Targeting Host Dependency Factors: A Paradigm Shift in Antiviral Strategy Against RNA Viruses
et al., International Journal of Molecular Sciences, doi:10.3390/ijms27010147, Dec 2025
Review of host dependency factors (HDFs) as therapeutic targets for RNA viruses including SARS-CoV-2. Authors systematically examine how RNA viruses hijack host cellular machinery throughout their life cycle - from entry and replication to assembly and release - and propose targeting these conserved host factors instead of mutable viral components. They argue this host-targeting approach offers advantages including broad-spectrum efficacy and higher barriers to resistance development compared to direct-acting antivirals.
Yang et al., 23 Dec 2025, peer-reviewed, 9 authors.
Contact: liyingying23@nudt.edu.cn (corresponding author), zhulvyun@nudt.edu.cn.
Targeting Host Dependency Factors: A Paradigm Shift in Antiviral Strategy Against RNA Viruses
International Journal of Molecular Sciences, doi:10.3390/ijms27010147
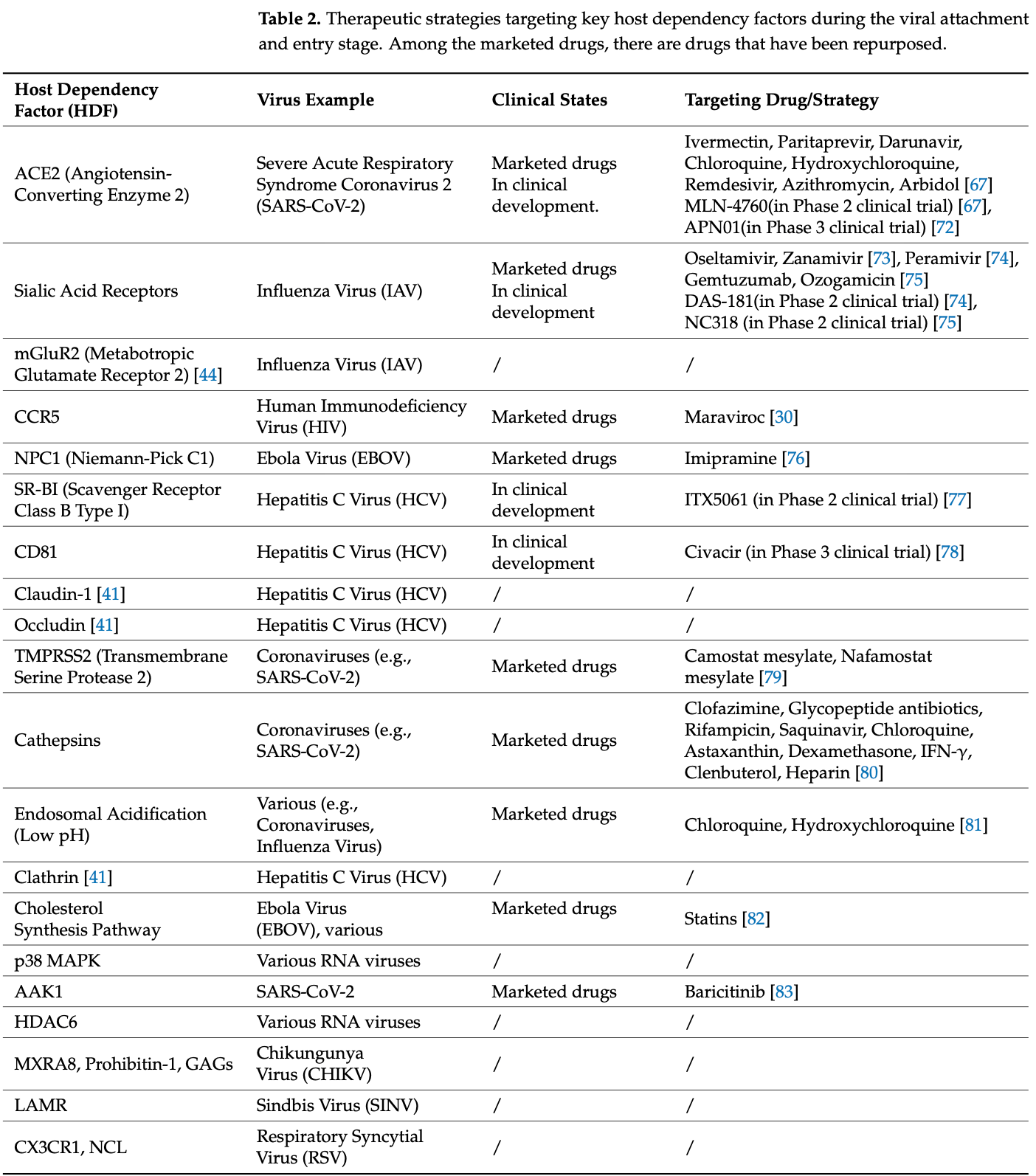
RNA viruses, such as SARS-CoV-2 and influenza, pose a persistent threat to global public health. Their high mutation rates undermine the effectiveness of conventional direct-acting antivirals (DAAs) and facilitate drug resistance. As obligate intracellular parasites, RNA viruses rely extensively on host cellular machinery and metabolic pathways throughout their life cycle. This dependency has prompted a strategic shift in antiviral research-from targeting the mutable virus to targeting relatively conserved host dependency factors (HDFs). In this review, we systematically analyze how RNA viruses exploit HDFs at each stage of infection: utilizing host receptors for entry; remodeling endomembrane systems to establish replication organelles; hijacking transcriptional, translational, and metabolic systems for genome replication and protein synthesis; and co-opting trafficking and budding machinery for assembly and egress. By comparing strategies across diverse RNA viruses, we highlight the broad-spectrum potential of HDF-targeting approaches, which offer a higher genetic barrier to resistance, providing a rational framework for developing host-targeting antiviral therapies.
In conclusion, viral assembly and release constitute an interconnected process that is profoundly dependent on the host. Therefore, targeting these critical host factors-such as the IP6 binding site, Rab proteins, nSMase2, or the CRM1 protein-has emerged as one of the most promising and cutting-edge directions for the development of next-generation, broad-spectrum antiviral drugs.
Conflicts of Interest: The authors declare no competing financial interest.
References
Ahmad, Fatemi, Ghaheri, Rezvani, Khezri et al., An overview of the role of Niemann-pick C1 (NPC1) in viral infections and inhibition of viral infections through NPC1 inhibitor, Cell Commun. Signal, doi:10.1186/s12964-023-01376-x
Ahmad, Pawara, Surana, Patel, The repurposed ACE2 inhibitors: SARS-CoV-2 entry blockers of COVID-19, Top. Curr. Chem, doi:10.1007/s41061-021-00353-7
Alenquer, Vale-Costa, Etibor, Ferreira, Sousa et al., Influenza A virus ribonucleoproteins form liquid organelles at endoplasmic reticulum exit sites, Nat. Commun, doi:10.1038/s41467-019-09549-4
Ali, Prasad, Alasmari, Alharbi, Rashid et al., Genomics-guided targeting of stress granule proteins G3BP1/2 to inhibit SARS-CoV-2 propagation, Int. J. Biol. Macromol, doi:10.1016/j.ijbiomac.2021.09.018
Avilov, Moisy, Naffakh, Cusack, Influenza A virus progeny vRNP trafficking in live infected cells studied with the virus-encoded fluorescently tagged PB2 protein, Vaccine, doi:10.1016/j.vaccine.2012.09.077
Bajimaya, Hayashi, Frankl, Bryk, Ward et al., Cholesterol reducing agents inhibit assembly of type I parainfluenza viruses, Virology, doi:10.1016/j.virol.2016.11.011
Banerjee, Miyake, Nobs, Schneider, Horvath et al., Influenza A virus uses the aggresome processing machinery for host cell entry, Science, doi:10.1126/science.1257037
Barnes, Lund-Andersen, Patel, Ytreberg, The effect of mutations on binding interactions between the SARS-CoV-2 receptor binding domain and neutralizing antibodies B38 and CB6, Sci. Rep, doi:10.1038/s41598-022-23482-5
Benjamin, Uhl, Grimont, Doane Ashley, Cohen et al., The NF-κB Transcriptional Footprint Is Essential for SARS-CoV-2 Replication, J. Virol, doi:10.1128/JVI.01257-21
Biedenkopf, Hartlieb, Hoenen, Becker, Phosphorylation of Ebola virus VP30 influences the composition of the viral nucleocapsid complex: Impact on viral transcription and replication, J. Biol. Chem, doi:10.1074/jbc.M113.461285
Bojkova, Klann, Koch, Widera, Krause et al., Proteomics of SARS-CoV-2-infected host cells reveals therapy targets, Nature, doi:10.1038/s41586-020-2332-7
Brandt, Wendt, Bodmer, Mettenleiter, Hoenen, The cellular protein CAD is recruited into Ebola virus inclusion bodies by the nucleoprotein NP to facilitate genome replication and transcription, Cells, doi:10.3390/cells9051126
Cabrera-Rodríguez, Pérez-Yanes, Montelongo, Lorenzo-Salazar, Estévez-Herrera et al., Transactive Response DNA-Binding Protein (TARDBP/TDP-43) Regulates Cell Permissivity to HIV-1 Infection by Acting on HDAC6, Int. J. Mol. Sci, doi:10.3390/ijms23116180
Cao, Jian, Wang, Yu, Song et al., Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution, Nature, doi:10.1038/s41586-022-05644-7
Carter, Iqbal, The influenza a virus replication cycle: A comprehensive review, Viruses, doi:10.3390/v16020316
Cavalli, Vilbois, Corti, Marcote, Tamura et al., The stress-induced MAP kinase p38 regulates endocytic trafficking via the GDI:Rab5 complex, Mol. Cell, doi:10.1016/S1097-2765(01)00189-7
Chander, Kumar, Khandelwal, Singh, Shringi et al., Role of p38 mitogen-activated protein kinase signalling in virus replication and potential for developing broad spectrum antiviral drugs, Rev. Med. Virol, doi:10.1002/rmv.2217
Chen, Wang, Luo, Su, Li et al., Histone Deacetylase 1 Plays an Acetylation-Independent Role in Influenza A Virus Replication, Front. Immunol, doi:10.3389/fimmu.2017.01757
Chen, Zhang, Deng, Liang, Ping, Virus-host interaction networks as new antiviral drug targets for IAV and SARS-CoV-2, Emerg. Microbes Infect, doi:10.1080/22221751.2022.2071175
Choi, Lee, Kim, Park, Kim et al., HDAC6 regulates cellular viral RNA sensing by deacetylation of RIG-I, EMBO J, doi:10.15252/embj.201592586
Ci, Shi, Compartmentalized replication organelle of flavivirus at the ER and the factors involved, Cell. Mol. Life Sci, doi:10.1007/s00018-021-03834-6
Colpitts, Tsai, Zeisel, Hepatitis C virus entry: An intriguingly complex and highly regulated process, Int. J. Mol. Sci, doi:10.3390/ijms21062091
Crawford, Yan, Zaher, Hardy, Host-Dependent Modifications of Packaged Alphavirus Genomic RNA Influence Virus Replication in Mammalian Cells, Viruses, doi:10.3390/v14122606
Cui, Lee, Kumar, Xu, Lu et al., Serum metabolome and lipidome changes in adult patients with primary dengue infection, PLoS Negl. Trop. Dis, doi:10.1371/journal.pntd.0002373
Daniloski, Jordan, Wessels, Hoagland, Kasela et al., Identification of required host factors for SARS-CoV-2 infection in human cells, Cell, doi:10.1016/j.cell.2020.10.030
De Lima Cavalcanti, Pereira, De Paula, Franca, A Review on Chikungunya Virus Epidemiology, Pathogenesis and Current Vaccine Development, Viruses, doi:10.3390/v14050969
Den Boon, Nishikiori, Zhan, Ahlquist, Positive-strand RNA virus genome replication organelles: Structure, assembly, control, Trends Genet, doi:10.1016/j.tig.2024.04.003
Deng, Cao, Wang, Li, Dai et al., Viral replication organelles: The highly complex and programmed replication machinery, Front. Microbiol, doi:10.3389/fmicb.2024.1450060
Dixit, Boulant, Zhang, Lee, Odendall et al., Peroxisomes are signaling platforms for antiviral innate immunity, Cell, doi:10.1016/j.cell.2010.04.018
Dolnik, Becker, Assembly and transport of filovirus nucleocapsids, PLoS Pathog, doi:10.1371/journal.ppat.1010616
Domingo, García-Crespo, Lobo-Vega, Perales, Mutation rates, mutation frequencies, and proofreading-repair activities in RNA virus genetics, Viruses, doi:10.3390/v13091882
Drucker, Diabetes, obesity, metabolism, and SARS-CoV-2 infection: The end of the beginning, Cell Metab, doi:10.1016/j.cmet.2021.01.016
Ehrhardt, Rückle, Hrincius, Haasbach, Anhlan et al., The NF-κB inhibitor SC75741 efficiently blocks influenza virus propagation and confers a high barrier for development of viral resistance, Cell. Microbiol, doi:10.1111/cmi.12108
Esparza, Bhat, Fontoura, Viral-host interactions during splicing and nuclear export of influenza virus mRNAs, Curr. Opin. Virol, doi:10.1016/j.coviro.2022.101254
Etibor, Yamauchi, Amorim, Liquid Biomolecular Condensates and Viral Lifecycles: Review and Perspectives, Viruses, doi:10.3390/v13030366
Farley, Kyle, Leier, Bramer, Weinstein et al., A global lipid map reveals host dependency factors conserved across SARS-CoV-2 variants, Nat. Commun, doi:10.1038/s41467-022-31097-7
Flint, Chatterjee, Lin, Mcmullan, Shrivastava-Ranjan et al., A genome-wide CRISPR screen identifies N-acetylglucosamine-1-phosphate transferase as a potential antiviral target for Ebola virus, Nat. Commun, doi:10.1038/s41467-018-08135-4
Fodor, Te, Velthuis, Structure and function of the influenza virus transcription and replication machinery, Cold Spring Harb. Perspect. Med, doi:10.1101/cshperspect.a038398
Frasson, Diamante, Zangrossi, Carbognin, Pietà et al., Identification of druggable host dependency factors shared by multiple SARS-CoV-2 variants of concern, J. Mol. Cell Biol, doi:10.1093/jmcb/mjae004
Freed, HIV-1 assembly, release and maturation, Nat. Rev. Microbiol, doi:10.1038/nrmicro3490
Frericks, Kloehn, Lange, Pottkämper, Carpentier et al., Host-targeting antivirals for chronic viral infections of the liver, Antivir. Res, doi:10.1016/j.antiviral.2024.106062
Gales, Kubina, Geldreich, Dimitrova, Strength in diversity: Nuclear export of viral RNAs, Viruses, doi:10.3390/v12091014
Garmaroudi, Marchant, Hendry, Luo, Yang et al., Coxsackievirus B3 replication and pathogenesis, Future Microbiol, doi:10.2217/fmb.15.5
Guedán, Caroe, Barr, Bishop, The role of capsid in HIV-1 nuclear entry, Viruses, doi:10.3390/v13081425
Gullberg, Steel, Pujari, Rovnak, Crick et al., Stearoly-CoA desaturase 1 differentiates early and advanced dengue virus infections and determines virus particle infectivity, PLoS Pathog, doi:10.1371/journal.ppat.1007261
Halfmann, Neumann, Kawaoka, The Ebolavirus VP24 protein blocks phosphorylation of p38 mitogen-activated protein kinase, J. Infect. Dis, doi:10.1093/infdis/jir325
Hashemian, Pourhanifeh, Hamblin, Shahrzad, Mirzaei, RdRp inhibitors and COVID-19: Is molnupiravir a good option?, Biomed. Pharmacother, doi:10.1016/j.biopha.2021.112517
Hashemian, Sheida, Taghizadieh, Memar, Hamblin et al., Nirmatrelvir/Ritonavir): A new approach to COVID-19 therapy?, Biomed. Pharmacother, doi:10.1016/j.biopha.2023.114367
He, Wang, Wang, Zhou, Yang et al., Liquid-liquid phase separation is essential for reovirus viroplasm formation and immune evasion, J. Virol, doi:10.1128/jvi.01028-24
Heath, Lloyd, Kulinski, Fromuth, Dembowski, Camptothecin, a topoisomerase I inhibitor, impedes productive herpes simplex virus type 1 infection, J. Virol, doi:10.1128/jvi.01276-25
Heaton, Randall, Dengue virus-induced autophagy regulates lipid metabolism, Cell Host Microbe, doi:10.1016/j.chom.2010.10.006
Heida, Bhide, Gasbarri, Kocabiyik, Stellacci et al., Advances in the development of entry inhibitors for sialic-acid-targeting viruses, Drug Discov. Today, doi:10.1016/j.drudis.2020.10.009
Hiatt, Hultquist, Mcgregor, Bouhaddou, Leenay et al., A functional map of HIV-host interactions in primary human T cells, Nat. Commun, doi:10.1038/s41467-022-29346-w
Hillis, Martin, Manchester, Högström, Zhang et al., Targeting cholesterol biosynthesis with statins synergizes with AKT inhibitors in triple-negative breast cancer, Cancer Res, doi:10.1158/0008-5472.CAN-24-0970
Hoffmann, Kleine-Weber, Schroeder, Krüger, Herrler et al., SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor, Cell, doi:10.1016/j.cell.2020.02.052
Horner, Liu, Park, Briley, Gale et al., Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus, Proc. Natl. Acad. Sci, doi:10.1073/pnas.1110133108
Hou, Wang, Wang, Wang, Yu et al., The ORF7a protein of SARS-CoV-2 initiates autophagy and limits autophagosome-lysosome fusion via degradation of SNAP29 to promote virus replication, Autophagy, doi:10.1080/15548627.2022.2084686
Huang, Wang, Zhong, Zhang, Zhang et al., Molecular architecture of coronavirus doublemembrane vesicle pore complex, Nature, doi:10.1038/s41586-024-07817-y
Hurley, Coyne, Mi Ączy Ńska, Stenmark, The expanding repertoire of ESCRT functions in cell biology and disease, Nature, doi:10.1038/s41586-025-08950-y
Hurley, Hanson, Membrane budding and scission by the ESCRT machinery: It's all in the neck, Nat. Rev. Mol. Cell Biol, doi:10.1038/nrm2937
Husby, Amiar, Prugar, David, Plescia et al., Phosphatidylserine clustering by the Ebola virus matrix protein is a critical step in viral budding, EMBO Rep, doi:10.15252/embr.202051709
Jackson, Farzan, Chen, Choe, Mechanisms of SARS-CoV-2 entry into cells, Nat. Rev. Mol. Cell Biol, doi:10.1038/s41580-021-00418-x
Jalily, Duncan, Fedida, Wang, Tietjen, Put a cork in it: Plugging the M2 viral ion channel to sink influenza, Antivir. Res, doi:10.1016/j.antiviral.2020.104780
Jastrz Ąb, Narejko, Car, Wielgat, Cell membrane sialome: Sialic acids as therapeutic targets and regulators of drug resistance in human cancer management, Cancers, doi:10.3390/cancers15205103
Ji, Li, Sun, Zhao, Li et al., VMP1 and TMEM41B are essential for DMV formation during β-coronavirus infection, J. Cell Biol, doi:10.1083/jcb.202112081
Jiang, Hu, He, Jin, He, Statins: A repurposed drug to fight cancer, J. Exp. Clin. Cancer Res, doi:10.1186/s13046-021-02041-2
Jiang, Wang, Guo, Zheng, Liu et al., Phospho-proteomics identifies a critical role of ATF2 in pseudorabies virus replication, Virol. Sin, doi:10.1016/j.virs.2022.06.003
Jin, He, Dong, Li, Ma et al., Altered lipid profile is a risk factor for the poor progression of COVID-19: From two retrospective cohorts, Front. Cell. Infect. Microbiol, doi:10.3389/fcimb.2021.712530
Jo, Murai, Agama, Sun, Saha et al., TOP1-DNA trapping by exatecan and combination therapy with ATR inhibitor, Mol. Cancer Ther, doi:10.1158/1535-7163.MCT-21-1000
Jocher, Grass, Tschirner, Riepler, Breimann et al., ADAM10 and ADAM17 promote SARS-CoV-2 cell entry and spike protein-mediated lung cell fusion, EMBO Rep, doi:10.15252/embr.202154305
Jordan, Randall, Dengue Virus Activates the AMP Kinase-mTOR Axis To Stimulate a Proviral Lipophagy, J. Virol, doi:10.1128/JVI.02020-16
Kanno, Miyako, Endo, The diverse interaction of metabolism, immune response, and viral pathogens, Front. Immunol, doi:10.3389/fimmu.2025.1619926
Kanojia, Sharma, Shiraz, Tripathi, Flavivirus-host interaction landscape visualized through genome-wide CRISPR screens, Viruses, doi:10.3390/v14102164
Kumar, Roy, Repurposing drugs: An empowering approach to drug discovery and development, Drug Res, doi:10.1055/a-2095-0826
Kumar, Sharma, Kumar, Tripathi, Barua et al., Host-directed antiviral therapy, Clin. Microbiol. Rev, doi:10.1128/CMR.00168-19
Kumar, Xin, Liang, Ly, Liang, NF-kappaB signaling differentially regulates influenza virus RNA synthesis, J. Virol, doi:10.1128/JVI.00909-08
Kurhade, Kang, Biering, Hwang, Randall, CAPRIN1 is required for control of viral replication complexes by interferon gamma, mBio, doi:10.1128/mbio.00172-23
Kvaratskhelia, Sharma, Larue, Serrao, Engelman, Molecular mechanisms of retroviral integration site selection, Nucleic Acids Res, doi:10.1093/nar/gku769
Kyle, Burnum-Johnson, Wendler, Eisfeld, Halfmann et al., Plasma lipidome reveals critical illness and recovery from human Ebola virus disease, Proc. Natl. Acad. Sci, doi:10.1073/pnas.1815356116
Labeau, Simon-Loriere, Hafirassou, Bonnet-Madin, Tessier et al., A genome-wide CRISPR-Cas9 screen identifies the dolichol-phosphate mannose synthase complex as a host dependency factor for dengue virus infection, J. Virol, doi:10.1128/JVI.01751-19
Lamers, Haagmans, SARS-CoV-2 pathogenesis, Nat. Rev. Microbiol, doi:10.1038/s41579-022-00713-0
Lee, Kreutzberger, Odongo, Nelson, Nyenhuis et al., Ebola virus glycoprotein interacts with cholesterol to enhance membrane fusion and cell entry, Biophys. J, doi:10.1016/j.bpj.2020.11.1317
Li, Clohisey, Chia, Wang, Cui et al., Genome-wide CRISPR screen identifies host dependency factors for influenza A virus infection, Nat. Commun, doi:10.1038/s41467-019-13965-x
Li, Gu, Zheng, Gao, Liu, Packaging signal of influenza A virus, Virol. J, doi:10.1186/s12985-021-01504-4
Li, Hou, Zhou, Yang, Zhao et al., LLPS of FXR proteins drives replication organelle clustering for β-coronaviral proliferation, J. Cell Biol, doi:10.1083/jcb.202309140
Li, Li, Tian, Su, Sun et al., SREBP2-dependent lipid droplet formation enhances viral replication and deteriorates lung injury in mice following IAV infection, Emerg. Microbes Infect, doi:10.1080/22221751.2025.2470371
Li, Liu, Hao, Liu, Wang et al., Rational design of an influenza-COVID-19 chimeric protective vaccine with HA-stalk and S-RBD, Emerg. Microbes Infect, doi:10.1080/22221751.2023.2231573
Liang, Pathogenicity and virulence of influenza, Virulence, doi:10.1080/21505594.2023.2223057
Ling, Li, Khan, Lundkvist, Li, Is heparan sulfate a target for inhibition of RNA virus infection?, Am. J. Physiol. Cell Physiol, doi:10.1152/ajpcell.00028.2022
Liu, Luo, Libby, Shi, Cathepsin L-selective inhibitors: A potentially promising treatment for COVID-19 patients, Pharmacol. Ther, doi:10.1016/j.pharmthera.2020.107587
Liu, Xia, Martynowycz, Gonen, Zhou, Molecular sociology of virus-induced cellular condensates supporting reovirus assembly and replication, Nat. Commun, doi:10.1038/s41467-024-54968-7
Liu, Yao, Lian, Yang, Biomolecular phase separation in stress granule assembly and virus infection, Acta Biochim. Biophys. Sin, doi:10.3724/abbs.2023117
Loperena González, Karthigeyan, Corry, Krishna, Hackenberg et al., Mammalian fatty acid synthase: A commonly used viral host dependency factor and a putative target for host-targeted broad-spectrum antiviral therapeutic development, mBio, doi:10.1128/mbio.03954-24
Lopez, Camporeale, Salgueiro, Borkosky, Visentín et al., Deconstructing virus condensation, PLoS Pathog, doi:10.1371/journal.ppat.1009926
Lu, Su, Yang, Jiang, Antivirals with common targets against highly pathogenic viruses, Cell, doi:10.1016/j.cell.2021.02.013
Lungu, Putz, SARS-CoV-2 spike protein interaction space, Int. J. Mol. Sci, doi:10.3390/ijms241512058
Luo, Li, Zhao, Ju, Ma et al., SARS-CoV-2 nucleocapsid protein phase separates with G3BPs to disassemble stress granules and facilitate viral production, Sci. Bull
Läubli, Nalle, Maslyar, Targeting the siglec-sialic acid immune axis in cancer: Current and future approaches, Cancer Immunol. Res, doi:10.1158/2326-6066.CIR-22-0366
Magazine, Zhang, Wu, Mcgee, Veggiani et al., Mutations and evolution of the SARS-CoV-2 spike protein, Correction in Viruses, doi:10.3390/v14030640
Malik, Properties of coronavirus and SARS-CoV-2, J. Pathol
Marchant, Singhera, Utokaparch, Hackett, Boyd et al., Toll-like receptor 4-mediated activation of p38 mitogen-activated protein kinase is a determinant of respiratory virus entry and tropism, J. Virol, doi:10.1128/JVI.00804-10
Martin-Serrano, Neil, Host factors involved in retroviral budding and release, Nat. Rev. Microbiol, doi:10.1038/nrmicro2596
Martín-Acebes, Vázquez-Calvo, Saiz, Lipids and flaviviruses, present and future perspectives for the control of dengue, Zika, and West Nile viruses, Prog. Lipid Res, doi:10.1016/j.plipres.2016.09.005
Mathew, Ghildyal, CRM1 inhibitors for antiviral therapy, Front. Microbiol, doi:10.3389/fmicb.2017.01171
Matsuyama, Nao, Shirato, Kawase, Saito et al., Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells, Proc. Natl. Acad. Sci, doi:10.1073/pnas.2002589117
Mcgraw, Hillmer, Medehincu, Hikichi, Gagliardi et al., Exploring HIV-1 maturation: A new frontier in antiviral development, Viruses, doi:10.3390/v16091423
Michieletto, Lusic, Marenduzzo, Orlandini, Physical principles of retroviral integration in the human genome, Nat. Commun, doi:10.1038/s41467-019-08333-8
Mishra, Rathore, RNA dependent RNA polymerase (RdRp) as a drug target for SARS-CoV2, J. Biomol. Struct. Dyn, doi:10.1080/07391102.2021.1875886
Miyake, Farley, Neubauer, Beddow, Hoenen et al., Ebola virus inclusion body formation and RNA synthesis are controlled by a novel domain of nucleoprotein interacting with VP35, J. Virol, doi:10.1128/JVI.02100-19
Monteil, Eaton, Postnikova, Murphy, Braunsfeld et al., Clinical grade ACE2 as a universal agent to block SARS-CoV-2 variants, EMBO Mol. Med, doi:10.15252/emmm.202115230
Moreira, Yamauchi, Matthias, How Influenza Virus Uses Host Cell Pathways during Uncoating, Cells, doi:10.3390/cells10071722
Mtambo, Amoako, Somboro, Agoni, Lawal et al., Influenza viruses: Harnessing the crucial role of the M2 ion-channel and neuraminidase toward inhibitor design, Molecules, doi:10.3390/molecules26040880
Nagy, Co-opted membranes, lipids, and host proteins: What have we learned from tombusviruses?, Curr. Opin. Virol, doi:10.1016/j.coviro.2022.101258
Ni, Wang, Yu, Wang, Wang et al., Influenza virus uses mGluR2 as an endocytic receptor to enter cells, Nat. Microbiol, doi:10.1038/s41564-024-01713-x
Ohlson, Eitson, Wells, Kumar, Jang et al., Genome-scale CRISPR screening reveals host factors required for ribosome formation and viral replication, mBio, doi:10.1128/mbio.00127-23
Oláh, Szénási, Lehotzky, Norris, Ovádi, Challenges in Discovering Drugs That Target the Protein-Protein Interactions of Disordered Proteins, Int. J. Mol. Sci, doi:10.3390/ijms23031550
Papa, Albecka, Mallery, Vaysburd, Renner et al., IP6-stabilised HIV capsids evade cGAS/STING-mediated host immune sensing, EMBO Rep, doi:10.15252/embr.202256275
Petrova, Russell, The evolution of seasonal influenza viruses, Nat. Rev. Microbiol, doi:10.1038/nrmicro.2017.118
Pizzorno, Padey, Terrier, Rosa-Calatrava, Drug repurposing approaches for the treatment of influenza viral infection: Reviving old drugs to fight against a long-lived enemy, Front. Immunol, doi:10.3389/fimmu.2019.00531
Poulakou, Barakat, Israel, Bacci, Abril et al., Ribavirin aerosol in hospitalized adults with respiratory distress and COVID-19: An open-label trial, Clin. Transl. Sci, doi:10.1111/cts.13436
Pérez-Yanes, Lorenzo-Sánchez, Cabrera-Rodríguez, García-Luis, Trujillo-González et al., The ZIKV NS5 Protein Aberrantly Alters the Tubulin Cytoskeleton, Induces the Accumulation of Autophagic p62 and Affects IFN Production: HDAC6 Has Emerged as an Anti-NS5/ZIKV Factor, Cells, doi:10.3390/cells13070598
Qin, Wang, Zheng, Wan, Fan, Current perspectives in drug targeting intrinsically disordered proteins and biomolecular condensates, BMC Biol, doi:10.1186/s12915-025-02214-x
Qu, Wei, Ke, Cheng, Ma et al., Histone deacetylase 6 inhibits STING-dependent antiviral immunity via site-specific deacetylation, J. Biol. Chem, doi:10.1016/j.jbc.2025.110841
Randall, Lipid Droplet Metabolism during Dengue Virus Infection, Trends Microbiol, doi:10.1016/j.tim.2018.05.010
Rashid, Xie, Suleman, Shah, Khan et al., Roles and functions of SARS-CoV-2 proteins in host immune evasion, Front. Immunol, doi:10.3389/fimmu.2022.940756
Rebensburg, Wei, Larue, Lindenberger, Francis et al., Sec24C is an HIV-1 host dependency factor crucial for virus replication, Nat. Microbiol, doi:10.1038/s41564-021-00868-1
Richardson, Griffin, Tucker, Smith, Oechsle et al., Baricitinib as potential treatment for 2019-nCoV acute respiratory disease, Lancet
Rossman, Lamb, Influenza virus assembly and budding, Virology, doi:10.1016/j.virol.2010.12.003
San Felipe, Batra, Muralidharan, Malpotra, Anand et al., Coupled equilibria of dimerization and lipid binding modulate SARS Cov 2 Orf9b interactions and interferon response, Elife, doi:10.7554/eLife.106484
Sanyal, Watson, Shah, Lee, Liang et al., Valproic acid use is associated with diminished risk of contracting COVID-19, and diminished disease severity: Epidemiologic and in vitro analysis reveal mechanistic insights, PLoS ONE, doi:10.1371/journal.pone.0307154
Saraste, Prydz, Assembly and Cellular Exit of Coronaviruses: Hijacking an Unconventional Secretory Pathway from the Pre-Golgi Intermediate Compartment via the Golgi Ribbon to the Extracellular Space, Cells, doi:10.3390/cells10030503
Schalkwijk, Snoeck, Andrei, Acyclovir resistance in herpes simplex viruses: Prevalence and therapeutic alternatives, Biochem. Pharmacol, doi:10.1016/j.bcp.2022.115322
Shang, Zhuang, Zhang, Li, Zhu et al., Inhibitors of endosomal acidification suppress SARS-CoV-2 replication and relieve viral pneumonia in hACE2 transgenic mice, Virol. J, doi:10.1186/s12985-021-01515-1
Shimotohno, HCV Assembly and Egress via Modifications in Host Lipid Metabolic Systems, Cold Spring Harb. Perspect. Med, doi:10.1101/cshperspect.a036814
Shiraki, Daikoku, Favipiravir, an anti-influenza drug against life-threatening RNA virus infections, Pharmacol. Ther, doi:10.1016/j.pharmthera.2020.107512
Shiraki, Yasumoto, Toyama, Fukuda, Amenamevir, a helicase-primase inhibitor, for the optimal treatment of herpes zoster, Viruses, doi:10.3390/v13081547
Silverberg, Boguniewicz, Quintana, Clark, Gross et al., Tapinarof validates the aryl hydrocarbon receptor as a therapeutic target: A clinical review, J. Allergy Clin. Immunol, doi:10.1016/j.jaci.2023.12.013
Simoneau, Chen, Xing, Hayashi, Chen et al., NF-κB inhibitor alpha controls SARS-CoV-2 infection in ACE2-overexpressing human airway organoids, Sci. Rep, doi:10.1038/s41598-024-66003-2
Sirohi, Kuhn, Zika, Virus Structure, Maturation, and Receptors, J. Infect. Dis, doi:10.1093/infdis/jix515
Sonawane, Barale, Dhanavade, Waghmare, Nadaf et al., Structural insights and inhibition mechanism of TMPRSS2 by experimentally known inhibitors Camostat mesylate, Nafamostat and Bromhexine hydrochloride to control SARS-coronavirus-2: A molecular modeling approach, Inform. Med. Unlocked, doi:10.1016/j.imu.2021.100597
Stalder, Gershlick, Direct trafficking pathways from the Golgi apparatus to the plasma membrane, Semin. Cell Dev. Biol, doi:10.1016/j.semcdb.2020.04.001
Su, Wilson, Samuel, Ma, Formation and Function of Liquid-Like Viral Factories in Negative-Sense Single-Stranded RNA Virus Infections, Viruses, doi:10.3390/v13010126
Sulkowski, Kang, Matining, Wyles, Johnson et al., Safety and antiviral activity of the HCV entry inhibitor ITX5061 in treatment-naive HCV-infected adults: A randomized, double-blind, phase 1b study, J. Infect. Dis, doi:10.1093/infdis/jit503
Takahashi, Halfmann, Oyama, Kozuka-Hata, Noda et al., DNA topoisomerase 1 facilitates the transcription and replication of the Ebola virus genome, J. Virol, doi:10.1128/JVI.03544-12
Tallon, Hollinger, Pal, Bell, Rais et al., Nipping disease in the bud: nSMase2 inhibitors as therapeutics in extracellular vesicle-mediated diseases, Drug Discov. Today, doi:10.1016/j.drudis.2021.03.025
Talukdar, Dutta, Ghosh, Bose, Bhattacharjee, Molecular Pathogenesis of Nipah Virus, Appl. Biochem. Biotechnol, doi:10.1007/s12010-022-04300-0
Tammaro, Guida, Appetecchia, Biava, Consalvi et al., Direct-acting antivirals and host-targeting approaches against enterovirus B infections: Recent advances, Pharmaceuticals, doi:10.3390/ph16020203
Tawar, Heydmann, Bach, Schüttrumpf, Chavan et al., Broad neutralization of hepatitis C virus-resistant variants by Civacir hepatitis C immunoglobulin, Hepatology, doi:10.1002/hep.28767
Titanji, Farley, Mehta, Connor-Schuler, Moanna et al., Use of Baricitinib in Patients With Moderate to Severe Coronavirus Disease, Clin. Infect. Dis, doi:10.1093/cid/ciaa879
Van Heuvel, Schatz, Rosengarten, Stitz, Infectious RNA: Human immunodeficiency virus (HIV) biology, therapeutic intervention, and the quest for a vaccine, Toxins, doi:10.3390/toxins14020138
Vazquez, Horner, Mavs, Coordination of Antiviral Innate Immunity, J. Virol, doi:10.1128/JVI.01918-14
Wali, Karbiener, Chou, Kovtunyk, Adonyi et al., Host-directed therapy with 2-deoxy-D-glucose inhibits human rhinoviruses, endemic coronaviruses, and SARS-CoV-2, J. Virus Erad, doi:10.1016/j.jve.2022.100305
Wang, Cao, Zhang, Wang, Man et al., COVID-19 metabolism: Mechanisms and therapeutic targets, MedComm, doi:10.1002/mco2.157
Wang, Zhang, Huang, Sha, Song et al., Plant negative-strand RNA virus phosphoprotein condensates exploit host trafficking and lipid synthesis for viral factory assembly, Sci. Adv, doi:10.1126/sciadv.adx7905
Wei, Alfajaro, Deweirdt, Hanna, Lu-Culligan et al., Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection, Cell, doi:10.1016/j.cell.2020.10.028
Wei, Bai, Yan, Meng, Wang et al., When liquid-liquid phase separation meets viral infections, Front. Immunol, doi:10.3389/fimmu.2022.985622
Wei, Liu, Hu, He, Yao et al., Recent Advances in Enterovirus A71 Infection and Antiviral Agents, Lab. Investig, doi:10.1016/j.labinv.2023.100298
Weller, Kuchta, The DNA helicase-primase complex as a target for herpes viral infection, Expert Opin. Ther. Targets, doi:10.1517/14728222.2013.827663
Williams-Noonan, Todorova, Kulkarni, Aguilar, Yarovsky, An active site inhibitor induces conformational penalties for ACE2 recognition by the spike protein of SARS-CoV-2, J. Phys. Chem. B, doi:10.1021/acs.jpcb.0c11321
Wong, Xu, Hou, Limonta, Kumar et al., Interplay between Zika Virus and Peroxisomes during Infection, Cells, doi:10.3390/cells8070725
Wu, Wagner, Moyle, Feng, Sharma et al., Disruption of Ebola NP0VP35 inclusion body-like structures reduce viral infection, J. Mol. Biol, doi:10.1016/j.jmb.2023.168241
Wu, Wilson, Influenza hemagglutinin structures and antibody recognition, Cold Spring Harb. Perspect. Med, doi:10.1101/cshperspect.a038778
Xia, Jiang, p53 promotes antiviral innate immunity by driving hexosamine metabolism, Cell Rep
Xiao, Yang, Xiong, Dong, The implications of FASN in immune cell biology and related diseases, Cell Death Dis, doi:10.1038/s41419-024-06463-6
Xu, Luo, Huang, Li, Ye et al., Influenza neuraminidase mutations and resistance to neuraminidase inhibitors, Emerg. Microbes Infect, doi:10.1080/22221751.2024.2429627
Yang, Shen, Hu, Cai, Zhang et al., Molecular mechanisms and cellular functions of liquid-liquid phase separation during antiviral immune responses, Front. Immunol, doi:10.3389/fimmu.2023.1162211
Yant, Mulato, Hansen, Tse, Niedziela-Majka et al., A highly potent long-acting small-molecule HIV-1 capsid inhibitor with efficacy in a humanized mouse model, Nat. Med, doi:10.1038/s41591-019-0560-x
Yao, Zhang, Duan, Tang, Lu, Molecular insights into the adaptive evolution of SARS-CoV-2 spike protein, J. Infect, doi:10.1016/j.jinf.2024.106121
Yeo, Goh, Su, Gan, The determination of HIV-1 RT mutation rate, its possible allosteric effects, and its implications on drug resistance, Viruses, doi:10.3390/v12030297
Zhang, Cao, Wang, Sun, Ji et al., Influenza virus infection reprograms cholesterol biosynthesis to facilitate virus replication by the TAK1-RORγ axis, PLoS Pathog, doi:10.1371/journal.ppat.1013646
Zhang, Tang, Jia, Zhou, RNA viruses: From RNA processing and interaction mechanisms to new prevention and control strategies, Sci. Sin. Vitae, doi:10.1360/SSV-2024-0100
Zhang, Wu, Wang, Liu, Liu et al., Influenza A virus infection activates STAT3 to enhance SREBP2 expression, cholesterol biosynthesis, and virus replication, iScience, doi:10.1016/j.isci.2024.110424
Zhang, Xiao, Cai, Chen, Structure of SARS-CoV-2 Spike Protein, Curr. Opin. Virol, doi:10.1016/j.coviro.2021.08.010
Zhang, Xu, Xie, Xu, Fu et al., Natural product sennoside B disrupts liquid-liquid phase separation of SARS-CoV-2 nucleocapsid protein by inhibiting its RNA-binding activity, J. Enzym. Inhib. Med. Chem, doi:10.1080/14756366.2025.2501743
Zhang, Zheng, Li, Ma, Liquid-liquid Phase Separation in Viral Function, J. Mol. Biol, doi:10.1016/j.jmb.2023.167955
Zhao, Zou, Gao, Xie, Cao et al., CMAS and ST3GAL4 play an important role in the adsorption of influenza virus by affecting the synthesis of sialic acid receptors, Int. J. Mol. Sci, doi:10.3390/ijms22116081
Zheng, Li, Song, Ye, Li et al., A broad antiviral strategy: Inhibitors of human DHODH pave the way for host-targeting antivirals against emerging and re-emerging viruses, Viruses, doi:10.3390/v14050928
Zhou, Yang, Wang, Hu, Zhang et al., A pneumonia outbreak associated with a new coronavirus of probable bat origin, Nature, doi:10.1038/s41586-020-2012-7
Zhu, Feng, Wang, Zheng, Jiang et al., New insights into the non-enzymatic function of HDAC6, Biomed. Pharmacother, doi:10.1016/j.biopha.2023.114438
Zhu, Zhang, Lin, Lyu, Lu et al., Progress on SARS-CoV-2 3CLpro inhibitors: Inspiration from SARS-CoV 3CLpro peptidomimetics and small-molecule anti-inflammatory compounds, Drug Des. Devel. Ther, doi:10.2147/DDDT.S359009
Zwillenberg, Tang, Quaas, Neuraminidase inhibitors for treatment of influenza, Acad. Emerg. Med, doi:10.1111/acem.14241
Álvarez-Fernández, Mingo-Casas, Blázquez, Caridi, Saiz et al., Allosteric inhibition of neutral sphingomyelinase 2 (nSMase2) by DPTIP: From antiflaviviral activity to deciphering its binding site through in silico studies and experimental validation, Int. J. Mol. Sci, doi:10.3390/ijms232213935
DOI record:
{
"DOI": "10.3390/ijms27010147",
"ISSN": [
"1422-0067"
],
"URL": "http://dx.doi.org/10.3390/ijms27010147",
"abstract": "<jats:p>RNA viruses, such as SARS-CoV-2 and influenza, pose a persistent threat to global public health. Their high mutation rates undermine the effectiveness of conventional direct-acting antivirals (DAAs) and facilitate drug resistance. As obligate intracellular parasites, RNA viruses rely extensively on host cellular machinery and metabolic pathways throughout their life cycle. This dependency has prompted a strategic shift in antiviral research—from targeting the mutable virus to targeting relatively conserved host dependency factors (HDFs). In this review, we systematically analyze how RNA viruses exploit HDFs at each stage of infection: utilizing host receptors for entry; remodeling endomembrane systems to establish replication organelles; hijacking transcriptional, translational, and metabolic systems for genome replication and protein synthesis; and co-opting trafficking and budding machinery for assembly and egress. By comparing strategies across diverse RNA viruses, we highlight the broad-spectrum potential of HDF-targeting approaches, which offer a higher genetic barrier to resistance, providing a rational framework for developing host-targeting antiviral therapies.</jats:p>",
"alternative-id": [
"ijms27010147"
],
"author": [
{
"affiliation": [
{
"name": "College of Science, National University of Defense Technology, Changsha 410073, China"
}
],
"family": "Yang",
"given": "Junru",
"sequence": "first"
},
{
"ORCID": "https://orcid.org/0009-0004-2735-8755",
"affiliation": [
{
"name": "College of Science, National University of Defense Technology, Changsha 410073, China"
}
],
"authenticated-orcid": false,
"family": "Qu",
"given": "Ying",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "College of Science, National University of Defense Technology, Changsha 410073, China"
}
],
"family": "Yuan",
"given": "Zhixiang",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "College of Science, National University of Defense Technology, Changsha 410073, China"
}
],
"family": "Lun",
"given": "Yufei",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "College of Science, National University of Defense Technology, Changsha 410073, China"
}
],
"family": "Kuang",
"given": "Jingyu",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "College of Science, National University of Defense Technology, Changsha 410073, China"
}
],
"family": "Shao",
"given": "Tong",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "College of Science, National University of Defense Technology, Changsha 410073, China"
}
],
"family": "Qi",
"given": "Yanhua",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-8803-0284",
"affiliation": [
{
"name": "College of Science, National University of Defense Technology, Changsha 410073, China"
}
],
"authenticated-orcid": false,
"family": "Li",
"given": "Yingying",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-2288-5981",
"affiliation": [
{
"name": "College of Science, National University of Defense Technology, Changsha 410073, China"
}
],
"authenticated-orcid": false,
"family": "Zhu",
"given": "Lvyun",
"sequence": "additional"
}
],
"container-title": "International Journal of Molecular Sciences",
"container-title-short": "IJMS",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2025,
12,
23
]
],
"date-time": "2025-12-23T10:57:18Z",
"timestamp": 1766487438000
},
"deposited": {
"date-parts": [
[
2025,
12,
25
]
],
"date-time": "2025-12-25T05:17:03Z",
"timestamp": 1766639823000
},
"funder": [
{
"DOI": "10.13039/501100001809",
"award": [
"32171429"
],
"award-info": [
{
"award-number": [
"32171429"
]
}
],
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/501100001809",
"id-type": "DOI"
}
],
"name": "National Natural Science Foundation of China"
},
{
"DOI": "10.13039/501100004735",
"award": [
"2022JJ30672"
],
"award-info": [
{
"award-number": [
"2022JJ30672"
]
}
],
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/501100004735",
"id-type": "DOI"
}
],
"name": "Natural Science Foundation of Hunan Province"
},
{
"DOI": "10.13039/501100004735",
"award": [
"2024JJ9218"
],
"award-info": [
{
"award-number": [
"2024JJ9218"
]
}
],
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/501100004735",
"id-type": "DOI"
}
],
"name": "Natural Science Foundation of Hunan Province"
}
],
"indexed": {
"date-parts": [
[
2025,
12,
25
]
],
"date-time": "2025-12-25T05:21:09Z",
"timestamp": 1766640069492,
"version": "3.48.0"
},
"is-referenced-by-count": 0,
"issue": "1",
"issued": {
"date-parts": [
[
2025,
12,
23
]
]
},
"journal-issue": {
"issue": "1",
"published-online": {
"date-parts": [
[
2026,
1
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
12,
23
]
],
"date-time": "2025-12-23T00:00:00Z",
"timestamp": 1766448000000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/1422-0067/27/1/147/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "147",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2025,
12,
23
]
]
},
"published-online": {
"date-parts": [
[
2025,
12,
23
]
]
},
"publisher": "MDPI AG",
"reference": [
{
"DOI": "10.1038/s41579-022-00713-0",
"article-title": "SARS-CoV-2 pathogenesis",
"author": "Lamers",
"doi-asserted-by": "crossref",
"first-page": "270",
"journal-title": "Nat. Rev. Microbiol.",
"key": "ref_1",
"volume": "20",
"year": "2022"
},
{
"DOI": "10.1080/21505594.2023.2223057",
"article-title": "Pathogenicity and virulence of influenza",
"author": "Liang",
"doi-asserted-by": "crossref",
"first-page": "2223057",
"journal-title": "Virulence",
"key": "ref_2",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.3390/toxins14020138",
"doi-asserted-by": "crossref",
"key": "ref_3",
"unstructured": "Van Heuvel, Y., Schatz, S., Rosengarten, J.F., and Stitz, J. (2022). Infectious RNA: Human immunodeficiency virus (HIV) biology, therapeutic intervention, and the quest for a vaccine. Toxins, 14."
},
{
"DOI": "10.3390/ijms241512058",
"doi-asserted-by": "crossref",
"key": "ref_4",
"unstructured": "Lungu, C.N., and Putz, M.V. (2023). SARS-CoV-2 spike protein interaction space. Int. J. Mol. Sci., 24."
},
{
"DOI": "10.1016/j.coviro.2021.08.010",
"article-title": "Structure of SARS-CoV-2 Spike Protein",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "173",
"journal-title": "Curr. Opin. Virol.",
"key": "ref_5",
"volume": "50",
"year": "2021"
},
{
"DOI": "10.1186/s12985-021-01504-4",
"article-title": "Packaging signal of influenza A virus",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "36",
"journal-title": "Virol. J.",
"key": "ref_6",
"volume": "18",
"year": "2021"
},
{
"DOI": "10.3390/v13091882",
"doi-asserted-by": "crossref",
"key": "ref_7",
"unstructured": "Domingo, E., García-Crespo, C., Lobo-Vega, R., and Perales, C. (2021). Mutation rates, mutation frequencies, and proofreading-repair activities in RNA virus genetics. Viruses, 13."
},
{
"DOI": "10.20944/preprints202002.0099.v1",
"doi-asserted-by": "crossref",
"key": "ref_8",
"unstructured": "Yeo, J.Y., Goh, G.-R., Su, C.T.-T., and Gan, S.K.-E. (2020). The determination of HIV-1 RT mutation rate, its possible allosteric effects, and its implications on drug resistance. Viruses, 12."
},
{
"DOI": "10.1038/s41598-022-23482-5",
"article-title": "The effect of mutations on binding interactions between the SARS-CoV-2 receptor binding domain and neutralizing antibodies B38 and CB6",
"author": "Barnes",
"doi-asserted-by": "crossref",
"first-page": "18819",
"journal-title": "Sci. Rep.",
"key": "ref_9",
"volume": "12",
"year": "2022"
},
{
"DOI": "10.1080/22221751.2023.2231573",
"article-title": "Rational design of an influenza-COVID-19 chimeric protective vaccine with HA-stalk and S-RBD",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "2231573",
"journal-title": "Emerg. Microbes Infect.",
"key": "ref_10",
"volume": "12",
"year": "2023"
},
{
"DOI": "10.1101/cshperspect.a038778",
"article-title": "Influenza hemagglutinin structures and antibody recognition",
"author": "Wu",
"doi-asserted-by": "crossref",
"first-page": "a038778",
"journal-title": "Cold Spring Harb. Perspect. Med.",
"key": "ref_11",
"volume": "10",
"year": "2020"
},
{
"DOI": "10.3389/fimmu.2022.940756",
"doi-asserted-by": "crossref",
"key": "ref_12",
"unstructured": "Rashid, F., Xie, Z., Suleman, M., Shah, A., Khan, S., and Luo, S. (2022). Roles and functions of SARS-CoV-2 proteins in host immune evasion. Front. Immunol., 13."
},
{
"DOI": "10.1016/j.cell.2021.02.013",
"article-title": "Antivirals with common targets against highly pathogenic viruses",
"author": "Lu",
"doi-asserted-by": "crossref",
"first-page": "1604",
"journal-title": "Cell",
"key": "ref_13",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.3390/v13081547",
"doi-asserted-by": "crossref",
"key": "ref_14",
"unstructured": "Shiraki, K., Yasumoto, S., Toyama, N., and Fukuda, H. (2021). Amenamevir, a helicase-primase inhibitor, for the optimal treatment of herpes zoster. Viruses, 13."
},
{
"DOI": "10.2147/DDDT.S359009",
"article-title": "Progress on SARS-CoV-2 3CLpro inhibitors: Inspiration from SARS-CoV 3CLpro peptidomimetics and small-molecule anti-inflammatory compounds",
"author": "Zhu",
"doi-asserted-by": "crossref",
"first-page": "1067",
"journal-title": "Drug Des. Devel. Ther.",
"key": "ref_15",
"volume": "16",
"year": "2022"
},
{
"DOI": "10.1080/07391102.2021.1875886",
"article-title": "RNA dependent RNA polymerase (RdRp) as a drug target for SARS-CoV2",
"author": "Mishra",
"doi-asserted-by": "crossref",
"first-page": "6039",
"journal-title": "J. Biomol. Struct. Dyn.",
"key": "ref_16",
"volume": "40",
"year": "2022"
},
{
"DOI": "10.1016/j.pharmthera.2020.107512",
"article-title": "Favipiravir, an anti-influenza drug against life-threatening RNA virus infections",
"author": "Shiraki",
"doi-asserted-by": "crossref",
"first-page": "107512",
"journal-title": "Pharmacol. Ther.",
"key": "ref_17",
"volume": "209",
"year": "2020"
},
{
"DOI": "10.1016/j.biopha.2021.112517",
"article-title": "RdRp inhibitors and COVID-19: Is molnupiravir a good option?",
"author": "Hashemian",
"doi-asserted-by": "crossref",
"first-page": "112517",
"journal-title": "Biomed. Pharmacother.",
"key": "ref_18",
"volume": "146",
"year": "2022"
},
{
"DOI": "10.1016/j.biopha.2023.114367",
"article-title": "Paxlovid (Nirmatrelvir/Ritonavir): A new approach to COVID-19 therapy?",
"author": "Hashemian",
"doi-asserted-by": "crossref",
"first-page": "114367",
"journal-title": "Biomed. Pharmacother.",
"key": "ref_19",
"volume": "162",
"year": "2023"
},
{
"DOI": "10.1111/acem.14241",
"article-title": "Neuraminidase inhibitors for treatment of influenza",
"author": "Zwillenberg",
"doi-asserted-by": "crossref",
"first-page": "1195",
"journal-title": "Acad. Emerg. Med.",
"key": "ref_20",
"volume": "28",
"year": "2021"
},
{
"DOI": "10.3390/molecules26040880",
"doi-asserted-by": "crossref",
"key": "ref_21",
"unstructured": "Mtambo, S.E., Amoako, D.G., Somboro, A.M., Agoni, C., Lawal, M.M., Gumede, N.S., Khan, R.B., and Kumalo, H.M. (2021). Influenza viruses: Harnessing the crucial role of the M2 ion-channel and neuraminidase toward inhibitor design. Molecules, 26."
},
{
"DOI": "10.1016/j.jinf.2024.106121",
"article-title": "Molecular insights into the adaptive evolution of SARS-CoV-2 spike protein",
"author": "Yao",
"doi-asserted-by": "crossref",
"first-page": "106121",
"journal-title": "J. Infect.",
"key": "ref_22",
"volume": "88",
"year": "2024"
},
{
"DOI": "10.3390/v15091787",
"doi-asserted-by": "crossref",
"key": "ref_23",
"unstructured": "Magazine, N., Zhang, T., Wu, Y., McGee, M.C., Veggiani, G., and Huang, W. (2022). Mutations and evolution of the SARS-CoV-2 spike protein. Viruses, 14, Correction in Viruses 2023, 15, 1787."
},
{
"DOI": "10.1080/22221751.2024.2429627",
"article-title": "Influenza neuraminidase mutations and resistance to neuraminidase inhibitors",
"author": "Xu",
"doi-asserted-by": "crossref",
"first-page": "2429627",
"journal-title": "Emerg. Microbes Infect.",
"key": "ref_24",
"volume": "13",
"year": "2024"
},
{
"DOI": "10.1016/j.antiviral.2020.104780",
"article-title": "Put a cork in it: Plugging the M2 viral ion channel to sink influenza",
"author": "Jalily",
"doi-asserted-by": "crossref",
"first-page": "104780",
"journal-title": "Antivir. Res.",
"key": "ref_25",
"volume": "178",
"year": "2020"
},
{
"DOI": "10.1038/nrmicro.2017.118",
"article-title": "The evolution of seasonal influenza viruses",
"author": "Petrova",
"doi-asserted-by": "crossref",
"first-page": "47",
"journal-title": "Nat. Rev. Microbiol.",
"key": "ref_26",
"volume": "16",
"year": "2018"
},
{
"article-title": "Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution",
"author": "Cao",
"first-page": "521",
"journal-title": "Nature",
"key": "ref_27",
"volume": "614",
"year": "2023"
},
{
"DOI": "10.1038/s41586-020-2332-7",
"article-title": "Proteomics of SARS-CoV-2-infected host cells reveals therapy targets",
"author": "Bojkova",
"doi-asserted-by": "crossref",
"first-page": "469",
"journal-title": "Nature",
"key": "ref_28",
"volume": "583",
"year": "2020"
},
{
"DOI": "10.1016/j.jve.2022.100305",
"article-title": "Host-directed therapy with 2-deoxy-D-glucose inhibits human rhinoviruses, endemic coronaviruses, and SARS-CoV-2",
"author": "Wali",
"doi-asserted-by": "crossref",
"first-page": "100305",
"journal-title": "J. Virus Erad.",
"key": "ref_29",
"volume": "8",
"year": "2022"
},
{
"DOI": "10.1038/s41467-022-29346-w",
"article-title": "A functional map of HIV-host interactions in primary human T cells",
"author": "Hiatt",
"doi-asserted-by": "crossref",
"first-page": "1752",
"journal-title": "Nat. Commun.",
"key": "ref_30",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.1038/s41591-019-0560-x",
"article-title": "A highly potent long-acting small-molecule HIV-1 capsid inhibitor with efficacy in a humanized mouse model",
"author": "Yant",
"doi-asserted-by": "crossref",
"first-page": "1377",
"journal-title": "Nat. Med.",
"key": "ref_31",
"volume": "25",
"year": "2019"
},
{
"DOI": "10.1016/j.antiviral.2024.106062",
"article-title": "Host-targeting antivirals for chronic viral infections of the liver",
"author": "Frericks",
"doi-asserted-by": "crossref",
"first-page": "106062",
"journal-title": "Antivir. Res.",
"key": "ref_32",
"volume": "234",
"year": "2025"
},
{
"DOI": "10.3390/ph16020203",
"doi-asserted-by": "crossref",
"key": "ref_33",
"unstructured": "Tammaro, C., Guida, M., Appetecchia, F., Biava, M., Consalvi, S., and Poce, G. (2023). Direct-acting antivirals and host-targeting approaches against enterovirus B infections: Recent advances. Pharmaceuticals, 16."
},
{
"DOI": "10.3390/v14050928",
"doi-asserted-by": "crossref",
"key": "ref_34",
"unstructured": "Zheng, Y., Li, S., Song, K., Ye, J., Li, W., Zhong, Y., Feng, Z., Liang, S., Cai, Z., and Xu, K. (2022). A broad antiviral strategy: Inhibitors of human DHODH pave the way for host-targeting antivirals against emerging and re-emerging viruses. Viruses, 14."
},
{
"DOI": "10.1016/j.cell.2020.10.030",
"article-title": "Identification of required host factors for SARS-CoV-2 infection in human cells",
"author": "Daniloski",
"doi-asserted-by": "crossref",
"first-page": "92",
"journal-title": "Cell",
"key": "ref_35",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2020.10.028",
"article-title": "Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection",
"author": "Wei",
"doi-asserted-by": "crossref",
"first-page": "76",
"journal-title": "Cell",
"key": "ref_36",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1038/s41467-019-13965-x",
"article-title": "Genome-wide CRISPR screen identifies host dependency factors for influenza A virus infection",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "164",
"journal-title": "Nat. Commun.",
"key": "ref_37",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1038/s41467-018-08135-4",
"article-title": "A genome-wide CRISPR screen identifies N-acetylglucosamine-1-phosphate transferase as a potential antiviral target for Ebola virus",
"author": "Flint",
"doi-asserted-by": "crossref",
"first-page": "285",
"journal-title": "Nat. Commun.",
"key": "ref_38",
"volume": "10",
"year": "2019"
},
{
"DOI": "10.1360/SSV-2024-0100",
"article-title": "RNA viruses: From RNA processing and interaction mechanisms to new prevention and control strategies",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "1851",
"journal-title": "Sci. Sin. Vitae",
"key": "ref_39",
"volume": "55",
"year": "2025"
},
{
"DOI": "10.1016/j.bpj.2020.11.1317",
"article-title": "Ebola virus glycoprotein interacts with cholesterol to enhance membrane fusion and cell entry",
"author": "Lee",
"doi-asserted-by": "crossref",
"first-page": "191A",
"journal-title": "Biophys. J.",
"key": "ref_40",
"volume": "120",
"year": "2021"
},
{
"DOI": "10.3390/ijms21062091",
"doi-asserted-by": "crossref",
"key": "ref_41",
"unstructured": "Colpitts, C.C., Tsai, P.-L., and Zeisel, M.B. (2020). Hepatitis C virus entry: An intriguingly complex and highly regulated process. Int. J. Mol. Sci., 21."
},
{
"DOI": "10.1038/s41580-021-00418-x",
"article-title": "Mechanisms of SARS-CoV-2 entry into cells",
"author": "Jackson",
"doi-asserted-by": "crossref",
"first-page": "3",
"journal-title": "Nat. Rev. Mol. Cell Biol.",
"key": "ref_42",
"volume": "23",
"year": "2022"
},
{
"DOI": "10.1038/s41586-020-2012-7",
"article-title": "A pneumonia outbreak associated with a new coronavirus of probable bat origin",
"author": "Zhou",
"doi-asserted-by": "crossref",
"first-page": "270",
"journal-title": "Nature",
"key": "ref_43",
"volume": "579",
"year": "2020"
},
{
"DOI": "10.1038/s41564-024-01713-x",
"article-title": "Influenza virus uses mGluR2 as an endocytic receptor to enter cells",
"author": "Ni",
"doi-asserted-by": "crossref",
"first-page": "1764",
"journal-title": "Nat. Microbiol.",
"key": "ref_44",
"volume": "9",
"year": "2024"
},
{
"DOI": "10.3390/v14050969",
"doi-asserted-by": "crossref",
"key": "ref_45",
"unstructured": "de Lima Cavalcanti, T.Y.V., Pereira, M.R., de Paula, S.O., and Franca, R.F.O. (2022). A Review on Chikungunya Virus Epidemiology, Pathogenesis and Current Vaccine Development. Viruses, 14."
},
{
"DOI": "10.1093/infdis/jix515",
"article-title": "Zika Virus Structure, Maturation, and Receptors",
"author": "Sirohi",
"doi-asserted-by": "crossref",
"first-page": "S935",
"journal-title": "J. Infect. Dis.",
"key": "ref_46",
"volume": "216",
"year": "2017"
},
{
"DOI": "10.1016/j.labinv.2023.100298",
"article-title": "Recent Advances in Enterovirus A71 Infection and Antiviral Agents",
"author": "Wei",
"doi-asserted-by": "crossref",
"first-page": "100298",
"journal-title": "Lab. Investig.",
"key": "ref_47",
"volume": "104",
"year": "2024"
},
{
"DOI": "10.2217/fmb.15.5",
"article-title": "Coxsackievirus B3 replication and pathogenesis",
"author": "Garmaroudi",
"doi-asserted-by": "crossref",
"first-page": "629",
"journal-title": "Future Microbiol.",
"key": "ref_48",
"volume": "10",
"year": "2015"
},
{
"DOI": "10.1007/s12010-022-04300-0",
"article-title": "Molecular Pathogenesis of Nipah Virus",
"author": "Talukdar",
"doi-asserted-by": "crossref",
"first-page": "2451",
"journal-title": "Appl. Biochem. Biotechnol.",
"key": "ref_49",
"volume": "195",
"year": "2023"
},
{
"DOI": "10.1073/pnas.2002589117",
"article-title": "Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells",
"author": "Matsuyama",
"doi-asserted-by": "crossref",
"first-page": "7001",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_50",
"volume": "117",
"year": "2020"
},
{
"DOI": "10.15252/embr.202154305",
"article-title": "ADAM10 and ADAM17 promote SARS-CoV-2 cell entry and spike protein-mediated lung cell fusion",
"author": "Jocher",
"doi-asserted-by": "crossref",
"first-page": "e54305",
"journal-title": "EMBO Rep.",
"key": "ref_51",
"volume": "23",
"year": "2022"
},
{
"DOI": "10.1016/S0140-6736(20)30304-4",
"article-title": "Baricitinib as potential treatment for 2019-nCoV acute respiratory disease",
"author": "Richardson",
"doi-asserted-by": "crossref",
"first-page": "e30",
"journal-title": "Lancet",
"key": "ref_52",
"volume": "395",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.02.052",
"article-title": "SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor",
"author": "Hoffmann",
"doi-asserted-by": "crossref",
"first-page": "271",
"journal-title": "Cell",
"key": "ref_53",
"volume": "181",
"year": "2020"
},
{
"DOI": "10.1128/JVI.00804-10",
"article-title": "Toll-like receptor 4-mediated activation of p38 mitogen-activated protein kinase is a determinant of respiratory virus entry and tropism",
"author": "Marchant",
"doi-asserted-by": "crossref",
"first-page": "11359",
"journal-title": "J. Virol.",
"key": "ref_54",
"volume": "84",
"year": "2010"
},
{
"DOI": "10.1016/S1097-2765(01)00189-7",
"article-title": "The stress-induced MAP kinase p38 regulates endocytic trafficking via the GDI:Rab5 complex",
"author": "Cavalli",
"doi-asserted-by": "crossref",
"first-page": "421",
"journal-title": "Mol. Cell",
"key": "ref_55",
"volume": "7",
"year": "2001"
},
{
"DOI": "10.1002/rmv.2217",
"article-title": "Role of p38 mitogen-activated protein kinase signalling in virus replication and potential for developing broad spectrum antiviral drugs",
"author": "Chander",
"doi-asserted-by": "crossref",
"first-page": "1",
"journal-title": "Rev. Med. Virol.",
"key": "ref_56",
"volume": "31",
"year": "2021"
},
{
"DOI": "10.1093/infdis/jir325",
"article-title": "The Ebolavirus VP24 protein blocks phosphorylation of p38 mitogen-activated protein kinase",
"author": "Halfmann",
"doi-asserted-by": "crossref",
"first-page": "S953",
"journal-title": "J. Infect. Dis.",
"key": "ref_57",
"volume": "204",
"year": "2011"
},
{
"DOI": "10.1016/j.virs.2022.06.003",
"article-title": "Phospho-proteomics identifies a critical role of ATF2 in pseudorabies virus replication",
"author": "Jiang",
"doi-asserted-by": "crossref",
"first-page": "591",
"journal-title": "Virol. Sin.",
"key": "ref_58",
"volume": "37",
"year": "2022"
},
{
"DOI": "10.3390/cells10071722",
"doi-asserted-by": "crossref",
"key": "ref_59",
"unstructured": "Moreira, E.A., Yamauchi, Y., and Matthias, P. (2021). How Influenza Virus Uses Host Cell Pathways during Uncoating. Cells, 10."
},
{
"DOI": "10.1126/science.1257037",
"article-title": "Influenza A virus uses the aggresome processing machinery for host cell entry",
"author": "Banerjee",
"doi-asserted-by": "crossref",
"first-page": "473",
"journal-title": "Science",
"key": "ref_60",
"volume": "346",
"year": "2014"
},
{
"DOI": "10.20944/preprints202402.1747.v1",
"doi-asserted-by": "crossref",
"key": "ref_61",
"unstructured": "Pérez-Yanes, S., Lorenzo-Sánchez, I., Cabrera-Rodríguez, R., García-Luis, J., Trujillo-González, R., Estévez-Herrera, J., and Valenzuela-Fernández, A. (2024). The ZIKV NS5 Protein Aberrantly Alters the Tubulin Cytoskeleton, Induces the Accumulation of Autophagic p62 and Affects IFN Production: HDAC6 Has Emerged as an Anti-NS5/ZIKV Factor. Cells, 13."
},
{
"DOI": "10.1016/j.jbc.2025.110841",
"article-title": "Histone deacetylase 6 inhibits STING-dependent antiviral immunity via site-specific deacetylation",
"author": "Qu",
"doi-asserted-by": "crossref",
"first-page": "110841",
"journal-title": "J. Biol. Chem.",
"key": "ref_62",
"volume": "301",
"year": "2025"
},
{
"DOI": "10.3390/ijms23116180",
"doi-asserted-by": "crossref",
"key": "ref_63",
"unstructured": "Cabrera-Rodríguez, R., Pérez-Yanes, S., Montelongo, R., Lorenzo-Salazar, J.M., Estévez-Herrera, J., García-Luis, J., Íñigo-Campos, A., Rubio-Rodríguez, L.A., Muñoz-Barrera, A., and Trujillo-González, R. (2022). Transactive Response DNA-Binding Protein (TARDBP/TDP-43) Regulates Cell Permissivity to HIV-1 Infection by Acting on HDAC6. Int. J. Mol. Sci., 23."
},
{
"DOI": "10.3390/ijms22116081",
"doi-asserted-by": "crossref",
"key": "ref_64",
"unstructured": "Zhao, Y., Zou, J., Gao, Q., Xie, S., Cao, J., and Zhou, H. (2021). CMAS and ST3GAL4 play an important role in the adsorption of influenza virus by affecting the synthesis of sialic acid receptors. Int. J. Mol. Sci., 22."
},
{
"DOI": "10.1021/acs.jpcb.0c11321",
"article-title": "An active site inhibitor induces conformational penalties for ACE2 recognition by the spike protein of SARS-CoV-2",
"author": "Todorova",
"doi-asserted-by": "crossref",
"first-page": "2533",
"journal-title": "J. Phys. Chem. B",
"key": "ref_65",
"volume": "125",
"year": "2021"
},
{
"DOI": "10.1038/s41598-024-66003-2",
"article-title": "NF-κB inhibitor alpha controls SARS-CoV-2 infection in ACE2-overexpressing human airway organoids",
"author": "Simoneau",
"doi-asserted-by": "crossref",
"first-page": "15351",
"journal-title": "Sci. Rep.",
"key": "ref_66",
"volume": "14",
"year": "2024"
},
{
"DOI": "10.1007/s41061-021-00353-7",
"article-title": "The repurposed ACE2 inhibitors: SARS-CoV-2 entry blockers of COVID-19",
"author": "Ahmad",
"doi-asserted-by": "crossref",
"first-page": "40",
"journal-title": "Top. Curr. Chem.",
"key": "ref_67",
"volume": "379",
"year": "2021"
},
{
"DOI": "10.1016/j.biopha.2023.114438",
"article-title": "New insights into the non-enzymatic function of HDAC6",
"author": "Zhu",
"doi-asserted-by": "crossref",
"first-page": "114438",
"journal-title": "Biomed. Pharmacother.",
"key": "ref_68",
"volume": "161",
"year": "2023"
},
{
"DOI": "10.15252/embj.201592586",
"article-title": "HDAC6 regulates cellular viral RNA sensing by deacetylation of RIG-I",
"author": "Choi",
"doi-asserted-by": "crossref",
"first-page": "429",
"journal-title": "EMBO J.",
"key": "ref_69",
"volume": "35",
"year": "2016"
},
{
"DOI": "10.1080/22221751.2022.2071175",
"article-title": "Virus-host interaction networks as new antiviral drug targets for IAV and SARS-CoV-2",
"author": "Chen",
"doi-asserted-by": "crossref",
"first-page": "1371",
"journal-title": "Emerg. Microbes Infect.",
"key": "ref_70",
"volume": "11",
"year": "2022"
},
{
"DOI": "10.3389/fimmu.2019.00531",
"doi-asserted-by": "crossref",
"key": "ref_71",
"unstructured": "Pizzorno, A., Padey, B., Terrier, O., and Rosa-Calatrava, M. (2019). Drug repurposing approaches for the treatment of influenza viral infection: Reviving old drugs to fight against a long-lived enemy. Front. Immunol., 10."
},
{
"DOI": "10.15252/emmm.202115230",
"article-title": "Clinical grade ACE2 as a universal agent to block SARS-CoV-2 variants",
"author": "Monteil",
"doi-asserted-by": "crossref",
"first-page": "e15230",
"journal-title": "EMBO Mol. Med.",
"key": "ref_72",
"volume": "14",
"year": "2022"
},
{
"DOI": "10.3390/cancers15205103",
"doi-asserted-by": "crossref",
"key": "ref_73",
"unstructured": "Jastrząb, P., Narejko, K., Car, H., and Wielgat, P. (2023). Cell membrane sialome: Sialic acids as therapeutic targets and regulators of drug resistance in human cancer management. Cancers, 15."
},
{
"DOI": "10.1016/j.drudis.2020.10.009",
"article-title": "Advances in the development of entry inhibitors for sialic-acid-targeting viruses",
"author": "Heida",
"doi-asserted-by": "crossref",
"first-page": "122",
"journal-title": "Drug Discov. Today",
"key": "ref_74",
"volume": "26",
"year": "2021"
},
{
"DOI": "10.1158/2326-6066.CIR-22-0366",
"article-title": "Targeting the siglec–sialic acid immune axis in cancer: Current and future approaches",
"author": "Nalle",
"doi-asserted-by": "crossref",
"first-page": "1423",
"journal-title": "Cancer Immunol. Res.",
"key": "ref_75",
"volume": "10",
"year": "2022"
},
{
"DOI": "10.1186/s12964-023-01376-x",
"article-title": "An overview of the role of Niemann-pick C1 (NPC1) in viral infections and inhibition of viral infections through NPC1 inhibitor",
"author": "Ahmad",
"doi-asserted-by": "crossref",
"first-page": "352",
"journal-title": "Cell Commun. Signal.",
"key": "ref_76",
"volume": "21",
"year": "2023"
},
{
"DOI": "10.1093/infdis/jit503",
"article-title": "Safety and antiviral activity of the HCV entry inhibitor ITX5061 in treatment-naive HCV-infected adults: A randomized, double-blind, phase 1b study",
"author": "Sulkowski",
"doi-asserted-by": "crossref",
"first-page": "658",
"journal-title": "J. Infect. Dis.",
"key": "ref_77",
"volume": "209",
"year": "2014"
},
{
"DOI": "10.1002/hep.28767",
"article-title": "Broad neutralization of hepatitis C virus-resistant variants by Civacir hepatitis C immunoglobulin",
"author": "Tawar",
"doi-asserted-by": "crossref",
"first-page": "1495",
"journal-title": "Hepatology",
"key": "ref_78",
"volume": "64",
"year": "2016"
},
{
"DOI": "10.1016/j.imu.2021.100597",
"article-title": "Structural insights and inhibition mechanism of TMPRSS2 by experimentally known inhibitors Camostat mesylate, Nafamostat and Bromhexine hydrochloride to control SARS-coronavirus-2: A molecular modeling approach",
"author": "Sonawane",
"doi-asserted-by": "crossref",
"first-page": "100597",
"journal-title": "Inform. Med. Unlocked",
"key": "ref_79",
"volume": "24",
"year": "2021"
},
{
"DOI": "10.1016/j.pharmthera.2020.107587",
"article-title": "Cathepsin L-selective inhibitors: A potentially promising treatment for COVID-19 patients",
"author": "Liu",
"doi-asserted-by": "crossref",
"first-page": "107587",
"journal-title": "Pharmacol. Ther.",
"key": "ref_80",
"volume": "213",
"year": "2020"
},
{
"DOI": "10.1186/s12985-021-01515-1",
"article-title": "Inhibitors of endosomal acidification suppress SARS-CoV-2 replication and relieve viral pneumonia in hACE2 transgenic mice",
"author": "Shang",
"doi-asserted-by": "crossref",
"first-page": "46",
"journal-title": "Virol. J.",
"key": "ref_81",
"volume": "18",
"year": "2021"
},
{
"DOI": "10.1158/0008-5472.CAN-24-0970",
"article-title": "Targeting cholesterol biosynthesis with statins synergizes with AKT inhibitors in triple-negative breast cancer",
"author": "Hillis",
"doi-asserted-by": "crossref",
"first-page": "3250",
"journal-title": "Cancer Res.",
"key": "ref_82",
"volume": "84",
"year": "2024"
},
{
"DOI": "10.1093/cid/ciaa879",
"article-title": "Use of Baricitinib in Patients With Moderate to Severe Coronavirus Disease 2019",
"author": "Titanji",
"doi-asserted-by": "crossref",
"first-page": "1247",
"journal-title": "Clin. Infect. Dis.",
"key": "ref_83",
"volume": "72",
"year": "2021"
},
{
"DOI": "10.1152/ajpcell.00028.2022",
"article-title": "Is heparan sulfate a target for inhibition of RNA virus infection?",
"author": "Ling",
"doi-asserted-by": "crossref",
"first-page": "C605",
"journal-title": "Am. J. Physiol. Cell Physiol.",
"key": "ref_84",
"volume": "322",
"year": "2022"
},
{
"DOI": "10.3389/fmicb.2024.1450060",
"doi-asserted-by": "crossref",
"key": "ref_85",
"unstructured": "Deng, H., Cao, H., Wang, Y., Li, J., Dai, J., Li, L.-F., Qiu, H.-J., and Li, S. (2024). Viral replication organelles: The highly complex and programmed replication machinery. Front. Microbiol., 15."
},
{
"DOI": "10.3390/v13030366",
"doi-asserted-by": "crossref",
"key": "ref_86",
"unstructured": "Etibor, T.A., Yamauchi, Y., and Amorim, M.J. (2021). Liquid Biomolecular Condensates and Viral Lifecycles: Review and Perspectives. Viruses, 13."
},
{
"DOI": "10.3390/v13010126",
"doi-asserted-by": "crossref",
"key": "ref_87",
"unstructured": "Su, J.M., Wilson, M.Z., Samuel, C.E., and Ma, D. (2021). Formation and Function of Liquid-Like Viral Factories in Negative-Sense Single-Stranded RNA Virus Infections. Viruses, 13."
},
{
"DOI": "10.1083/jcb.202309140",
"article-title": "LLPS of FXR proteins drives replication organelle clustering for β-coronaviral proliferation",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "e202309140",
"journal-title": "J. Cell Biol.",
"key": "ref_88",
"volume": "223",
"year": "2024"
},
{
"DOI": "10.1126/sciadv.adx7905",
"article-title": "Plant negative-strand RNA virus phosphoprotein condensates exploit host trafficking and lipid synthesis for viral factory assembly",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "eadx7905",
"journal-title": "Sci. Adv.",
"key": "ref_89",
"volume": "11",
"year": "2025"
},
{
"DOI": "10.3389/fimmu.2022.985622",
"doi-asserted-by": "crossref",
"key": "ref_90",
"unstructured": "Wei, W., Bai, L., Yan, B., Meng, W., Wang, H., Zhai, J., Si, F., and Zheng, C. (2022). When liquid-liquid phase separation meets viral infections. Front. Immunol., 13."
},
{
"DOI": "10.1038/s41467-024-54968-7",
"article-title": "Molecular sociology of virus-induced cellular condensates supporting reovirus assembly and replication",
"author": "Liu",
"doi-asserted-by": "crossref",
"first-page": "10638",
"journal-title": "Nat. Commun.",
"key": "ref_91",
"volume": "15",
"year": "2024"
},
{
"DOI": "10.1007/s00018-021-03834-6",
"article-title": "Compartmentalized replication organelle of flavivirus at the ER and the factors involved",
"author": "Ci",
"doi-asserted-by": "crossref",
"first-page": "4939",
"journal-title": "Cell. Mol. Life Sci.",
"key": "ref_92",
"volume": "78",
"year": "2021"
},
{
"DOI": "10.1083/jcb.202112081",
"article-title": "VMP1 and TMEM41B are essential for DMV formation during β-coronavirus infection",
"author": "Ji",
"doi-asserted-by": "crossref",
"first-page": "e202112081",
"journal-title": "J. Cell Biol.",
"key": "ref_93",
"volume": "221",
"year": "2022"
},
{
"DOI": "10.1038/s41586-024-07817-y",
"article-title": "Molecular architecture of coronavirus double-membrane vesicle pore complex",
"author": "Huang",
"doi-asserted-by": "crossref",
"first-page": "224",
"journal-title": "Nature",
"key": "ref_94",
"volume": "633",
"year": "2024"
},
{
"DOI": "10.1371/journal.ppat.1010616",
"doi-asserted-by": "crossref",
"key": "ref_95",
"unstructured": "Dolnik, O., and Becker, S. (2022). Assembly and transport of filovirus nucleocapsids. PLoS Pathog., 18."
},
{
"DOI": "10.1074/jbc.M113.461285",
"article-title": "Phosphorylation of Ebola virus VP30 influences the composition of the viral nucleocapsid complex: Impact on viral transcription and replication",
"author": "Biedenkopf",
"doi-asserted-by": "crossref",
"first-page": "11165",
"journal-title": "J. Biol. Chem.",
"key": "ref_96",
"volume": "288",
"year": "2013"
},
{
"DOI": "10.1128/JVI.02100-19",
"article-title": "Ebola virus inclusion body formation and RNA synthesis are controlled by a novel domain of nucleoprotein interacting with VP35",
"author": "Miyake",
"doi-asserted-by": "crossref",
"first-page": "e02100-19",
"journal-title": "J. Virol.",
"key": "ref_97",
"volume": "94",
"year": "2020"
},
{
"DOI": "10.3390/cells9051126",
"doi-asserted-by": "crossref",
"key": "ref_98",
"unstructured": "Brandt, J., Wendt, L., Bodmer, B.S., Mettenleiter, T.C., and Hoenen, T. (2020). The cellular protein CAD is recruited into Ebola virus inclusion bodies by the nucleoprotein NP to facilitate genome replication and transcription. Cells, 9."
},
{
"DOI": "10.1371/journal.ppat.1009926",
"doi-asserted-by": "crossref",
"key": "ref_99",
"unstructured": "Lopez, N., Camporeale, G., Salgueiro, M., Borkosky, S.S., Visentín, A., Peralta-Martinez, R., Loureiro, M.E., and de Prat-Gay, G. (2021). Deconstructing virus condensation. PLoS Pathog., 17."
},
{
"DOI": "10.1016/j.scib.2021.01.013",
"article-title": "SARS-CoV-2 nucleocapsid protein phase separates with G3BPs to disassemble stress granules and facilitate viral production",
"author": "Luo",
"doi-asserted-by": "crossref",
"first-page": "1194",
"journal-title": "Sci. Bull.",
"key": "ref_100",
"volume": "66",
"year": "2021"
},
{
"DOI": "10.3389/fimmu.2023.1162211",
"doi-asserted-by": "crossref",
"key": "ref_101",
"unstructured": "Yang, S., Shen, W., Hu, J., Cai, S., Zhang, C., Jin, S., Guan, X., Wu, J., Wu, Y., and Cui, J. (2023). Molecular mechanisms and cellular functions of liquid-liquid phase separation during antiviral immune responses. Front. Immunol., 14."
},
{
"DOI": "10.3724/abbs.2023117",
"article-title": "Biomolecular phase separation in stress granule assembly and virus infection",
"author": "Liu",
"doi-asserted-by": "crossref",
"first-page": "1099",
"journal-title": "Acta Biochim. Biophys. Sin.",
"key": "ref_102",
"volume": "55",
"year": "2023"
},
{
"DOI": "10.1016/j.jmb.2023.167955",
"article-title": "Liquid-liquid Phase Separation in Viral Function",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "167955",
"journal-title": "J. Mol. Biol.",
"key": "ref_103",
"volume": "435",
"year": "2023"
},
{
"DOI": "10.1080/14756366.2025.2501743",
"article-title": "Natural product sennoside B disrupts liquid-liquid phase separation of SARS-CoV-2 nucleocapsid protein by inhibiting its RNA-binding activity",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "2501743",
"journal-title": "J. Enzym. Inhib. Med. Chem.",
"key": "ref_104",
"volume": "40",
"year": "2025"
},
{
"DOI": "10.1016/j.ijbiomac.2021.09.018",
"article-title": "Genomics-guided targeting of stress granule proteins G3BP1/2 to inhibit SARS-CoV-2 propagation",
"author": "Ali",
"doi-asserted-by": "crossref",
"first-page": "636",
"journal-title": "Int. J. Biol. Macromol.",
"key": "ref_105",
"volume": "190",
"year": "2021"
},
{
"DOI": "10.1016/j.jmb.2023.168241",
"article-title": "Disruption of Ebola NP0VP35 inclusion body-like structures reduce viral infection",
"author": "Wu",
"doi-asserted-by": "crossref",
"first-page": "168241",
"journal-title": "J. Mol. Biol.",
"key": "ref_106",
"volume": "435",
"year": "2023"
},
{
"DOI": "10.1128/mbio.00172-23",
"article-title": "CAPRIN1 is required for control of viral replication complexes by interferon gamma",
"author": "Kurhade",
"doi-asserted-by": "crossref",
"first-page": "e00172-23",
"journal-title": "mBio",
"key": "ref_107",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.3390/v14102164",
"doi-asserted-by": "crossref",
"key": "ref_108",
"unstructured": "Kanojia, A., Sharma, M., Shiraz, R., and Tripathi, S. (2022). Flavivirus–host interaction landscape visualized through genome-wide CRISPR screens. Viruses, 14."
},
{
"DOI": "10.1371/journal.pntd.0002373",
"doi-asserted-by": "crossref",
"key": "ref_109",
"unstructured": "Cui, L., Lee, Y.H., Kumar, Y., Xu, F., Lu, K., Ooi, E.E., Tannenbaum, S.R., and Ong, C.N. (2013). Serum metabolome and lipidome changes in adult patients with primary dengue infection. PLoS Negl. Trop. Dis., 7."
},
{
"DOI": "10.1073/pnas.1815356116",
"article-title": "Plasma lipidome reveals critical illness and recovery from human Ebola virus disease",
"author": "Kyle",
"doi-asserted-by": "crossref",
"first-page": "3919",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_110",
"volume": "116",
"year": "2019"
},
{
"DOI": "10.3389/fcimb.2021.712530",
"doi-asserted-by": "crossref",
"key": "ref_111",
"unstructured": "Jin, H., He, J., Dong, C., Li, B., Ma, Z., Li, B., Huang, T., Fan, J., He, G., and Zhao, X. (2021). Altered lipid profile is a risk factor for the poor progression of COVID-19: From two retrospective cohorts. Front. Cell. Infect. Microbiol., 11."
},
{
"DOI": "10.1016/j.tig.2024.04.003",
"article-title": "Positive-strand RNA virus genome replication organelles: Structure, assembly, control",
"author": "Nishikiori",
"doi-asserted-by": "crossref",
"first-page": "681",
"journal-title": "Trends Genet.",
"key": "ref_112",
"volume": "40",
"year": "2024"
},
{
"DOI": "10.1016/j.coviro.2022.101258",
"article-title": "Co-opted membranes, lipids, and host proteins: What have we learned from tombusviruses?",
"author": "Nagy",
"doi-asserted-by": "crossref",
"first-page": "101258",
"journal-title": "Curr. Opin. Virol.",
"key": "ref_113",
"volume": "56",
"year": "2022"
},
{
"DOI": "10.3390/v13081425",
"doi-asserted-by": "crossref",
"key": "ref_114",
"unstructured": "Guedán, A., Caroe, E.R., Barr, G.C., and Bishop, K.N. (2021). The role of capsid in HIV-1 nuclear entry. Viruses, 13."
},
{
"DOI": "10.1038/s41467-022-31097-7",
"article-title": "A global lipid map reveals host dependency factors conserved across SARS-CoV-2 variants",
"author": "Farley",
"doi-asserted-by": "crossref",
"first-page": "3487",
"journal-title": "Nat. Commun.",
"key": "ref_115",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.1016/j.cmet.2021.01.016",
"article-title": "Diabetes, obesity, metabolism, and SARS-CoV-2 infection: The end of the beginning",
"author": "Drucker",
"doi-asserted-by": "crossref",
"first-page": "479",
"journal-title": "Cell Metab.",
"key": "ref_116",
"volume": "33",
"year": "2021"
},
{
"DOI": "10.1002/mco2.157",
"article-title": "COVID-19 metabolism: Mechanisms and therapeutic targets",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "e157",
"journal-title": "MedComm",
"key": "ref_117",
"volume": "3",
"year": "2022"
},
{
"DOI": "10.7554/eLife.106484",
"article-title": "Coupled equilibria of dimerization and lipid binding modulate SARS Cov 2 Orf9b interactions and interferon response",
"author": "Batra",
"doi-asserted-by": "crossref",
"first-page": "RP106484",
"journal-title": "Elife",
"key": "ref_118",
"volume": "14",
"year": "2025"
},
{
"DOI": "10.1080/15548627.2022.2084686",
"article-title": "The ORF7a protein of SARS-CoV-2 initiates autophagy and limits autophagosome-lysosome fusion via degradation of SNAP29 to promote virus replication",
"author": "Hou",
"doi-asserted-by": "crossref",
"first-page": "551",
"journal-title": "Autophagy",
"key": "ref_119",
"volume": "19",
"year": "2023"
},
{
"DOI": "10.1016/j.chom.2010.10.006",
"article-title": "Dengue virus-induced autophagy regulates lipid metabolism",
"author": "Heaton",
"doi-asserted-by": "crossref",
"first-page": "422",
"journal-title": "Cell Host Microbe",
"key": "ref_120",
"volume": "8",
"year": "2010"
},
{
"DOI": "10.1128/JVI.02020-16",
"article-title": "Dengue Virus Activates the AMP Kinase-mTOR Axis To Stimulate a Proviral Lipophagy",
"author": "Jordan",
"doi-asserted-by": "crossref",
"first-page": "e02020-16",
"journal-title": "J. Virol.",
"key": "ref_121",
"volume": "91",
"year": "2017"
},
{
"DOI": "10.1016/j.tim.2018.05.010",
"article-title": "Lipid Droplet Metabolism during Dengue Virus Infection",
"author": "Randall",
"doi-asserted-by": "crossref",
"first-page": "640",
"journal-title": "Trends Microbiol.",
"key": "ref_122",
"volume": "26",
"year": "2018"
},
{
"DOI": "10.1016/j.plipres.2016.09.005",
"article-title": "Lipids and flaviviruses, present and future perspectives for the control of dengue, Zika, and West Nile viruses",
"author": "Saiz",
"doi-asserted-by": "crossref",
"first-page": "123",
"journal-title": "Prog. Lipid Res.",
"key": "ref_123",
"volume": "64",
"year": "2016"
},
{
"DOI": "10.1371/journal.ppat.1007261",
"doi-asserted-by": "crossref",
"key": "ref_124",
"unstructured": "Gullberg, R.C., Steel, J.J., Pujari, V., Rovnak, J., Crick, D.C., and Perera, R. (2018). Stearoly-CoA desaturase 1 differentiates early and advanced dengue virus infections and determines virus particle infectivity. PLoS Pathog., 14."
},
{
"DOI": "10.1073/pnas.1110133108",
"article-title": "Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus",
"author": "Horner",
"doi-asserted-by": "crossref",
"first-page": "14590",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_125",
"volume": "108",
"year": "2011"
},
{
"DOI": "10.1128/JVI.01918-14",
"article-title": "MAVS Coordination of Antiviral Innate Immunity",
"author": "Vazquez",
"doi-asserted-by": "crossref",
"first-page": "6974",
"journal-title": "J. Virol.",
"key": "ref_126",
"volume": "89",
"year": "2015"
},
{
"DOI": "10.1016/j.celrep.2024.113724",
"article-title": "p53 promotes antiviral innate immunity by driving hexosamine metabolism",
"author": "Xia",
"doi-asserted-by": "crossref",
"first-page": "113724",
"journal-title": "Cell Rep.",
"key": "ref_127",
"volume": "43",
"year": "2024"
},
{
"DOI": "10.3389/fimmu.2025.1619926",
"doi-asserted-by": "crossref",
"key": "ref_128",
"unstructured": "Kanno, T., Miyako, K., and Endo, Y. (2025). The diverse interaction of metabolism, immune response, and viral pathogens. Front. Immunol., 16."
},
{
"DOI": "10.1016/j.cell.2010.04.018",
"article-title": "Peroxisomes are signaling platforms for antiviral innate immunity",
"author": "Dixit",
"doi-asserted-by": "crossref",
"first-page": "668",
"journal-title": "Cell",
"key": "ref_129",
"volume": "141",
"year": "2010"
},
{
"DOI": "10.3390/cells8070725",
"doi-asserted-by": "crossref",
"key": "ref_130",
"unstructured": "Wong, C.P., Xu, Z., Hou, S., Limonta, D., Kumar, A., Power, C., and Hobman, T.C. (2019). Interplay between Zika Virus and Peroxisomes during Infection. Cells, 8."
},
{
"DOI": "10.1093/jmcb/mjae004",
"article-title": "Identification of druggable host dependency factors shared by multiple SARS-CoV-2 variants of concern",
"author": "Frasson",
"doi-asserted-by": "crossref",
"first-page": "mjae004",
"journal-title": "J. Mol. Cell Biol.",
"key": "ref_131",
"volume": "16",
"year": "2024"
},
{
"DOI": "10.1101/cshperspect.a038398",
"article-title": "Structure and function of the influenza virus transcription and replication machinery",
"author": "Fodor",
"doi-asserted-by": "crossref",
"first-page": "a038398",
"journal-title": "Cold Spring Harb. Perspect. Med.",
"key": "ref_132",
"volume": "10",
"year": "2020"
},
{
"DOI": "10.1128/JVI.03544-12",
"article-title": "DNA topoisomerase 1 facilitates the transcription and replication of the Ebola virus genome",
"author": "Takahashi",
"doi-asserted-by": "crossref",
"first-page": "8862",
"journal-title": "J. Virol.",
"key": "ref_133",
"volume": "87",
"year": "2013"
},
{
"DOI": "10.1517/14728222.2013.827663",
"article-title": "The DNA helicase–primase complex as a target for herpes viral infection",
"author": "Weller",
"doi-asserted-by": "crossref",
"first-page": "1119",
"journal-title": "Expert Opin. Ther. Targets",
"key": "ref_134",
"volume": "17",
"year": "2013"
},
{
"DOI": "10.1111/cmi.12108",
"article-title": "The NF-κB inhibitor SC75741 efficiently blocks influenza virus propagation and confers a high barrier for development of viral resistance",
"author": "Ehrhardt",
"doi-asserted-by": "crossref",
"first-page": "1198",
"journal-title": "Cell. Microbiol.",
"key": "ref_135",
"volume": "15",
"year": "2013"
},
{
"DOI": "10.1128/JVI.00909-08",
"article-title": "NF-kappaB signaling differentially regulates influenza virus RNA synthesis",
"author": "Kumar",
"doi-asserted-by": "crossref",
"first-page": "9880",
"journal-title": "J. Virol.",
"key": "ref_136",
"volume": "82",
"year": "2008"
},
{
"DOI": "10.1128/JVI.01257-21",
"article-title": "The NF-κB Transcriptional Footprint Is Essential for SARS-CoV-2 Replication",
"author": "Uhl",
"doi-asserted-by": "crossref",
"first-page": "e0125721",
"journal-title": "J. Virol.",
"key": "ref_137",
"volume": "95",
"year": "2021"
},
{
"DOI": "10.1093/nar/gku769",
"article-title": "Molecular mechanisms of retroviral integration site selection",
"author": "Kvaratskhelia",
"doi-asserted-by": "crossref",
"first-page": "10209",
"journal-title": "Nucleic Acids Res.",
"key": "ref_138",
"volume": "42",
"year": "2014"
},
{
"DOI": "10.1038/s41467-019-08333-8",
"article-title": "Physical principles of retroviral integration in the human genome",
"author": "Michieletto",
"doi-asserted-by": "crossref",
"first-page": "575",
"journal-title": "Nat. Commun.",
"key": "ref_139",
"volume": "10",
"year": "2019"
},
{
"DOI": "10.3390/v14122606",
"doi-asserted-by": "crossref",
"key": "ref_140",
"unstructured": "Crawford, J.M., Yan, L.L., Zaher, H., and Hardy, R.W. (2022). Host-Dependent Modifications of Packaged Alphavirus Genomic RNA Influence Virus Replication in Mammalian Cells. Viruses, 14."
},
{
"DOI": "10.3389/fimmu.2017.01757",
"doi-asserted-by": "crossref",
"key": "ref_141",
"unstructured": "Chen, L., Wang, C., Luo, J., Su, W., Li, M., Zhao, N., Lyu, W., Attaran, H., He, Y., and Ding, H. (2017). Histone Deacetylase 1 Plays an Acetylation-Independent Role in Influenza A Virus Replication. Front. Immunol., 8."
},
{
"DOI": "10.1038/s41564-021-00868-1",
"article-title": "Sec24C is an HIV-1 host dependency factor crucial for virus replication",
"author": "Rebensburg",
"doi-asserted-by": "crossref",
"first-page": "435",
"journal-title": "Nat. Microbiol.",
"key": "ref_142",
"volume": "6",
"year": "2021"
},
{
"DOI": "10.1128/JVI.01751-19",
"article-title": "A genome-wide CRISPR-Cas9 screen identifies the dolichol-phosphate mannose synthase complex as a host dependency factor for dengue virus infection",
"author": "Labeau",
"doi-asserted-by": "crossref",
"first-page": "e01751-19",
"journal-title": "J. Virol.",
"key": "ref_143",
"volume": "94",
"year": "2020"
},
{
"DOI": "10.1128/mbio.00127-23",
"article-title": "Genome-scale CRISPR screening reveals host factors required for ribosome formation and viral replication",
"author": "Ohlson",
"doi-asserted-by": "crossref",
"first-page": "e0012723",
"journal-title": "mBio",
"key": "ref_144",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.1158/1535-7163.MCT-21-1000",
"article-title": "TOP1-DNA trapping by exatecan and combination therapy with ATR inhibitor",
"author": "Jo",
"doi-asserted-by": "crossref",
"first-page": "1090",
"journal-title": "Mol. Cancer Ther.",
"key": "ref_145",
"volume": "21",
"year": "2022"
},
{
"DOI": "10.1016/j.bcp.2022.115322",
"article-title": "Acyclovir resistance in herpes simplex viruses: Prevalence and therapeutic alternatives",
"author": "Schalkwijk",
"doi-asserted-by": "crossref",
"first-page": "115322",
"journal-title": "Biochem. Pharmacol.",
"key": "ref_146",
"volume": "206",
"year": "2022"
},
{
"DOI": "10.3390/ijms23031550",
"doi-asserted-by": "crossref",
"key": "ref_147",
"unstructured": "Oláh, J., Szénási, T., Lehotzky, A., Norris, V., and Ovádi, J. (2022). Challenges in Discovering Drugs That Target the Protein-Protein Interactions of Disordered Proteins. Int. J. Mol. Sci., 23."
},
{
"DOI": "10.1186/s12915-025-02214-x",
"doi-asserted-by": "crossref",
"key": "ref_148",
"unstructured": "Qin, C., Wang, Y.L., Zheng, J., Wan, X.B., and Fan, X.J. (2025). Current perspectives in drug targeting intrinsically disordered proteins and biomolecular condensates. BMC Biol., 23."
},
{
"DOI": "10.1128/CMR.00168-19",
"article-title": "Host-directed antiviral therapy",
"author": "Kumar",
"doi-asserted-by": "crossref",
"first-page": "e00168-19",
"journal-title": "Clin. Microbiol. Rev.",
"key": "ref_149",
"volume": "33",
"year": "2020"
},
{
"DOI": "10.1111/cts.13436",
"article-title": "Ribavirin aerosol in hospitalized adults with respiratory distress and COVID-19: An open-label trial",
"author": "Poulakou",
"doi-asserted-by": "crossref",
"first-page": "165",
"journal-title": "Clin. Transl. Sci.",
"key": "ref_150",
"volume": "16",
"year": "2023"
},
{
"DOI": "10.1128/jvi.01276-25",
"article-title": "Camptothecin, a topoisomerase I inhibitor, impedes productive herpes simplex virus type 1 infection",
"author": "Heath",
"doi-asserted-by": "crossref",
"first-page": "e0127625",
"journal-title": "J. Virol.",
"key": "ref_151",
"volume": "99",
"year": "2025"
},
{
"DOI": "10.1371/journal.pone.0307154",
"doi-asserted-by": "crossref",
"key": "ref_152",
"unstructured": "Sanyal, M., Watson, A., Shah, P., Lee, D., Liang, S., Joshi, G., Metitiri, E., Chowdhury, W.H., Bacich, D., and Dube, P. (2024). Valproic acid use is associated with diminished risk of contracting COVID-19, and diminished disease severity: Epidemiologic and in vitro analysis reveal mechanistic insights. PLoS ONE, 19."
},
{
"DOI": "10.1128/mbio.03954-24",
"article-title": "Mammalian fatty acid synthase: A commonly used viral host dependency factor and a putative target for host-targeted broad-spectrum antiviral therapeutic development",
"author": "Karthigeyan",
"doi-asserted-by": "crossref",
"first-page": "e0395424",
"journal-title": "mBio",
"key": "ref_153",
"volume": "16",
"year": "2025"
},
{
"DOI": "10.3390/v12091014",
"doi-asserted-by": "crossref",
"key": "ref_154",
"unstructured": "Gales, J.P., Kubina, J., Geldreich, A., and Dimitrova, M. (2020). Strength in diversity: Nuclear export of viral RNAs. Viruses, 12."
},
{
"DOI": "10.1016/j.coviro.2022.101254",
"article-title": "Viral–host interactions during splicing and nuclear export of influenza virus mRNAs",
"author": "Esparza",
"doi-asserted-by": "crossref",
"first-page": "101254",
"journal-title": "Curr. Opin. Virol.",
"key": "ref_155",
"volume": "55",
"year": "2022"
},
{
"DOI": "10.1128/jvi.01028-24",
"article-title": "Liquid-liquid phase separation is essential for reovirus viroplasm formation and immune evasion",
"author": "He",
"doi-asserted-by": "crossref",
"first-page": "e0102824",
"journal-title": "J. Virol.",
"key": "ref_156",
"volume": "98",
"year": "2024"
},
{
"DOI": "10.1101/cshperspect.a036814",
"article-title": "HCV Assembly and Egress via Modifications in Host Lipid Metabolic Systems",
"author": "Shimotohno",
"doi-asserted-by": "crossref",
"first-page": "a036814",
"journal-title": "Cold Spring Harb. Perspect. Med.",
"key": "ref_157",
"volume": "11",
"year": "2021"
},
{
"DOI": "10.3390/cells10030503",
"doi-asserted-by": "crossref",
"key": "ref_158",
"unstructured": "Saraste, J., and Prydz, K. (2021). Assembly and Cellular Exit of Coronaviruses: Hijacking an Unconventional Secretory Pathway from the Pre-Golgi Intermediate Compartment via the Golgi Ribbon to the Extracellular Space. Cells, 10."
},
{
"article-title": "Properties of coronavirus and SARS-CoV-2",
"author": "Malik",
"first-page": "3",
"journal-title": "Malays. J. Pathol.",
"key": "ref_159",
"volume": "42",
"year": "2020"
},
{
"DOI": "10.1016/j.vaccine.2012.09.077",
"article-title": "Influenza A virus progeny vRNP trafficking in live infected cells studied with the virus-encoded fluorescently tagged PB2 protein",
"author": "Avilov",
"doi-asserted-by": "crossref",
"first-page": "7411",
"journal-title": "Vaccine",
"key": "ref_160",
"volume": "30",
"year": "2012"
},
{
"DOI": "10.1038/s41467-019-09549-4",
"article-title": "Influenza A virus ribonucleoproteins form liquid organelles at endoplasmic reticulum exit sites",
"author": "Alenquer",
"doi-asserted-by": "crossref",
"first-page": "1629",
"journal-title": "Nat. Commun.",
"key": "ref_161",
"volume": "10",
"year": "2019"
},
{
"DOI": "10.1016/j.virol.2010.12.003",
"article-title": "Influenza virus assembly and budding",
"author": "Rossman",
"doi-asserted-by": "crossref",
"first-page": "229",
"journal-title": "Virology",
"key": "ref_162",
"volume": "411",
"year": "2011"
},
{
"DOI": "10.15252/embr.202256275",
"article-title": "IP6-stabilised HIV capsids evade cGAS/STING-mediated host immune sensing",
"author": "Papa",
"doi-asserted-by": "crossref",
"first-page": "e56275",
"journal-title": "EMBO Rep.",
"key": "ref_163",
"volume": "24",
"year": "2023"
},
{
"DOI": "10.3390/v16091423",
"doi-asserted-by": "crossref",
"key": "ref_164",
"unstructured": "McGraw, A., Hillmer, G., Medehincu, S.M., Hikichi, Y., Gagliardi, S., Narayan, K., Tibebe, H., Marquez, D., Mei Bose, L., and Keating, A. (2024). Exploring HIV-1 maturation: A new frontier in antiviral development. Viruses, 16."
},
{
"DOI": "10.1038/nrm2937",
"article-title": "Membrane budding and scission by the ESCRT machinery: It’s all in the neck",
"author": "Hurley",
"doi-asserted-by": "crossref",
"first-page": "556",
"journal-title": "Nat. Rev. Mol. Cell Biol.",
"key": "ref_165",
"volume": "11",
"year": "2010"
},
{
"DOI": "10.1038/nrmicro3490",
"article-title": "HIV-1 assembly, release and maturation",
"author": "Freed",
"doi-asserted-by": "crossref",
"first-page": "484",
"journal-title": "Nat. Rev. Microbiol.",
"key": "ref_166",
"volume": "13",
"year": "2015"
},
{
"DOI": "10.3390/v16020316",
"doi-asserted-by": "crossref",
"key": "ref_167",
"unstructured": "Carter, T., and Iqbal, M. (2024). The influenza a virus replication cycle: A comprehensive review. Viruses, 16."
},
{
"DOI": "10.3390/ijms232213935",
"doi-asserted-by": "crossref",
"key": "ref_168",
"unstructured": "Álvarez-Fernández, H., Mingo-Casas, P., Blázquez, A.-B., Caridi, F., Saiz, J.C., Pérez-Pérez, M.-J., Martín-Acebes, M.A., and Priego, E.-M. (2022). Allosteric inhibition of neutral sphingomyelinase 2 (nSMase2) by DPTIP: From antiflaviviral activity to deciphering its binding site through in silico studies and experimental validation. Int. J. Mol. Sci., 23."
},
{
"DOI": "10.15252/embr.202051709",
"article-title": "Phosphatidylserine clustering by the Ebola virus matrix protein is a critical step in viral budding",
"author": "Husby",
"doi-asserted-by": "crossref",
"first-page": "e51709",
"journal-title": "EMBO Rep.",
"key": "ref_169",
"volume": "23",
"year": "2022"
},
{
"DOI": "10.1016/j.isci.2024.110424",
"article-title": "Influenza A virus infection activates STAT3 to enhance SREBP2 expression, cholesterol biosynthesis, and virus replication",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "110424",
"journal-title": "iScience",
"key": "ref_170",
"volume": "27",
"year": "2024"
},
{
"DOI": "10.1371/journal.ppat.1013646",
"doi-asserted-by": "crossref",
"key": "ref_171",
"unstructured": "Zhang, J., Cao, R., Wang, Y., Sun, Y., Ji, X., Liu, P., Liu, K., Sun, J., Chen, X., and Cai, D. (2025). Influenza virus infection reprograms cholesterol biosynthesis to facilitate virus replication by the TAK1-RORγ axis. PLoS Pathog., 21."
},
{
"DOI": "10.1080/22221751.2025.2470371",
"article-title": "SREBP2-dependent lipid droplet formation enhances viral replication and deteriorates lung injury in mice following IAV infection",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "2470371",
"journal-title": "Emerg. Microbes Infect.",
"key": "ref_172",
"volume": "14",
"year": "2025"
},
{
"DOI": "10.1055/a-2095-0826",
"article-title": "Repurposing drugs: An empowering approach to drug discovery and development",
"author": "Kumar",
"doi-asserted-by": "crossref",
"first-page": "481",
"journal-title": "Drug Res.",
"key": "ref_173",
"volume": "73",
"year": "2023"
},
{
"DOI": "10.1038/s41419-024-06463-6",
"article-title": "The implications of FASN in immune cell biology and related diseases",
"author": "Xiao",
"doi-asserted-by": "crossref",
"first-page": "88",
"journal-title": "Cell Death Dis.",
"key": "ref_174",
"volume": "15",
"year": "2024"
},
{
"DOI": "10.1186/s13046-021-02041-2",
"article-title": "Statins: A repurposed drug to fight cancer",
"author": "Jiang",
"doi-asserted-by": "crossref",
"first-page": "241",
"journal-title": "J. Exp. Clin. Cancer Res.",
"key": "ref_175",
"volume": "40",
"year": "2021"
},
{
"DOI": "10.1016/j.drudis.2021.03.025",
"article-title": "Nipping disease in the bud: nSMase2 inhibitors as therapeutics in extracellular vesicle-mediated diseases",
"author": "Tallon",
"doi-asserted-by": "crossref",
"first-page": "1656",
"journal-title": "Drug Discov. Today",
"key": "ref_176",
"volume": "26",
"year": "2021"
},
{
"DOI": "10.1016/j.semcdb.2020.04.001",
"article-title": "Direct trafficking pathways from the Golgi apparatus to the plasma membrane",
"author": "Stalder",
"doi-asserted-by": "crossref",
"first-page": "112",
"journal-title": "Semin. Cell Dev. Biol.",
"key": "ref_177",
"volume": "107",
"year": "2020"
},
{
"DOI": "10.3389/fmicb.2017.01171",
"doi-asserted-by": "crossref",
"key": "ref_178",
"unstructured": "Mathew, C., and Ghildyal, R. (2017). CRM1 inhibitors for antiviral therapy. Front. Microbiol., 8."
},
{
"DOI": "10.1016/j.jaci.2023.12.013",
"article-title": "Tapinarof validates the aryl hydrocarbon receptor as a therapeutic target: A clinical review",
"author": "Silverberg",
"doi-asserted-by": "crossref",
"first-page": "1",
"journal-title": "J. Allergy Clin. Immunol.",
"key": "ref_179",
"volume": "154",
"year": "2024"
},
{
"DOI": "10.1038/nrmicro2596",
"article-title": "Host factors involved in retroviral budding and release",
"author": "Neil",
"doi-asserted-by": "crossref",
"first-page": "519",
"journal-title": "Nat. Rev. Microbiol.",
"key": "ref_180",
"volume": "9",
"year": "2011"
},
{
"DOI": "10.1038/s41586-025-08950-y",
"article-title": "The expanding repertoire of ESCRT functions in cell biology and disease",
"author": "Hurley",
"doi-asserted-by": "crossref",
"first-page": "877",
"journal-title": "Nature",
"key": "ref_181",
"volume": "642",
"year": "2025"
},
{
"DOI": "10.1016/j.virol.2016.11.011",
"article-title": "Cholesterol reducing agents inhibit assembly of type I parainfluenza viruses",
"author": "Bajimaya",
"doi-asserted-by": "crossref",
"first-page": "127",
"journal-title": "Virology",
"key": "ref_182",
"volume": "501",
"year": "2017"
}
],
"reference-count": 182,
"references-count": 182,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.mdpi.com/1422-0067/27/1/147"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Targeting Host Dependency Factors: A Paradigm Shift in Antiviral Strategy Against RNA Viruses",
"type": "journal-article",
"update-policy": "https://doi.org/10.3390/mdpi_crossmark_policy",
"volume": "27"
}