",
"year": ""
},
{
"DOI": "10.1016/j.chom.2020.11.004",
"article-title": "Cryo-EM structures of SARS-CoV-2 spike without and with ACE2 reveal a pH-dependent switch to mediate endosomal positioning of receptor-binding domains",
"author": "Zhou",
"doi-asserted-by": "publisher",
"first-page": "867",
"journal-title": "Cell Host Microbe",
"key": "B42",
"volume": "28",
"year": ""
},
{
"DOI": "10.1056/NEJMoa2001017",
"article-title": "A novel coronavirus from patients with pneumonia in China, 2019",
"author": "Zhu",
"doi-asserted-by": "publisher",
"first-page": "727",
"journal-title": "N. Engl. J. Med.",
"key": "B43",
"volume": "382",
"year": "2020"
}
],
"reference-count": 43,
"references-count": 43,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.frontiersin.org/articles/10.3389/fphar.2025.1745277/full"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Bromhexine inhibits SARS-CoV-2 Omicron and variant pseudovirus infection via ACE2-targeted mechanisms",
"type": "journal-article",
"update-policy": "https://doi.org/10.3389/crossmark-policy",
"volume": "16"
}