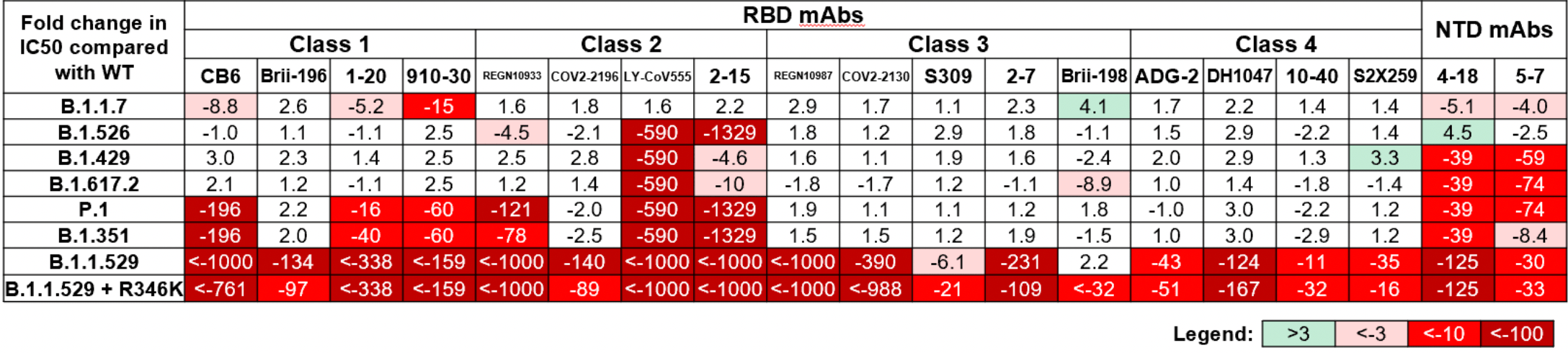
Striking Antibody Evasion Manifested by the Omicron Variant of SARS-CoV-2
et al., bioRxiv, doi:10.1101/2021.12.14.472719, Dec 2021
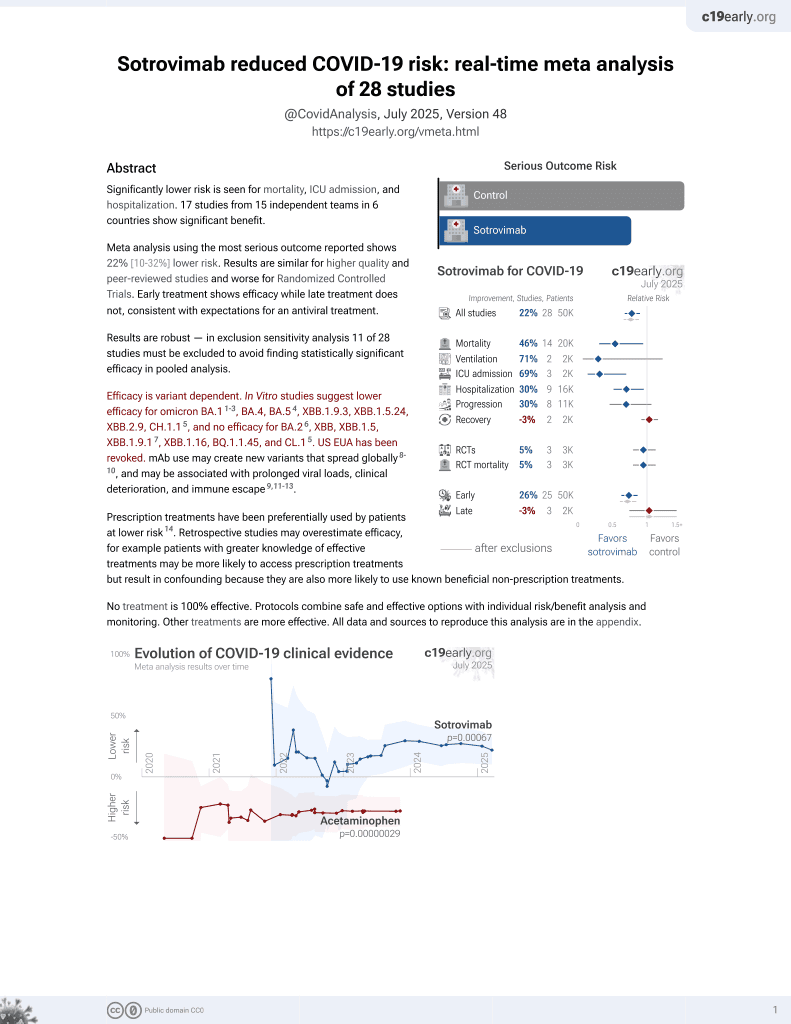
Sotrovimab for COVID-19
45th treatment shown to reduce risk in
August 2022, now with p = 0.00048 from 29 studies, recognized in 42 countries.
Efficacy is variant dependent.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
In vitro study (Vero-E6-TMPRSS2) showing 18 of 19 monoclonal antibodies were no longer effective or significantly impaired with B.1.1.529 omicron.
Study covers casirivimab/imdevimab, bamlanivimab/etesevimab, and sotrovimab.
Liu et al., 15 Dec 2021, preprint, 23 authors.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Striking Antibody Evasion Manifested by the Omicron Variant of SARS-CoV-2
doi:10.1101/2021.12.14.472719
The Omicron (B.1.1.529) variant of SARS-CoV-2 was only recently detected in southern Africa, but its subsequent spread has been extensive, both regionally and globally 1 . It is expected to become dominant in the coming weeks 2 , probably due to enhanced transmissibility. A striking feature of this variant is the large number of spike mutations 3 that pose a threat to the efficacy of current COVID-19 vaccines and antibody therapies 4 . This concern is amplified by the findings from our study. We found B.1.1.529 to be markedly resistant to neutralization by serum not only from convalescent patients, but also from individuals vaccinated with one of the four widely used COVID-19 vaccines. Even serum from persons vaccinated and boosted with mRNA-based vaccines exhibited substantially diminished neutralizing activity against B.1.1.529. By evaluating a panel of monoclonal antibodies to all known epitope clusters on the spike protein, we noted that the activity of 18 of the 19 antibodies tested were either abolished or impaired, including ones currently authorized or approved for use in patients. In addition, we also identified four new spike mutations (S371L, N440K, G446S, and Q493R) that confer greater antibody resistance to B.1.1.529. The Omicron variant presents a serious threat to many existing COVID-19 vaccines and therapies, compelling the development of new interventions that anticipate the evolutionary trajectory of SARS-CoV-2.
Methods
Serum samples Convalescent plasma samples were obtained from patients with documented SARS-CoV-2 infection approximately one month after recovery or later. These samples were collected at the beginning of the pandemic in early 2020 at Columbia University Irving Medical Center, and therefore are assumed to be infection by the wild-type strain of SARS-CoV-2 4 . Sera from individuals who received two or three doses of mRNA-1273 or BNT162b2 vaccine were collected at Columbia University Irving Medical Center at least two weeks after the final dose. Sera from individuals who received one dose of Ad26.COV2.S or two doses of ChAdOx1 nCov-19 were obtained from BEI Resources. Some individuals were also infected by SARS-CoV-2 in addition to the vaccinations they received. Note that, whenever possible, we specifically chose samples with high titers against the wild-type strain of SARS-CoV-2 such that the loss in activity against B.1.1.529 could be better quantified, and therefore the titers observed here should be considered in that context. All collections were conducted under protocols reviewed and approved by the Institutional Review Board of Columbia University. Additional information for the vaccinee samples can be found in Extended Data Table 1 .
Monoclonal antibodies Antibodies were expressed as previously described 22 , by synthesis of VH and VL genes (GenScript), transfection of Expi293 cells (Thermo Fisher), and affinity purification from the supernatant by..
References
Abu-Raddad, Chemaitelly, Butt, Study Group For, Effectiveness of the BNT162b2 Covid-19 Vaccine against the B, and B
Andrews, Effectiveness of COVID-19 vaccines against the Omicron, medRxiv, doi:10.1101/2021.12.14.21267615
Annavajhala, Emergence and expansion of SARS-CoV-2 B.1.526 after identification in New York, Nature, doi:10.1038/s41586-021-03908-2
Banach, Paired heavy-and light-chain signatures contribute to potent SARS-CoV-2 neutralization in public antibody responses, Cell Rep, doi:10.1016/j.celrep.2021.109771
Barnes, SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies, Nature, doi:10.1038/s41586-020-2852-1
Baym, Inexpensive multiplexed library preparation for megabase-sized genomes, PLoS One, doi:10.1371/journal.pone.0128036
Cerutti, Neutralizing antibody 5-7 defines a distinct site of vulnerability in SARS-CoV-2 spike N-terminal domain, Cell Rep, doi:10.1016/j.celrep.2021.109928
Cerutti, Potent SARS-CoV-2 neutralizing antibodies directed against spike Nterminal domain target a single supersite, Cell Host Microbe, doi:10.1016/j.chom.2021.03.005
Chu, Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study, Lancet Microbe, doi:10.1016/S2666-5247(20)30004-5
Cromer, Neutralising antibody titres as predictors of protection against SARS-CoV-2 variants and the impact of boosting: a meta-analysis, Lancet Microbe, doi:10.1016/S2666-5247(21)00267-6
Davies, Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England, Science, doi:10.1126/science.abg3055
Faria, Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil, Science, doi:10.1126/science.abh2644
Garcia-Beltran, mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant, medRxiv, doi:10.1101/2021.12.14.21267755
Grabowski, Kochańczyk, Lipniacki, Omicron strain spreads with the doubling time of 3.2-3.6 days in South Africa province of Gauteng that achieved herd immunity to Delta variant, medRxiv, doi:10.1101/2021.12.08.21267494
Hadfield, Nextstrain: real-time tracking of pathogen evolution, Bioinformatics, doi:10.1093/bioinformatics/bty407
Hansen, Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail, Science
Jones, The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates, Sci Transl Med, doi:10.1126/scitranslmed.abf1906
Ju, Human neutralizing antibodies elicited by SARS-CoV-2 infection, Nature, doi:10.1038/s41586-020-2380-z
Krissinel, Henrick, Inference of macromolecular assemblies from crystalline state, J Mol Biol, doi:10.1016/j.jmb.2007.05.022
Kuhlmann, Breakthrough Infections with SARS-CoV-2 Omicron Variant Despite Booster Dose of mRNA Vaccine, SSRN, doi:10.2139/ssrn.3981711
Langmead, Salzberg, Fast gapped-read alignment with Bowtie 2, Nat Methods
Li, In vitro and in vivo functions of SARS-CoV-2 infection-enhancing and neutralizing antibodies, Cell, doi:10.1016/j.cell.2021.06.021
Liu, Isolation and comparative analysis of antibodies that broadly neutralize sarbecoviruses, bioRxiv, doi:10.1101/2021.12.11.472236
Liu, Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike, Nature, doi:10.1038/s41586-020-2571-7
Madhi, Efficacy of the ChAdOx1 nCoV-19 Covid-19 Vaccine against the B.1.351 Variant, N Engl J Med, doi:10.1056/NEJMoa2102214
Martin, Cutadapt Removes Adapter Sequences From High-Throughput Sequencing Reads, EMBnet.journal, doi:10.14806/ej.17.1.200
Mlcochova, SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion, Nature, doi:10.1038/s41586-021-03944-y
Pinto, Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody, Nature, doi:10.1038/s41586-020-2349-y
Pulliam, Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa, medRxiv, doi:10.1101/2021.11.11.21266068
Rappazzo, Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody, Science, doi:10.1126/science.abf4830
Robinson, Integrative genomics viewer, Nat Biotechnol, doi:10.1038/nbt.1754
Sadoff, Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against Covid-19, N Engl J Med, doi:10.1056/NEJMoa2101544
Scott, Track Omicron's spread with molecular data, Science, doi:10.1126/science.abn4543
Shi, A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2, Nature, doi:10.1038/s41586-020-2381-y
Shu, Mccauley, Gisaid, Global initiative on sharing all influenza data -from vision to reality, Euro Surveill, doi:10.2807/1560-7917.ES.2017.22.13.30494
Tegally, Detection of a SARS-CoV-2 variant of concern in South Africa, Nature, doi:10.1038/s41586-021-03402-9
Tortorici, Broad sarbecovirus neutralization by a human monoclonal antibody, Nature, doi:10.1038/s41586-021-03817-4
Uriu, Neutralization of the SARS-CoV-2 Mu Variant by Convalescent and Vaccine Serum, N Engl J Med, doi:10.1056/NEJMc2114706
Wang, Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7, Nature, doi:10.1038/s41586-021-03398-2
Wroughton, The Washington Post
Zhang, Emergence of a Novel SARS-CoV-2 Variant in Southern California, JAMA, doi:10.1001/jama.2021.1612
DOI record:
{
"DOI": "10.1101/2021.12.14.472719",
"URL": "http://dx.doi.org/10.1101/2021.12.14.472719",
"abstract": "<jats:p>The Omicron (B.1.1.529) variant of SARS-CoV-2 was only recently detected in southern Africa, but its subsequent spread has been extensive, both regionally and globally. It is expected to become dominant in the coming weeks, probably due to enhanced transmissibility. A striking feature of this variant is the large number of spike mutations that pose a threat to the efficacy of current COVID-19 vaccines and antibody therapies. This concern is amplified by the findings from our study. We found B.1.1.529 to be markedly resistant to neutralization by serum not only from convalescent patients, but also from individuals vaccinated with one of the four widely used COVID-19 vaccines. Even serum from persons vaccinated and boosted with mRNA-based vaccines exhibited substantially diminished neutralizing activity against B.1.1.529. By evaluating a panel of monoclonal antibodies to all known epitope clusters on the spike protein, we noted that the activity of 18 of the 19 antibodies tested were either abolished or impaired, including ones currently authorized or approved for use in patients. In addition, we also identified four new spike mutations (S371L, N440K, G446S, and Q493R) that confer greater antibody resistance to B.1.1.529. The Omicron variant presents a serious threat to many existing COVID-19 vaccines and therapies, compelling the development of new interventions that anticipate the evolutionary trajectory of SARS-CoV-2.</jats:p>",
"accepted": {
"date-parts": [
[
2021,
12,
17
]
]
},
"author": [
{
"affiliation": [],
"family": "Liu",
"given": "Lihong",
"sequence": "first"
},
{
"ORCID": "http://orcid.org/0000-0002-3733-9556",
"affiliation": [],
"authenticated-orcid": false,
"family": "Iketani",
"given": "Sho",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Guo",
"given": "Yicheng",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Chan",
"given": "Jasper Fuk-Woo",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Wang",
"given": "Maple",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Liu",
"given": "Liyuan",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Luo",
"given": "Yang",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Chu",
"given": "Hin",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Huang",
"given": "Yiming",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Nair",
"given": "Manoj S.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Yu",
"given": "Jian",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Chik",
"given": "Kenn Ka-Heng",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Yuen",
"given": "Terrence Tsz-Tai",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Yoon",
"given": "Chaemin",
"sequence": "additional"
},
{
"affiliation": [],
"family": "To",
"given": "Kelvin Kai-Wang",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Chen",
"given": "Honglin",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Yin",
"given": "Michael T.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Sobieszczyk",
"given": "Magdalena E.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Huang",
"given": "Yaoxing",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Wang",
"given": "Harris H.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-3253-3309",
"affiliation": [],
"authenticated-orcid": false,
"family": "Sheng",
"given": "Zizhang",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Yuen",
"given": "Kwok-Yung",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Ho",
"given": "David D.",
"sequence": "additional"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2021,
12,
15
]
],
"date-time": "2021-12-15T22:45:13Z",
"timestamp": 1639608313000
},
"deposited": {
"date-parts": [
[
2021,
12,
18
]
],
"date-time": "2021-12-18T05:55:12Z",
"timestamp": 1639806912000
},
"group-title": "Microbiology",
"indexed": {
"date-parts": [
[
2021,
12,
19
]
],
"date-time": "2021-12-19T06:03:29Z",
"timestamp": 1639893809447
},
"institution": [
{
"name": "bioRxiv"
}
],
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2021,
12,
15
]
]
},
"link": [
{
"URL": "https://syndication.highwire.org/content/doi/10.1101/2021.12.14.472719",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "246",
"original-title": [],
"posted": {
"date-parts": [
[
2021,
12,
15
]
]
},
"prefix": "10.1101",
"published": {
"date-parts": [
[
2021,
12,
15
]
]
},
"publisher": "Cold Spring Harbor Laboratory",
"reference": [
{
"DOI": "10.2807/1560-7917.ES.2017.22.13.30494",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.1"
},
{
"DOI": "10.1101/2021.12.08.21267494",
"doi-asserted-by": "crossref",
"key": "2021121721000709000_2021.12.14.472719v1.2",
"unstructured": "Grabowski, F. , Kochańczyk, M. & Lipniacki, T. Omicron strain spreads with the doubling time of 3.2—3.6 days in South Africa province of Gauteng that achieved herd immunity to Delta variant. medRxiv, doi:https://doi.org/10.1101/2021.12.08.21267494 (2021)."
},
{
"DOI": "10.1126/science.abn4543",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.3"
},
{
"DOI": "10.1038/s41586-021-03398-2",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.4"
},
{
"DOI": "10.1056/NEJMc2104974",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.5"
},
{
"DOI": "10.1056/NEJMoa2102214",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.6"
},
{
"DOI": "10.1056/NEJMoa2101544",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.7"
},
{
"DOI": "10.1126/science.abg3055",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.8"
},
{
"DOI": "10.1038/s41586-021-03944-y",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.9"
},
{
"DOI": "10.1093/bioinformatics/bty407",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.10"
},
{
"DOI": "10.1101/2021.11.11.21266068",
"doi-asserted-by": "crossref",
"key": "2021121721000709000_2021.12.14.472719v1.11",
"unstructured": "Pulliam, J. R. C. et al. Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa. medRxiv, doi:https://doi.org/10.1101/2021.11.11.21266068 (2021)."
},
{
"DOI": "10.1126/science.abd0827",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.12"
},
{
"DOI": "10.1038/s41586-020-2548-6",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.13"
},
{
"DOI": "10.1126/scitranslmed.abf1906",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.14"
},
{
"DOI": "10.1038/s41586-020-2381-y",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.15"
},
{
"DOI": "10.1038/s41586-020-2380-z",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.16"
},
{
"DOI": "10.1038/s41586-020-2349-y",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.17"
},
{
"DOI": "10.1016/j.celrep.2021.109771",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.18"
},
{
"DOI": "10.1126/science.abf4830",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.19"
},
{
"DOI": "10.1016/j.cell.2021.06.021",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.20"
},
{
"DOI": "10.1038/s41586-021-03817-4",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.21"
},
{
"DOI": "10.1038/s41586-020-2571-7",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.22"
},
{
"DOI": "10.1016/j.celrep.2021.109928",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.23"
},
{
"DOI": "10.1101/2021.12.11.472236",
"doi-asserted-by": "crossref",
"key": "2021121721000709000_2021.12.14.472719v1.24",
"unstructured": "Liu, L. et al. Isolation and comparative analysis of antibodies that broadly neutralize sarbecoviruses. bioRxiv, doi:https://doi.org/10.1101/2021.12.11.472236 (2021)."
},
{
"DOI": "10.1038/s41586-020-2852-1",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.25"
},
{
"DOI": "10.1016/j.chom.2021.03.005",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.26"
},
{
"DOI": "10.1056/NEJMc2114706",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.27"
},
{
"DOI": "10.1038/s41586-021-03908-2",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.28"
},
{
"DOI": "10.1001/jama.2021.1612",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.29"
},
{
"DOI": "10.1126/science.abh2644",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.30"
},
{
"DOI": "10.1038/s41586-021-03402-9",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.31"
},
{
"DOI": "10.1101/2021.12.14.21267615",
"doi-asserted-by": "crossref",
"key": "2021121721000709000_2021.12.14.472719v1.32",
"unstructured": "Andrews, N. et al. Effectiveness of COVID-19 vaccines against the Omicron (B.1.1.529) variant of concern. medRxiv, doi:https://doi.org/10.1101/2021.12.14.21267615 (2021)."
},
{
"key": "2021121721000709000_2021.12.14.472719v1.33",
"unstructured": "Wroughton, L. in The Washington Post (2021)."
},
{
"DOI": "10.2139/ssrn.3981711",
"doi-asserted-by": "crossref",
"key": "2021121721000709000_2021.12.14.472719v1.34",
"unstructured": "Kuhlmann, C. et al. Breakthrough Infections with SARS-CoV-2 Omicron Variant Despite Booster Dose of mRNA Vaccine. SSRN, doi:https://dx.doi.org/10.2139/ssrn.3981711 (2021)."
},
{
"DOI": "10.1101/2021.12.14.21267755",
"doi-asserted-by": "crossref",
"key": "2021121721000709000_2021.12.14.472719v1.35",
"unstructured": "Garcia-Beltran, W. F. et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. medRxiv, doi:https://doi.org/10.1101/2021.12.14.21267755 (2021)."
},
{
"DOI": "10.1016/S2666-5247(21)00267-6",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.36"
},
{
"DOI": "10.1371/journal.pone.0128036",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.37"
},
{
"DOI": "10.14806/ej.17.1.200",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.38"
},
{
"DOI": "10.1038/nmeth.1923",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.39"
},
{
"DOI": "10.1038/nbt.1754",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.40"
},
{
"DOI": "10.1016/S2666-5247(20)30004-5",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.41"
},
{
"DOI": "10.1016/j.jmb.2007.05.022",
"doi-asserted-by": "publisher",
"key": "2021121721000709000_2021.12.14.472719v1.42"
}
],
"reference-count": 42,
"references-count": 42,
"relation": {},
"score": 1,
"short-container-title": [],
"short-title": [],
"source": "Crossref",
"subtitle": [],
"subtype": "preprint",
"title": [
"Striking Antibody Evasion Manifested by the Omicron Variant of SARS-CoV-2"
],
"type": "posted-content"
}
liu3