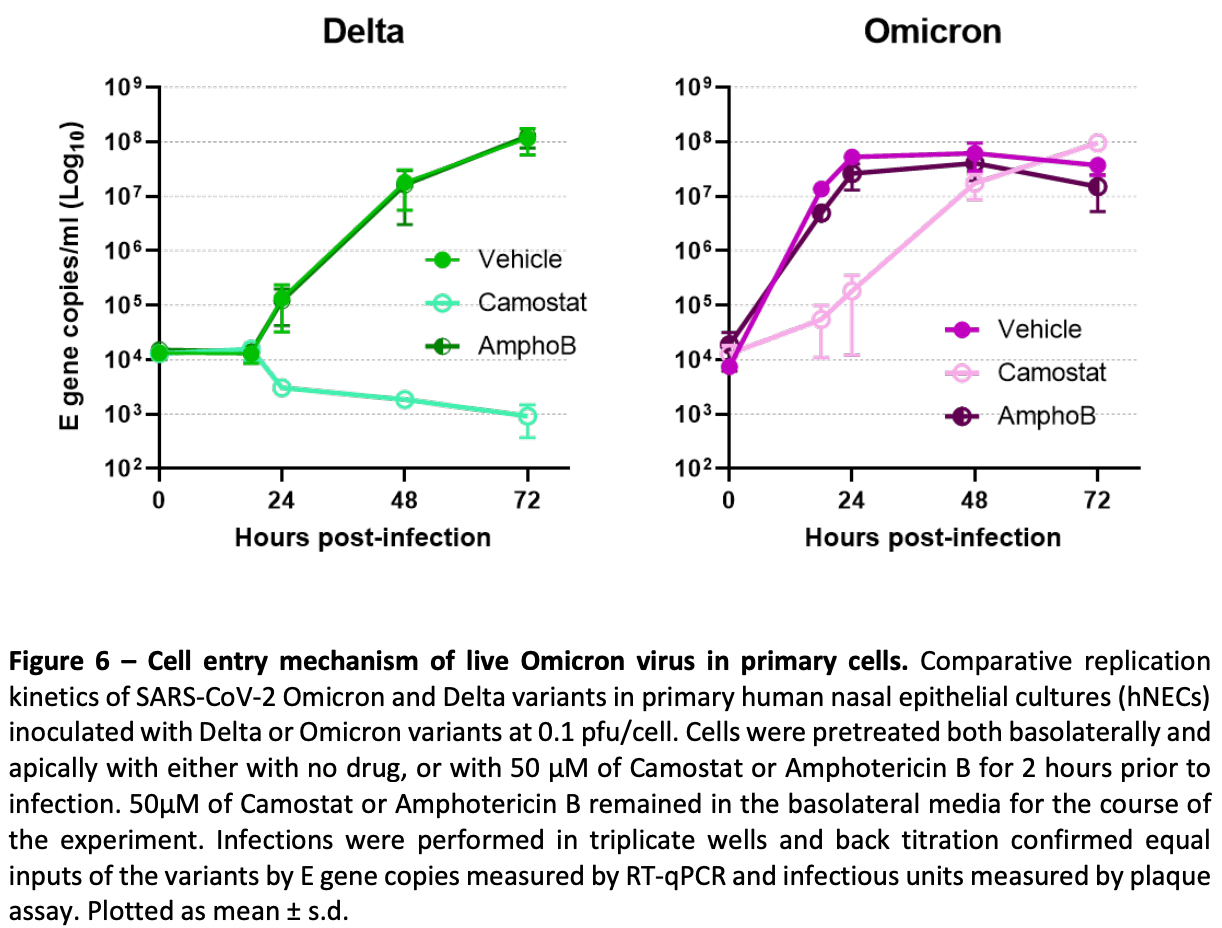
The SARS-CoV-2 variant, Omicron, shows rapid replication in human primary nasal epithelial cultures and efficiently uses the endosomal route of entry
et al., bioRxiv, doi:10.1101/2021.12.31.474653, Jan 2022
In vitro study showing that omicron can efficiently enter cells via the endosomal route, independent of TMPRSS2.
6 preclinical studies support the efficacy of bromhexine for COVID-19:
In silico studies predict inhibition of SARS-CoV-2 with bromhexine or metabolites via binding to the spikeA,1, MproB,1, RNA-dependent RNA polymeraseC,1, and TMPRSS2D,2 proteins.
In vitro studies demonstrate inhibition of the TMPRSS2D,4 and acid sphingomyelinaseE,5 proteins.
Bromhexine is a mucolytic agent that helps thin mucus
secretions in the respiratory tract and has been shown to have antiviral
properties against respiratory viruses.
Bromhexine inhibits the expression of TMPRSS2 which plays an important role in SARS-CoV-2 cell entry and replication2,4,6 , may prevent SARS-CoV-2 infection by disrupting spike-ACE2 binding through direct interaction with the ACE2 receptor3, and bromhexine metabolite ambroxol inhibits SARS-CoV-2 via inhibition of acid sphingomyelinase in epithelial cells5.
1.
Haque et al., Exploring potential therapeutic candidates against COVID-19: a molecular docking study, Discover Molecules, doi:10.1007/s44345-024-00005-5.
2.
Sgrignani et al., Computational Identification of a Putative Allosteric Binding Pocket in TMPRSS2, Frontiers in Molecular Biosciences, doi:10.3389/fmolb.2021.666626.
3.
Zúñiga et al., Bromhexine inhibits SARS-CoV-2 Omicron and variant pseudovirus infection via ACE2-targeted mechanisms, Frontiers in Pharmacology, doi:10.3389/fphar.2025.1745277.
4.
Martins et al., In Vitro Inhibition of SARS-CoV-2 Infection by Bromhexine hydrochloride, bioRxiv, doi:10.1101/2022.12.23.521817.
a.
The trimeric spike (S) protein is a glycoprotein that mediates viral entry by binding to the host ACE2 receptor, is critical for SARS-CoV-2's ability to infect host cells, and is a target of neutralizing antibodies. Inhibition of the spike protein prevents viral attachment, halting infection at the earliest stage.
b.
The main protease or Mpro, also known as 3CLpro or nsp5, is a cysteine protease that cleaves viral polyproteins into functional units needed for replication. Inhibiting Mpro disrupts the SARS-CoV-2 lifecycle within the host cell, preventing the creation of new copies.
c.
RNA-dependent RNA polymerase (RdRp), also called nsp12, is the core enzyme of the viral replicase-transcriptase complex that copies the positive-sense viral RNA genome into negative-sense templates for progeny RNA synthesis. Inhibiting RdRp blocks viral genome replication and transcription.
d.
Transmembrane protease serine 2 (TMPRSS2) is a host cell protease that primes the spike protein, facilitating cellular entry. TMPRSS2 activity helps enable cleavage of the spike protein required for membrane fusion and virus entry. Inhibition may especially protect respiratory epithelial cells, buy may have physiological effects.
e.
Acid sphingomyelinase (ASM) is a lysosomal enzyme that hydrolyzes sphingomyelin into ceramide and phosphorylcholine. ASM activity is upregulated by SARS-CoV-2 infection, leading to ceramide-enriched membrane domains that facilitate viral entry and replication. Inhibiting ASM may disrupt viral entry and reduce infection severity while potentially restoring membrane stability and immune homeostasis.
Peacock et al., 3 Jan 2022, preprint, 10 authors.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
The altered entry pathway and antigenic distance of the SARS-CoV-2 Omicron variant map to separate domains of spike protein
doi:10.1101/2021.12.31.474653
At the end of 2021 a new SARS-CoV-2 variant, Omicron, emerged and quickly spread across the world. It has been demonstrated that Omicron's high number of Spike mutations lead to partial immune evasion from even polyclonal antibody responses, allowing frequent re-infection and vaccine breakthroughs. However, it seems unlikely these antigenic differences alone explain its rapid growth; here we show Omicron replicates rapidly in human primary airway cultures, more so even than the previously dominant variant of concern, Delta. Omicron Spike continues to use human ACE2 as its primary receptor, to which it binds more strongly than other variants. Omicron Spike mediates enhanced entry into cells expressing several different animal ACE2s, including various domestic avian species, horseshoe bats and mice suggesting it has an increased propensity for reverse zoonosis and is more likely than previous variants to establish an animal reservoir of SARS-CoV-2. Unlike other SARS-CoV-2 variants, however, Omicron Spike has a diminished ability to induce syncytia formation. Furthermore, Omicron is capable of efficiently entering cells in a TMPRSS2-independent manner, via the endosomal route. We posit this enables Omicron to infect a greater number of cells in the respiratory epithelium, allowing it to be more infectious at lower exposure doses, and resulting in enhanced intrinsic transmissibility. .
Supplementary Figures Supplementary Figure S1
Supplementary Figure S2 -Different species ACE2 preference of different variants of concern. Receptor usage was screened using pseudoviruses expressing the indicated Spike proteins into 293Ts expressing the indicated ACE2 protein. Viral entry was measured by assaying luciferase activity (RLU) using the BrightGlo reagent (Promega).
References
Abdelnabi, SARS-CoV-2 variant of concern does not readily infect Syrian hamsters, The omicron, doi:10.1101/2021.12.24.474086
Bentley, SARS-CoV-2 Omicron-B.1.1.529 Variant leads to less severe disease than Pango B and Delta variants strains in a mouse model of severe COVID-19, bioRxiv, doi:10.1101/2021.12.26.474085
Braga, Drugs that inhibit TMEM16 proteins block SARS-CoV-2 spike-induced syncytia, Nature, doi:10.1038/s41586-021-03491-6
Cameroni, Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift, bioRxiv, doi:10.1101/2021.12.12.472269
Cao, Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies, bioRxiv, doi:10.1101/2021.12.07.470392
Cele, SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection, medRxiv, doi:10.1101/2021.12.08.21267417
Conceicao, The SARS-CoV-2 Spike protein has a broad tropism for mammalian ACE2 proteins, PLoS Biol, doi:10.1371/journal.pbio.3001016
Corman, Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR, Eurosurveillance, doi:10.2807/1560-7917.ES.2020.25.3.2000045
David, A common <em>TMPRSS2</em> variant protects against severe COVID-19, medRxiv, doi:10.1101/2021.03.04.21252931
Dinnon, A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures, Nature, doi:10.1038/s41586-020-2708-8
Edie, Survey of human chromosome 21 gene expression effects on early development in Danio rerio. G3: Genes, Genomes, Genetics
Ferguson, Report 49 -Growth, population distribution and immune escape of Omicron in England
Hale, SARS-CoV-2 infection in free-ranging white-tailed deer, Nature, doi:10.1038/s41586-021-04353-x
Heurich, TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein, J Virol, doi:10.1128/jvi.02202-13
Hoffmann, Kleine-Weber, Pöhlmann, A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells, Mol Cell, doi:10.1016/j.molcel.2020.04.022
Hoffmann, The Omicron variant is highly resistant against antibody-mediated neutralization -implications for control of the COVID-19 pandemic, Cell, doi:10.1016/j.cell.2021.12.032
Ishikawa, Meng, Kondo, Iwamoto, Matsuda, Generation of a dual-functional split-reporter protein for monitoring membrane fusion using self-associating split GFP, Protein Eng Des Sel, doi:10.1093/protein/gzs051
Johnson, Furin Cleavage Site Is Key to SARS-CoV-2 Pathogenesis, bioRxiv, doi:10.1101/2020.08.26.268854
Liu, Striking Antibody Evasion Manifested by the Omicron Variant of SARS-CoV-2, bioRxiv, doi:10.1101/2021.12.14.472719
Mckay, Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice, Nat Commun, doi:10.1038/s41467-020-17409-9
Meng, SARS-CoV-2 Omicron spike mediated immune escape, infectivity and cell-cell fusion, doi:10.1101/2021.12.17.473248
Mlcochova, SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion, Nature, doi:10.1038/s41586-021-03944-y
Muruato, Mouse Adapted SARS-CoV-2 protects animals from lethal SARS-CoV challenge, bioRxiv, doi:10.1101/2021.05.03.442357
Muus, Single-cell meta-analysis of SARS-CoV-2 entry genes across tissues and demographics, Nature Medicine, doi:10.1038/s41591-020-01227-z
Newman, Neutralising antibody activity against SARS-CoV-2 variants, including Omicron, in an elderly cohort vaccinated with BNT162b2, medRxiv, doi:10.1101/2021.12.23.21268293
Ni, Structural analysis of the Spike of the Omicron SARS-COV-2 variant by Cryo-EM and implications for immune evasion, bioRxiv, doi:10.1101/2021.12.27.474250
Peacock, The SARS-CoV-2 variants associated with infections in India, B.1.617, show enhanced spike cleavage by furin, bioRxiv, doi:10.1101/2021.05.28.446163
Peacock, The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets, Nat Microbiol, doi:10.1038/s41564-021-00908-w
Planas, Considerable escape of SARS-CoV-2 variant Omicron to antibody neutralization, bioRxiv, doi:10.1101/2021.12.14.472630
Pulliam, Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa, medRxiv, doi:10.1101/2021.11.11.21266068
Puray-Chavez, Systematic analysis of SARS-CoV-2 infection of an ACE2-negative human airway cell, Cell Reports, doi:10.1016/j.celrep.2021.109364
Rihn, A plasmid DNA-launched SARS-CoV-2 reverse genetics system and coronavirus toolkit for COVID-19 research, PLoS Biol, doi:10.1371/journal.pbio.3001091
Saito, Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation, Nature, doi:10.1038/s41586-021-04266-9
Thakur, Micro-fusion inhibition tests: quantifying antibody neutralization of virusmediated cell-cell fusion, Journal of General Virology, doi:10.1099/jgv.0.001506
Thakur, SARS-CoV-2 variants of concern Alpha, Beta, Gamma and Delta have extended ACE2 receptor host-ranges, bioRxiv, doi:10.1101/2021.11.23.469663
Viana, Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa, medRxiv, doi:10.1101/2021.12.19.21268028
Winstone, The polybasic cleavage site in the SARS-CoV-2 spike modulates viral sensitivity to Type I interferon and IFITM2, J Virol, doi:10.1128/JVI.02422-20
Yin, Structures of the Omicron Spike trimer with ACE2 and an anti-Omicron antibody, bioRxiv, doi:10.1101/2021.12.27.474273
Zhou, Investigating SARS-CoV-2 surface and air contamination in an acute healthcare setting during the peak of the COVID-19 pandemic in London, Clin Infect Dis, doi:10.1093/cid/ciaa905
Zhou, Mutations that adapt SARS-CoV-2 to mustelid hosts do not increase fitness in the human airway, bioRxiv, doi:10.1101/2021.08.20.456972
DOI record:
{
"DOI": "10.1101/2021.12.31.474653",
"URL": "http://dx.doi.org/10.1101/2021.12.31.474653",
"abstract": "<jats:p>At the end of 2021 a new SARS-CoV-2 variant, Omicron, emerged and quickly spread across the world. It has been demonstrated that Omicrons high number of Spike mutations lead to partial immune evasion from even polyclonal antibody responses, allowing frequent re-infection and vaccine breakthroughs. However, it seems unlikely these antigenic differences alone explain its rapid growth; here we show Omicron replicates rapidly in human primary airway cultures, more so even than the previously dominant variant of concern, Delta. Omicron Spike continues to use human ACE2 as its primary receptor, to which it binds more strongly than other variants. Omicron Spike mediates enhanced entry into cells expressing several different animal ACE2s, including various domestic avian species, horseshoe bats and mice suggesting it has an increased propensity for reverse zoonosis and is more likely than previous variants to establish an animal reservoir of SARS-CoV-2. Unlike other SARS-CoV-2 variants, however, Omicron Spike has a diminished ability to induce syncytia formation. Furthermore, Omicron is capable of efficiently entering cells in a TMPRSS2-independent manner, via the endosomal route. We posit this enables Omicron to infect a greater number of cells in the respiratory epithelium, allowing it to be more infectious at lower exposure doses, and resulting in enhanced intrinsic transmissibility.</jats:p>",
"accepted": {
"date-parts": [
[
2022,
1,
3
]
]
},
"author": [
{
"ORCID": "http://orcid.org/0000-0001-7077-2928",
"affiliation": [],
"authenticated-orcid": false,
"family": "Peacock",
"given": "Thomas P.",
"sequence": "first"
},
{
"ORCID": "http://orcid.org/0000-0001-6849-3962",
"affiliation": [],
"authenticated-orcid": false,
"family": "Brown",
"given": "Jonathan C",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-6413-2454",
"affiliation": [],
"authenticated-orcid": false,
"family": "Zhou",
"given": "Jie",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-4450-5911",
"affiliation": [],
"authenticated-orcid": false,
"family": "Thakur",
"given": "Nazia",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Newman",
"given": "Joseph",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-4591-6026",
"affiliation": [],
"authenticated-orcid": false,
"family": "Kugathasan",
"given": "Ruthiran",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Sukhova",
"given": "Ksenia",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-9878-4007",
"affiliation": [],
"authenticated-orcid": false,
"family": "Kaforou",
"given": "Myrsini",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-5640-2266",
"affiliation": [],
"authenticated-orcid": false,
"family": "Bailey",
"given": "Dalan",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-3948-0895",
"affiliation": [],
"authenticated-orcid": false,
"family": "Barclay",
"given": "Wendy S",
"sequence": "additional"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2022,
1,
4
]
],
"date-time": "2022-01-04T02:25:13Z",
"timestamp": 1641263113000
},
"deposited": {
"date-parts": [
[
2022,
1,
4
]
],
"date-time": "2022-01-04T02:25:13Z",
"timestamp": 1641263113000
},
"group-title": "Microbiology",
"indexed": {
"date-parts": [
[
2022,
1,
4
]
],
"date-time": "2022-01-04T05:54:12Z",
"timestamp": 1641275652490
},
"institution": [
{
"name": "bioRxiv"
}
],
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2022,
1,
3
]
]
},
"link": [
{
"URL": "https://syndication.highwire.org/content/doi/10.1101/2021.12.31.474653",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "246",
"original-title": [],
"posted": {
"date-parts": [
[
2022,
1,
3
]
]
},
"prefix": "10.1101",
"published": {
"date-parts": [
[
2022,
1,
3
]
]
},
"publisher": "Cold Spring Harbor Laboratory",
"reference-count": 0,
"references-count": 0,
"relation": {},
"score": 1,
"short-container-title": [],
"short-title": [],
"source": "Crossref",
"subtitle": [],
"subtype": "preprint",
"title": [
"The SARS-CoV-2 variant, Omicron, shows rapid replication in human primary nasal epithelial cultures and efficiently uses the endosomal route of entry."
],
"type": "posted-content"
}