Assessing the inhibition efficacy of clinical drugs against the main proteases of SARS‐CoV‐2 variants and other coronaviruses
et al., Quantitative Biology, doi:10.1002/qub2.60, Jul 2024
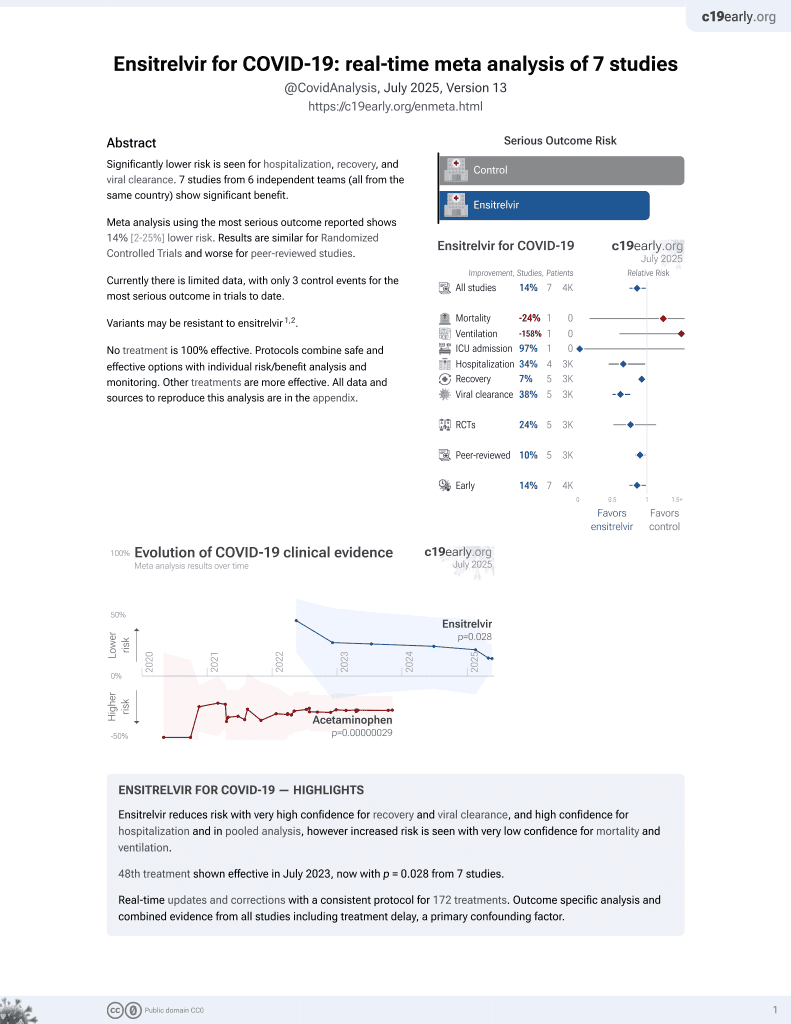
50th treatment shown to reduce risk in
July 2023, now with p = 0.015 from 8 studies.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
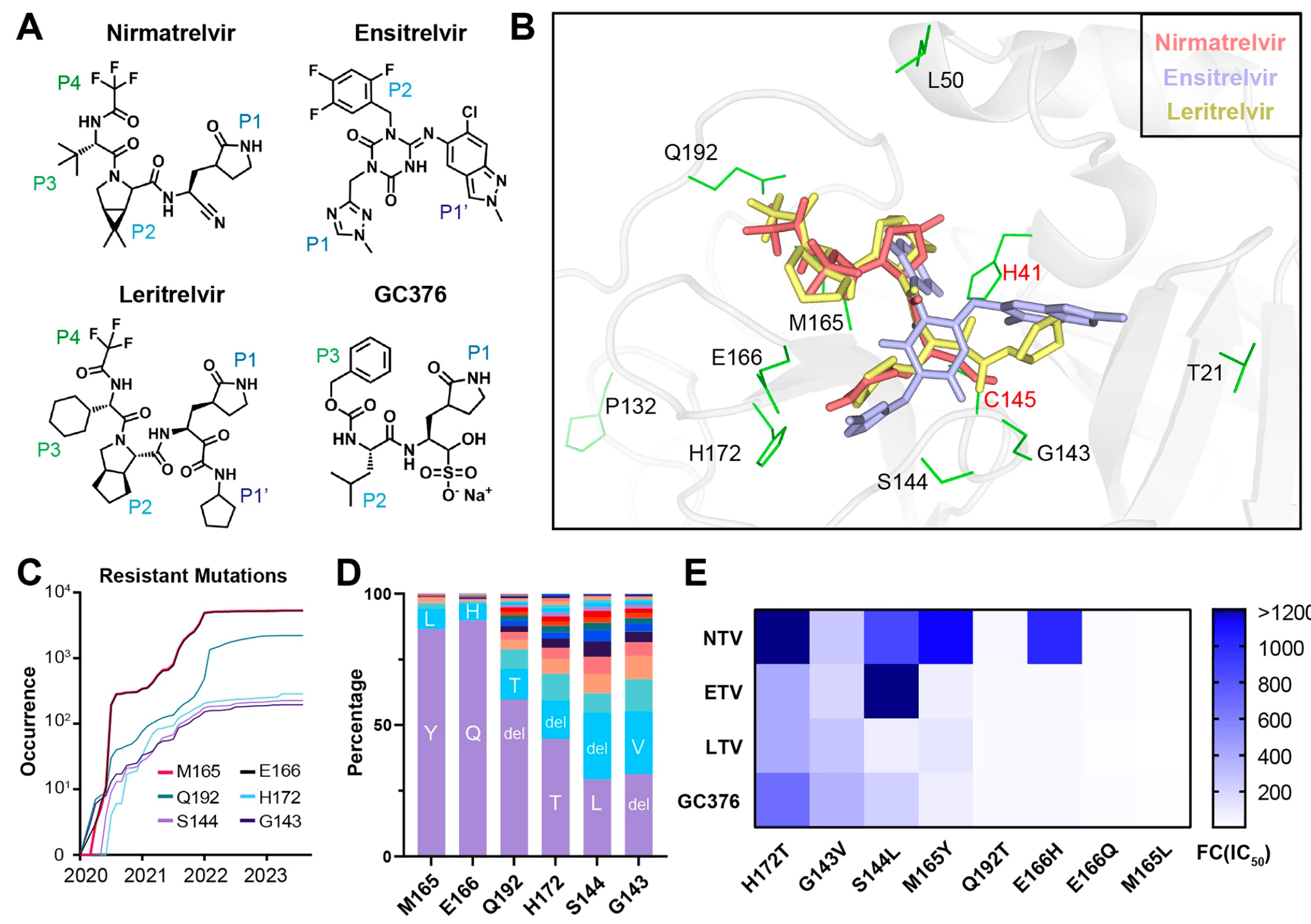
In vitro study showing that leritrelvir and GC376 remained effective against some nirmatrelvir- and ensitrelvir-resistant Mpro mutants. Leritrelvir showed better broad-spectrum activity against other pathogenic coronaviruses compared to ensitrelvir, nirmatrelvir, and GC376.
Potential mechanisms for improved efficacy with leritrelvir include:
Warhead differentiation: leritrelvir possesses an α-ketoamide warhead, which differs from the nitrile warhead of nirmatrelvir and the non-covalent nature of ensitrelvir. This warhead forms two hydrogen bonds with conserved active residues in the Mpro, specifically histidine and cysteine, which are crucial for the protease’s activity. This interaction likely enhances the binding affinity and stability of leritrelvir within the Mpro active site.
Binding pocket interactions: leritrelvir’s warhead interacts not only at the P1 position but also at the S1’ pocket. The α-ketoamide warhead of leritrelvir can form a hydrogen bond with H41 and a hydrophobic contact with L27, contributing to a stronger and more stable binding within the Mpro active site. This dual-site interaction increases its resilience against mutations that may affect other inhibitors.
Pharmacokinetics: leritrelvir exhibits improved pharmacokinetics, such as a longer half-life compared to nirmatrelvir and ensitrelvir. This longer half-life allows leritrelvir to maintain therapeutic levels in the body for a more extended period, providing a sustained antiviral effect even against resistant strains.
Slow-on, slow-off kinetics: leritrelvir shows "slow-on, slow-off" kinetic behavior, forming a stable enzyme-inhibitor complex. This characteristic prolongs the drug-target residence time, enhancing its inhibitory activity against Mpro mutants.
Broad-spectrum activity: the structure of leritrelvir allows it to exhibit broad-spectrum activity against various coronaviruses, which may involve targeting conserved regions within the Mpro of these viruses. This broad-spectrum efficacy suggests a robust interaction with the protease that is less susceptible to resistance mutations.
5 preclinical studies support the efficacy of ensitrelvir for COVID-19:
Study covers ensitrelvir, paxlovid, and leritrelvir.
1.
Nair et al., Persistence of an infectious form of SARS-CoV-2 post protease inhibitor treatment of permissive cells in vitro, The Journal of Infectious Diseases, doi:10.1093/infdis/jiae385.
2.
Moghadasi et al., Transmissible SARS-CoV-2 variants with resistance to clinical protease inhibitors, Science Advances, doi:10.1126/sciadv.ade8778.
a.
VeroE6/TMPRSS2 is a Vero E6 cell line engineered to express the human serine protease TMPRSS2, enabling SARS-CoV-2 S protein priming and entry.
b.
HEK293T/ACE2-TMPRSS2 is a human embryonic kidney cell line engineered to express human ACE2 and TMPRSS2, making it highly susceptible to SARS-CoV-2 infection.
c.
MucilAir cells are primary human nasal epithelial cells that mimic the structure and physiology of the human airway epithelium.
d.
A mouse model commonly used in infectious disease and cancer research due to higher immune response and susceptibility to infection.
e.
A rodent model used in SARS-CoV-2 research that replicates key aspects of human infection including efficient replication in the upper and lower respiratory tract.
f.
The original SARS-CoV-2 strain that emerged in Wuhan, China in late 2019. Also referred to as wild-type.
g.
A variant of concern first identified in India in late 2020, delta (B.1.617.2) transmitted more efficiently than previous variants. It contains spike mutations including L452R which increases binding to the ACE2 receptor.
h.
A highly transmissible variant of concern first detected in South Africa in late 2021. Omicron possesses many spike mutations which confer partial immune evasion, including deletions near the furin cleavage site.
Zhao et al., 6 Jul 2024, China, peer-reviewed, 8 authors, study period January 2020 - September 2023.
Contact: xuefei.li@siat.ac.cn, nan.li@siat.ac.cn.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Abstract: Received: 1 February 2024
DOI: 10.1002/qub2.60
- Revised: 10 April 2024
Accepted: 22 April 2024
COMMUNICATION
Assessing the inhibition efficacy of clinical drugs against
the main proteases of SARS‐CoV‐2 variants and other
coronaviruses
Wenlong Zhao1,2 | Cecylia S. Lupala1 | Shifeng Hou1 | Shuxin Yang1 |
Ziqi Yan1 | Shujie Liao1,2 | Xuefei Li1 | Nan Li1
1
Key Laboratory of Quantitative Synthetic Biology, Shenzhen Institute of Synthetic Biology, Shenzhen Institutes of Advanced Technology, Chinese Academy of
Sciences, Shenzhen, China
2
University of Chinese Academy of Sciences, Beijing, China
Correspondence
Xuefei Li and Nan Li.
Email: xuefei.li@siat.ac.cn and nan.li@siat.ac.cn
Funding information
National Key Research and Development Program of China, Grant/Award Number: 2023YFA0913900; National Natural Science Foundation of China, Grant/Award
Numbers: 31971354, 32100146, 32170672, 32271501
KEYWORDS
drug resistance, enzymatic activity, main protease, SARS‐CoV‐2
Dear Editor,
The rapid evolution of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) mainly due to its
high mutation rate and rapid viral replication, has led to
new variants resistant to the available vaccines and
monoclonal antibodies. In contrast, oral clinical drugs
targeting viral protease and RNA polymerase remain
effective against Omicron variants [1]. Main protease
(Mpro) plays a crucial role in the maturation and replication of viral strains, making it an attractive target for
developing antiviral drugs. Nirmatrelvir (NTV) is the
first‐in‐class Mpro peptidomimetic covalent inhibitor
known as “Paxlovid” approved in 2021 by the Food and
Drug Administration [2]. Nevertheless, NTV‐resistant
Mpro mutants particularly the E166V mutation, have
been characterized in the Global Initiative on Sharing
Avian Influenza Data (GISAID) database [3] and reported in COVID‐19 patients [4, 5]. Additionally, viral
passage experiments have identified other mutations
such as L50F and T21I, which can restore the viral
fitness reduced by E166V [6]. The second‐generation
Mpro drug, ensitrelvir (ETV), is a non‐covalent inhibitor
approved in 2022 with the brand name “Xocova” [7].
Besides, leritrelvir (LTV) is another covalent inhibitor
that was approved in China last year [8]. Preclinical
studies showed that ETV and LTV exhibited comparable antiviral activity as NTV and improved pharmacokinetics. However, the effectiveness of these clinical
drugs against NTV‐resistant Mpro mutants has yet to be
fully assessed. Here, we analyzed the inhibition efficiency of four inhibitors, NTV, ETV, LTV, and a veterinary drug, GC376 (Figure 1A), against the Mpro of
SARS‐CoV‐2
variants
and
other
pathogenic
coronaviruses.
The Mpro drugs interact tightly with the amino acids
of the active pocket (Figure 1B), and nonsynonymous
mutations of pocket residues have the potential to
induce severe resistance than mutations in other locations [3]. Particularly, six pocket residues, G143,
S144, M165, E166, H172, and Q192S have been reported to confer SARS‐CoV‐2 resistance to NTV [3].
Based on the GISAID database, we investigated the
occurrence and frequency of mutations at these six
residues. Results showed that all of these sites have
-
This is an open access article under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in any medium, provided
the original work is properly cited.
© 2024 The Author(s). Quantitative Biology published by John Wiley &..
DOI record:
{
"DOI": "10.1002/qub2.60",
"ISSN": [
"2095-4689",
"2095-4697"
],
"URL": "http://dx.doi.org/10.1002/qub2.60",
"alternative-id": [
"10.1002/qub2.60"
],
"assertion": [
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Received",
"name": "received",
"order": 0,
"value": "2024-02-01"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Accepted",
"name": "accepted",
"order": 1,
"value": "2024-04-22"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Published",
"name": "published",
"order": 2,
"value": "2024-07-06"
}
],
"author": [
{
"affiliation": [
{
"name": "Key Laboratory of Quantitative Synthetic Biology Shenzhen Institute of Synthetic Biology Shenzhen Institutes of Advanced Technology Chinese Academy of Sciences Shenzhen China"
},
{
"name": "University of Chinese Academy of Sciences Beijing China"
}
],
"family": "Zhao",
"given": "Wenlong",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Key Laboratory of Quantitative Synthetic Biology Shenzhen Institute of Synthetic Biology Shenzhen Institutes of Advanced Technology Chinese Academy of Sciences Shenzhen China"
}
],
"family": "Lupala",
"given": "Cecylia S.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Key Laboratory of Quantitative Synthetic Biology Shenzhen Institute of Synthetic Biology Shenzhen Institutes of Advanced Technology Chinese Academy of Sciences Shenzhen China"
}
],
"family": "Hou",
"given": "Shifeng",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Key Laboratory of Quantitative Synthetic Biology Shenzhen Institute of Synthetic Biology Shenzhen Institutes of Advanced Technology Chinese Academy of Sciences Shenzhen China"
}
],
"family": "Yang",
"given": "Shuxin",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Key Laboratory of Quantitative Synthetic Biology Shenzhen Institute of Synthetic Biology Shenzhen Institutes of Advanced Technology Chinese Academy of Sciences Shenzhen China"
}
],
"family": "Yan",
"given": "Ziqi",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Key Laboratory of Quantitative Synthetic Biology Shenzhen Institute of Synthetic Biology Shenzhen Institutes of Advanced Technology Chinese Academy of Sciences Shenzhen China"
},
{
"name": "University of Chinese Academy of Sciences Beijing China"
}
],
"family": "Liao",
"given": "Shujie",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Key Laboratory of Quantitative Synthetic Biology Shenzhen Institute of Synthetic Biology Shenzhen Institutes of Advanced Technology Chinese Academy of Sciences Shenzhen China"
}
],
"family": "Li",
"given": "Xuefei",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Key Laboratory of Quantitative Synthetic Biology Shenzhen Institute of Synthetic Biology Shenzhen Institutes of Advanced Technology Chinese Academy of Sciences Shenzhen China"
}
],
"family": "Li",
"given": "Nan",
"sequence": "additional"
}
],
"container-title": "Quantitative Biology",
"container-title-short": "Quant. Biol.",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"onlinelibrary.wiley.com"
]
},
"created": {
"date-parts": [
[
2024,
7,
6
]
],
"date-time": "2024-07-06T07:49:32Z",
"timestamp": 1720252172000
},
"deposited": {
"date-parts": [
[
2024,
7,
6
]
],
"date-time": "2024-07-06T07:49:38Z",
"timestamp": 1720252178000
},
"funder": [
{
"DOI": "10.13039/501100012166",
"award": [
"2023YFA0913900"
],
"doi-asserted-by": "publisher",
"name": "National Key Research and Development Program of China"
},
{
"DOI": "10.13039/501100001809",
"award": [
"31971354",
"32100146",
"32170672",
"32271501"
],
"doi-asserted-by": "publisher",
"name": "National Natural Science Foundation of China"
}
],
"indexed": {
"date-parts": [
[
2024,
7,
7
]
],
"date-time": "2024-07-07T00:19:45Z",
"timestamp": 1720311585189
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2024,
7,
6
]
]
},
"language": "en",
"license": [
{
"URL": "http://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
7,
6
]
],
"date-time": "2024-07-06T00:00:00Z",
"timestamp": 1720224000000
}
}
],
"link": [
{
"URL": "https://onlinelibrary.wiley.com/doi/pdf/10.1002/qub2.60",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "311",
"original-title": [],
"prefix": "10.1002",
"published": {
"date-parts": [
[
2024,
7,
6
]
]
},
"published-online": {
"date-parts": [
[
2024,
7,
6
]
]
},
"publisher": "Wiley",
"reference": [
{
"DOI": "10.1038/s41422-022-00618-w",
"article-title": "SARS‐CoV‐2 Omicron variant is highly sensitive to molnupiravir, nirmatrelvir, and the combination",
"author": "Li P",
"doi-asserted-by": "crossref",
"first-page": "322",
"issue": "3",
"journal-title": "Cell Res",
"key": "e_1_2_6_2_1",
"volume": "32",
"year": "2022"
},
{
"DOI": "10.1126/science.abl4784",
"article-title": "An oral SARS‐CoV‐2 Mpro inhibitor clinical candidate for the treatment of COVID‐19",
"author": "Owen DR",
"doi-asserted-by": "crossref",
"first-page": "1586",
"issue": "6575",
"journal-title": "Science",
"key": "e_1_2_6_3_1",
"volume": "374",
"year": "2021"
},
{
"DOI": "10.1021/acscentsci.3c00538",
"article-title": "Naturally occurring mutations of SARS‐CoV‐2 main protease confer drug resistance to nirmatrelvir",
"author": "Hu Y",
"doi-asserted-by": "crossref",
"first-page": "1658",
"issue": "8",
"journal-title": "ACS Central Sci",
"key": "e_1_2_6_4_1",
"volume": "9",
"year": "2023"
},
{
"DOI": "10.1016/j.medj.2023.08.001",
"article-title": "Multidrug‐resistant mutations to antiviral and antibody therapy in an immunocompromised patient infected with SARS‐CoV‐2",
"author": "Hirotsu Y",
"doi-asserted-by": "crossref",
"first-page": "813",
"issue": "11",
"journal-title": "Med",
"key": "e_1_2_6_5_1",
"volume": "4",
"year": "2023"
},
{
"DOI": "10.1093/cid/ciad494",
"article-title": "Nirmatrelvir resistance—de novo E166V/L50V mutations in an immunocompromised patient treated with prolonged nirmatrelvir/ritonavir monotherapy leading to clinical and virological treatment failure—a case report",
"author": "Zuckerman NS",
"doi-asserted-by": "crossref",
"first-page": "352",
"issue": "2",
"journal-title": "Clin Infect Dis",
"key": "e_1_2_6_6_1",
"volume": "78",
"year": "2024"
},
{
"DOI": "10.1038/s41586-022-05514-2",
"article-title": "Multiple pathways for SARS‐CoV‐2 resistance to nirmatrelvir",
"author": "Iketani S",
"doi-asserted-by": "crossref",
"first-page": "558",
"issue": "7944",
"journal-title": "Nature",
"key": "e_1_2_6_7_1",
"volume": "613",
"year": "2023"
},
{
"DOI": "10.1021/acs.jmedchem.2c00117",
"article-title": "Discovery of S‐217622, a noncovalent oral SARS‐CoV‐2 3CL protease inhibitor clinical candidate for treating COVID‐19",
"author": "Unoh Y",
"doi-asserted-by": "crossref",
"first-page": "6499",
"issue": "9",
"journal-title": "J Med Chem",
"key": "e_1_2_6_8_1",
"volume": "65",
"year": "2022"
},
{
"DOI": "10.1038/s41564-024-01618-9",
"article-title": "Preclinical evaluation of the SARS‐CoV‐2 Mpro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir",
"author": "Chen X",
"doi-asserted-by": "crossref",
"first-page": "1075",
"issue": "4",
"journal-title": "Nat Microbiol",
"key": "e_1_2_6_9_1",
"volume": "9",
"year": "2024"
},
{
"article-title": "Validation and invalidation of SARS‐CoV‐2 main protease inhibitors using the Flip‐GFP and Protease‐Glo luciferase assays",
"author": "Ma C",
"journal-title": "Acta Pharm Sin B",
"key": "e_1_2_6_10_1",
"year": "2021"
},
{
"DOI": "10.1038/s41422-022-00640-y",
"article-title": "The P132H mutation in the main protease of Omicron SARS‐CoV‐2 decreases thermal stability without compromising catalysis or small‐molecule drug inhibition",
"author": "Sacco MD",
"doi-asserted-by": "crossref",
"first-page": "498",
"issue": "5",
"journal-title": "Cell Res",
"key": "e_1_2_6_11_1",
"volume": "32",
"year": "2022"
},
{
"DOI": "10.1016/j.jbc.2023.103004",
"article-title": "Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS‐CoV‐2 main protease",
"author": "Noske GD",
"doi-asserted-by": "crossref",
"issue": "3",
"journal-title": "J Biol Chem",
"key": "e_1_2_6_12_1",
"volume": "299",
"year": "2023"
}
],
"reference-count": 11,
"references-count": 11,
"relation": {},
"resource": {
"primary": {
"URL": "https://onlinelibrary.wiley.com/doi/10.1002/qub2.60"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Assessing the inhibition efficacy of clinical drugs against the main proteases of SARS‐CoV‐2 variants and other coronaviruses",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1002/crossmark_policy"
}