Construction of Synthetic Probiotic Bacteria for In Situ Delivery of Anti-SARS-CoV-2 Nanobodies
et al., Probiotics and Antimicrobial Proteins, doi:10.1007/s12602-025-10758-1, Sep 2025
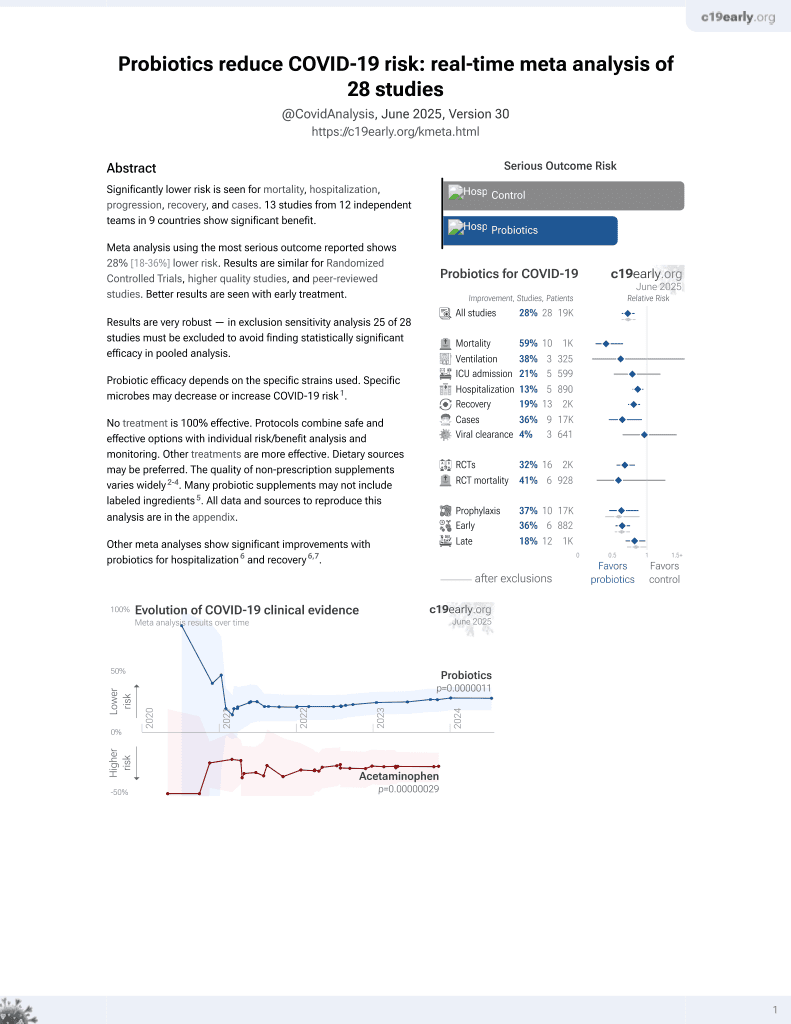
Probiotics for COVID-19
20th treatment shown to reduce risk in
March 2021, now with p = 0.00000044 from 29 studies.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
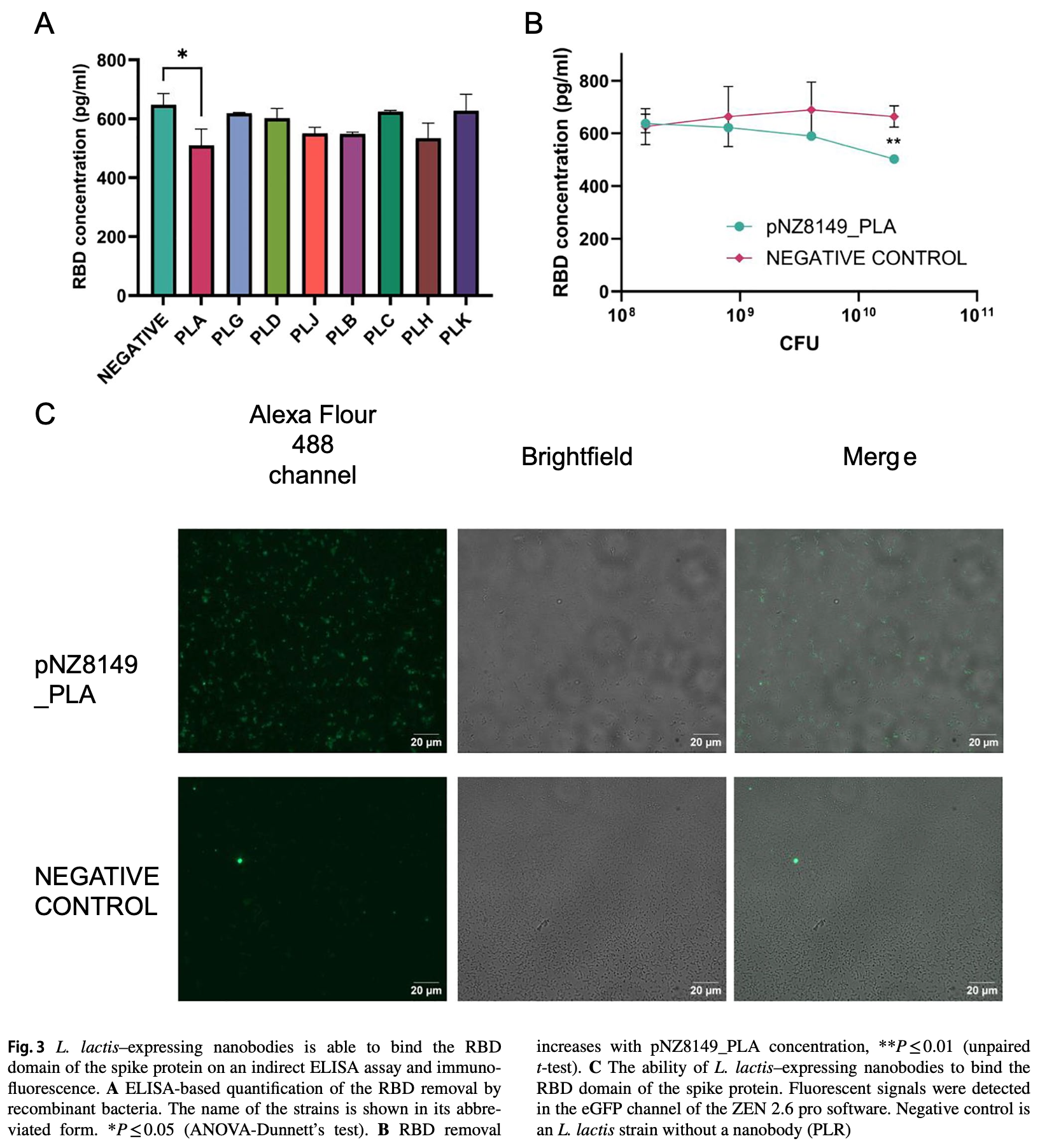
In vitro study showing that genetically modified Lactococcus lactis bacteria expressing anti-SARS-CoV-2 nanobodies can inhibit viral infection by blocking spike protein-ACE2 receptor interaction.
Probiotic efficacy depends on the specific strains used. Specific microbes may decrease or increase COVID-19 risk1.
2 preclinical studies support the efficacy of probiotics for COVID-19:
1.
Li et al., Large-scale genetic correlation studies explore the causal relationship and potential mechanism between gut microbiota and COVID-19-associated risks, BMC Microbiology, doi:10.1186/s12866-024-03423-0.
Portero et al., 11 Sep 2025, peer-reviewed, 6 authors.
Contact: hany@ohio.edu.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Construction of Synthetic Probiotic Bacteria for In Situ Delivery of Anti-SARS-CoV-2 Nanobodies
Probiotics and Antimicrobial Proteins, doi:10.1007/s12602-025-10758-1
SARS-CoV-2 viral infection can be inhibited by blocking the interaction between the viral spike protein and the human receptor angiotensin-converting enzyme 2 (hACE2). The development of specific spike inhibitors using nanobodies, the antigen-binding region of llamas' antibodies, arose as a promising therapeutic method against SARS-CoV-2. However, one limitation of nanobodies is that they cannot be used directly in the human body due to their susceptibility to degradation. Bacteria-based delivery systems provide site-specific targeted action that can circumvent nanobody degradation. Here, we report the development of a genetically modified bacterium expressing anti-SARS-CoV-2 nanobodies that can inhibit the interaction between the hACE2 receptor and the receptor-binding domain (RBD) of the spike protein. Lactococcus lactis, a human symbiont probiotic bacterium, was selected to express nanobodies attached to their cell surface. Our data shows that FLAG-tagged anti-SARS-CoV-2 nanobodies were detected on the cell surface of recombinant L. lactis strains by flow cytometry and immunofluorescence without permeabilization. Furthermore, nanobodies are functional and can bind the RBD region from the spike protein in a dose-dependent manner. Inhibition of the hACE2-RBD interaction in cellular assays was quantified using a pseudotype lentivirus that mimics SARS-CoV-2 in an adaptation of the neutralization assay. Our results suggested that the recombinant bacteria can inhibit viral infectivity in more than 50% compared with a control without bacteria in a neutralization assay. These outcomes suggest that the engineered strain can be used in the future as a new therapeutic tool in COVID-19 prevention.
Supplementary Information The online version contains supplementary material available at https:// doi. org/ 10. 1007/ s12602-025-10758-1 .
Author Contributions
Declarations Competing interests The authors declare no competing interests. Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . Publisher's Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Acharya, Pandey, Thurman, Discovery and evaluation of entry inhibitors for SARS-CoV-2 and its emerging variants, J Virol, doi:10.1128/JVI.01437-21
Adekiya, Sumaila, Aruleba, Choonara, Mucosal targeting strategies for antiviral drug delivery
Babamohamadi, Mohammadi, Faryadi, Anti-CTLA-4 nanobody as a promising approach in cancer immunotherapy, Cell Death Dis, doi:10.1038/s41419-023-06391-x
Badi, Tarashi, Fateh, From the role of microbiota in gut-lung axis to SARS-CoV-2 pathogenesis, Mediators Inflamm, doi:10.1155/2021/6611222
Bannas, Hambach, Koch-Nolte, Nanobodies and nanobody-based human heavy chain antibodies as antitumor therapeutics, Front Immunol, doi:10.3389/fimmu.2017.01603
Bermúdez-Humarán, Motta, Serine protease inhibitors protect better than IL-10 and TGF-β antiinflammatory cytokines against mouse colitis when delivered by recombinant lactococci, Microb Cell Fact, doi:10.1186/s12934-015-0198-4
Bhattacharya, Gupta, A natural food preservative peptide nisin can interact with the SARS-CoV-2 spike protein receptor human ACE2, Virology, doi:10.1016/j.virol.2020.10.002
Botic, Klingberg, Weingartl, Cencic, A novel eukaryotic cell culture model to study antiviral activity of potential probiotic bacteria, Int J Food Microbiol, doi:10.1016/j.ijfoodmicro.2006.10.044
Braat, Rottiers, Hommes, A phase I trial with transgenic bacteria expressing interleukin-10 in Crohn's disease, Clin Gastroenterol Hepatol, doi:10.1016/j.cgh.2006.03.028
Caluwaerts, Vandenbroucke, Steidler, AG013, a mouth rinse formulation of Lactococcus lactis secreting human trefoil factor 1, provides a safe and efficacious therapeutic tool for treating oral mucositis, Oral Oncol, doi:10.1016/j.oraloncology.2010.04.008
Charbonneau, Isabella, Li, Kurtz, Developing a new class of engineered live bacterial therapeutics to treat human diseases, Nat Commun, doi:10.1038/s41467-020-15508-1
Cho, Skinner, Lim, The impact of Lactococcus lactis (probiotic nasal rinse) co-culture on growth of patientderived strains of Pseudomonas aeruginosa, Int Forum Allergy Rhinol, doi:10.1002/alr.22521
Chowdhury, Castro, Coker, Programmable bacteria induce durable tumor regression and systemic antitumor immunity, Nat Med, doi:10.1038/s41591-019-0498-z
Cofas-Vargas, Olivos-Ramirez, Chwastyk, Nanomechanical footprint of SARS-CoV-2 variants in complex with a potent nanobody by molecular simulations, Nanoscale, doi:10.1039/D4NR02074J
Cook, Gysemans, Mathieu, Lactococcus lactis as a versatile vehicle for tolerogenic immunotherapy, Front Immunol, doi:10.3389/fimmu.2017.01961
Crawford, Eguia, Dingens, Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays, Viruses, doi:10.3390/v12050513
De Greve, Fioravanti, Single domain antibodies from camelids in the treatment of microbial infections, Front Immunol, doi:10.3389/fimmu.2024.1334829
Dosoky, Zhang, Davies, Engineering the gut microbiota to treat chronic diseases, Appl Microbiol Biotechnol, doi:10.1007/s00253-020-10771-0
Driessen, Nouwen, Protein translocation across the bacterial cytoplasmic membrane, Annu Rev Biochem, doi:10.1146/annurev.biochem.77.061606.160747
Endam, Alromaih, Gonzalez, Intranasal application of Lactococcus lactis W136 is safe in chronic rhinosinusitis patients with previous sinus surgery, Front Cell Infect Microbiol, doi:10.3389/fcimb.2020.00440
Frische, Brooks, Gybel-Brask, Optimization and evaluation of a live virus SARS-CoV-2 neutralization assay, PLoS ONE, doi:10.1371/journal.pone.0272298
Gentschev, Spreng, Sieber, Vivotif® -a 'magic shield' for protection against typhoid fever and delivery of heterologous antigens, Chemotherapy, doi:10.1159/000100515
Golcuk, Hacisuleyman, Erman, Binding mechanism of neutralizing nanobodies targeting SARS-CoV-2 spike glycoprotein, J Chem Inf Model, doi:10.1021/acs.jcim.1c00695
Gurbatri, Vincent, Engineered probiotics for local tumor delivery of checkpoint blockade nanobodies, Sci Transl Med, doi:10.1126/scitranslmed.aax0876
Gutierrez-Guerrero, Cosset, Verhoeyen, Lentiviral vector pseudotypes: precious tools to improve gene modification of hematopoietic cells for research and gene therapy, Viruses, doi:10.3390/v12091016
Hanke, Perez, Sheward, An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction, Nat Commun, doi:10.1038/s41467-020-18174-5
Hanson, Hixon, Oral delivery of IL-27 recombinant bacteria attenuates immune colitis in mice, Gastroenterology, doi:10.1053/j.gastro.2013.09.060
Held, Using phenol red to assess pH in tissue culture media
Hou, Okuda, Edwards, SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract, Cell, doi:10.1016/j.cell.2020.05.042
Huo, Bas, Ruza, Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2, Nat Struct Mol Biol, doi:10.1038/s41594-020-0469-6
Inda, Broset, Lu, De La Fuente-Nunez, Emerging frontiers in microbiome engineering, Trends Immunol, doi:10.1016/j.it.2019.08.007
Jackson, Farzan, Chen, Choe, Mechanisms of SARS-CoV-2 entry into cells, Nat Rev Mol Cell Biol, doi:10.1038/s41580-021-00418-x
Jin, Odongo, Radwanska, Magez, NANOBOD-IES®: a review of diagnostic and therapeutic applications, IJMS, doi:10.3390/ijms24065994
Klijn, Weerkamp, Vos, Genetic marking of Lactococcus lactis shows its survival in the human gastrointestinal tract, Appl Environ Microbiol, doi:10.1128/aem.61.7.2771-2774.1995
Kuipers, Wierenga, Rink, Sec-mediated transport of posttranslationally dehydrated peptides in Lactococcus lactis, Appl Environ Microbiol, doi:10.1128/AEM.01802-06
Lang=, The QIAexpressionist TM
Liang, Wang, Shao, Recent advances in bacteriamediated cancer therapy, Front Bioeng Biotechnol, doi:10.3389/fbioe.2022.1026248
Limaye, Haddad, Cilli, Phase 1b, multicenter, single blinded, placebo-controlled, sequential dose escalation study to assess the safety and tolerability of topically applied AG013 in subjects with locally advanced head and neck cancer receiving induction chemotherapy, Cancer, doi:10.1002/cncr.28365
Linares, Gómez, Renes, Lactic acid bacteria and bifidobacteria with potential to design natural biofunctional health-promoting dairy foods, Front Microbiol, doi:10.3389/fmicb.2017.00846
Liu, Lu, Cai, A broad neutralizing nanobody against SARS-CoV-2 engineered from an approved drug, Cell Death Dis, doi:10.1038/s41419-024-06802-7
Liu, Shao, Dong, Bai, Solution for promoting egl 3 gene of Trichoderma reesei high-efficiency secretory expression in Escherichia coli and Lactococcus lactis, Process Biochem, doi:10.1016/j.procbio.2017.07.031
Liu, Wei, Alvarez, Epithelial cells lining salivary gland ducts are early target cells of severe acute respiratory syndrome coronavirus infection in the upper respiratory tracts of rhesus macaques, J Virol, doi:10.1128/JVI.02292-10
Lu, Zhao, Li, Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding, Lancet, doi:10.1016/S0140-6736(20)30251-8
Luke, Piha-Paul, Medina, Phase I study of SYNB1891, an engineered E. coli Nissle strain expressing STING agonist, with and without atezolizumab in advanced malignancies, Clin Cancer Res, doi:10.1158/1078-0432.CCR-23-0118
Mast, Fridy, Ketaren, Highly synergistic combinations of nanobodies that target SARS-CoV-2 and are resistant to escape, Elife, doi:10.7554/eLife.73027
Mathiesen, Sveen, Brurberg, Genome-wide analysis of signal peptide functionality in Lactobacillus plantarum WCFS1, BMC Genomics, doi:10.1186/1471-2164-10-425
Mathieu, Wiedeman, Cerosaletti, A first-inhuman, open-label phase 1b and a randomised, double-blind phase 2a clinical trial in recent-onset type 1 diabetes with AG019 as monotherapy and in combination with teplizumab, Diabetologia, doi:10.1007/s00125-023-06014-2
Michon, Langella, Eijsink, Display of recombinant proteins at the surface of lactic acid bacteria: strategies and applications, Microb Cell Fact, doi:10.1186/s12934-016-0468-9
Mikolajek, Weckener, Brotzakis, Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes, Proc Natl Acad Sci, doi:10.1073/pnas.2205412119
Milone, Doherty, Clinical use of lentiviral vectors, Leukemia, doi:10.1038/s41375-018-0106-0
Mohseni, Keyvani, The first clinical use of a recombinant Lactococcus lactis expressing human papillomavirus type 16 E7 oncogene oral vaccine: a phase I safety and immunogenicity trial in healthy women volunteers, Mol Cancer Ther, doi:10.1158/1535-7163.MCT-19-0375
Morello, Bermúdez-Humarán, Llull, Lactococcus lactis, an efficient cell factory for recombinant protein production and secretion, Microb Physiol, doi:10.1159/000106082
Motta, Bermúdez-Humarán, Deraison, Food-grade bacteria expressing elafin protect against inflammation and restore colon homeostasis, Sci Transl Med, doi:10.1126/scitranslmed.3004212
Namai, Shigemori, Ogita, Microbial therapeutics for acute colitis based on genetically modified Lactococcus lactis hypersecreting IL-1Ra in mice, Exp Mol Med, doi:10.1038/s12276-020-00507-5
Natale, Brüser, Driessen, Sec-and Tat-mediated protein secretion across the bacterial cytoplasmic membranedistinct translocases and mechanisms, Biochimica et Biophysica Acta (BBA), doi:10.1016/j.bbamem.2007.07.015
Nguyen, Li, Antibody-nanobody combination increases their neutralizing activity against SARS-CoV-2 and nanobody H11-H4 is effective against alpha, kappa and delta variants, Sci Rep, doi:10.1038/s41598-022-14263-1
Nie, Li, Wu, Quantification of SARS-CoV-2 neutralizing antibody by a pseudotyped virus-based assay, Nat Protoc, doi:10.1038/s41596-020-0394-5
Okello, Moonens, Erume, Greve, Orally fed recombinant Lactococcus lactis displaying surface anti-fimbrial nanobodies protects piglets against Escherichia coli causing postweaning diarrhea, Agriculture, doi:10.3390/agriculture11030186
Plavec, Štrukelj, Berlec, Screening for new surface anchoring domains for Lactococcus lactis, Front Microbiol, doi:10.3389/fmicb.2019.01879
Rossotti, Van Faassen, Tran, Arsenal of nanobodies shows broad-spectrum neutralization against SARS-CoV-2 variants of concern in vitro and in vivo in hamster models, Commun Biol, doi:10.1038/s42003-022-03866-z
Rutherford, Mourez, Surface display of proteins by gram-negative bacterial autotransporters, Microb Cell Fact, doi:10.1186/1475-2859-5-22
Ruyter, Kuipers, Vos, Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin, Appl Environ Microbiol, doi:10.1128/aem.62.10.3662-3667.1996
Scott, Supramaniam, Idris, Engineered extracellular vesicles directed to the spike protein inhibit SARS-CoV-2, Mol Ther, doi:10.1016/j.omtm.2022.01.015
Shen, Aggarwal, Wun, Engineered microbial systems for advanced drug delivery, Adv Drug Deliv Rev, doi:10.1016/j.addr.2022.114364
Shuwen, Yifei, Xinyue, Advances in bacteriabased drug delivery systems for anti-tumor therapy, Clin Transl Immunol, doi:10.1002/cti2.1518
Sirichokchatchawan, Temeeyasen, Nilubol, Prapasarakul, Protective effects of cell-free supernatant and live lactic acid bacteria isolated from Thai pigs against a pandemic strain of porcine epidemic diarrhea virus, Probiotics Antimicrob Proteins, doi:10.1007/s12602-017-9281-y
Slastnikova, Ulasov, Rosenkranz, Sobolev, Targeted intracellular delivery of antibodies: the state of the art, Front Pharmacol, doi:10.3389/fphar.2018.01208
Song, Nikoloff, Zhang, Improving protein production on the level of regulation of both expression and secretion pathways in Bacillus subtilis, J Microbiol Biotechnol, doi:10.4014/jmb.1501.01028
Sonvico, Colombo, Quarta, Nasal delivery as a strategy for the prevention and treatment of COVID-19, Expert Opin Drug Deliv, doi:10.1080/17425247.2023.2263363
Steen, Buist, Horsburgh, AcmA of Lactococcus lactis is an N -acetylglucosaminidase with an optimal number of LysM domains for proper functioning, FEBS J, doi:10.1111/j.1742-4658.2005.04706.x
Steidler, Robinson, Chamberlain, Mucosal delivery of murine interleukin-2 (IL-2) and IL-6 by recombinant strains of Lactococcus lactis coexpressing antigen and cytokine, Infect Immun, doi:10.1128/IAI.66.7.3183-3189.1998
Tang, Owens, Naismith, Structural biology of nanobodies against the spike protein of SARS-CoV-2, Viruses, doi:10.3390/v13112214
Tavares, De Jesus, Da Silva, Novel strategies for efficient production and delivery of live biotherapeutics and biotechnological uses of Lactococcus lactis: the lactic acid bacterium model, Front Bioeng Biotechnol, doi:10.3389/fbioe.2020.517166
Teymennet-Ramírez, Martínez-Morales, Trejo-Hernández, Yeast surface display system: strategies for improvement and biotechnological applications, Front Bioeng Biotechnol, doi:10.3389/fbioe.2021.794742
Valenzuela-Nieto, Miranda-Chacon, Salinas-Rebolledo, Nanobodies: COVID-19 and future perspectives, Front Drug Discov, doi:10.3389/fddsv.2022.927164
Vandenbroucke, Haard, Beirnaert, Orally administered L. lactis secreting an anti-TNF nanobody demonstrate efficacy in chronic colitis, Mucosal Immunol, doi:10.1038/mi.2009.116
Vo, Lee, Synthetic bacteria for therapeutics, J Microbiol Biotechnol, doi:10.4014/jmb.1904.04016
Wang, Chai, Burwinkel, Inhibitory influence of Enterococcus faecium on the propagation of swine influenza A virus in vitro, PLoS ONE, doi:10.1371/journal.pone.0053043
Wang, Zhou, Guo, Oral immunization of chickens with Lactococcus lactis expressing cjaA temporarily reduces Campylobacter jejuni colonization, Foodborne Pathog Dis, doi:10.1089/fpd.2019.2727
Wessels, Axelsson, Hansen, The lactic acid bacteria, the food chain, and their regulation, Trends Food Sci Technol, doi:10.1016/j.tifs.2004.03.003
Wiggins, Nguyen, Lactic acid bacterial surface display of scytovirin inhibitors for anti-ebolavirus infection, Front Microbiol, doi:10.3389/fmicb.2023.1269869
Yang, Gu, Xu, Recombinant Lactococcus lactis secreting FliC protein nanobodies for resistance against Salmonella enteritidis invasion in the intestinal tract, J Nanobiotechnol, doi:10.1186/s12951-024-02904-8
Yang, Li, Du, Therapeutic nanobodies against SARS-CoV-2 and other pathogenic human coronaviruses, J Nanobiotechnol, doi:10.1186/s12951-024-02573-7
Yoda, Takase, Suzuki, Development of engineered IL-36γ-hypersecreting Lactococcus lactis to improve the intestinal environment, World J Microbiol Biotechnol, doi:10.1007/s11274-024-04157-x
Yumoto, Sato, Nakashima, Nasally administered Lactococcus lactis secreting heme oxygenase-1 attenuates murine emphysema, Antioxidants, doi:10.3390/antiox9111049
Zahirović, Berlec, Targeting IL-6 by engineered Lactococcus lactis via surface-displayed affibody, Microb Cell Fact, doi:10.1186/s12934-022-01873-7
Zahirović, Plavec, Berlec, Dual functionalized Lactococcus lactis shows tumor antigen targeting and cytokine binding in vitro, Front Bioeng Biotechnol, doi:10.3389/fbioe.2022.822823
Zare, Aghamollaei, Hosseindokht, Nanobodies, the potent agents to detect and treat the coronavirus infections: a systematic review, Mol Cell Probes, doi:10.1016/j.mcp.2020.101692
Zhang, Dong, Xu, Recombinant Lactococcus lactis expressing human LL-37 prevents deaths from viral infections in piglets and chicken, Probiotics Antimicrob Proteins, doi:10.1007/s12602-023-10155-6
Zhang, Ni, Zhang, Recombinant L. lactis vaccine LL-plSAM-WAE targeting four virulence factors provides mucosal immunity against H. pylori infection, Microb Cell Fact, doi:10.1186/s12934-024-02321-4
Zhang, Wang, Wu, A SARS-CoV-2 nanobody displayed on the surface of human ferritin with high neutralization activity, Int J Nanomedicine, doi:10.2147/IJN.S450829
Zhou, Li, Ma, Pan, The nisin-controlled gene expression system: construction, application and improvements, Biotechnol Adv, doi:10.1016/j.biotechadv.2005.11.001
DOI record:
{
"DOI": "10.1007/s12602-025-10758-1",
"ISSN": [
"1867-1306",
"1867-1314"
],
"URL": "http://dx.doi.org/10.1007/s12602-025-10758-1",
"abstract": "<jats:title>Abstract</jats:title>\n <jats:p>SARS-CoV-2 viral infection can be inhibited by blocking the interaction between the viral spike protein and the human receptor angiotensin-converting enzyme 2 (hACE2). The development of specific spike inhibitors using nanobodies, the antigen-binding region of llamas’ antibodies, arose as a promising therapeutic method against SARS-CoV-2. However, one limitation of nanobodies is that they cannot be used directly in the human body due to their susceptibility to degradation. Bacteria-based delivery systems provide site-specific targeted action that can circumvent nanobody degradation. Here, we report the development of a genetically modified bacterium expressing anti-SARS-CoV-2 nanobodies that can inhibit the interaction between the hACE2 receptor and the receptor-binding domain (RBD) of the spike protein. <jats:italic>Lactococcus lactis</jats:italic>, a human symbiont probiotic bacterium, was selected to express nanobodies attached to their cell surface. Our data shows that FLAG-tagged anti-SARS-CoV-2 nanobodies were detected on the cell surface of recombinant <jats:italic>L. lactis </jats:italic>strains by flow cytometry and immunofluorescence without permeabilization. Furthermore, nanobodies are functional and can bind the RBD region from the spike protein in a dose-dependent manner. Inhibition of the hACE2-RBD interaction in cellular assays was quantified using a pseudotype lentivirus that mimics SARS-CoV-2 in an adaptation of the neutralization assay. Our results suggested that the recombinant bacteria can inhibit viral infectivity in more than 50% compared with a control without bacteria in a neutralization assay. These outcomes suggest that the engineered strain can be used in the future as a new therapeutic tool in COVID-19 prevention.\n</jats:p>",
"alternative-id": [
"10758"
],
"assertion": [
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Received",
"name": "received",
"order": 1,
"value": "6 May 2025"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "27 August 2025"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "First Online",
"name": "first_online",
"order": 3,
"value": "11 September 2025"
},
{
"group": {
"label": "Declarations",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 1
},
{
"group": {
"label": "Competing interests",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 2,
"value": "The authors declare no competing interests."
}
],
"author": [
{
"affiliation": [],
"family": "Portero",
"given": "Carolina E.",
"sequence": "first"
},
{
"affiliation": [],
"family": "Smith",
"given": "Claire",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Zhou",
"given": "Yuxi",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Marchán-Rivadeneira",
"given": "M. Raquel",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Wu",
"given": "Shiyong",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Han",
"given": "Yong",
"sequence": "additional"
}
],
"container-title": "Probiotics and Antimicrobial Proteins",
"container-title-short": "Probiotics & Antimicro. Prot.",
"content-domain": {
"crossmark-restriction": false,
"domain": [
"link.springer.com"
]
},
"created": {
"date-parts": [
[
2025,
9,
11
]
],
"date-time": "2025-09-11T10:20:25Z",
"timestamp": 1757586025000
},
"deposited": {
"date-parts": [
[
2025,
9,
11
]
],
"date-time": "2025-09-11T10:20:28Z",
"timestamp": 1757586028000
},
"funder": [
{
"award": [
"UT22117"
],
"name": "I- Corps"
},
{
"name": "Konneker Research Foundation Endowment"
}
],
"indexed": {
"date-parts": [
[
2025,
9,
16
]
],
"date-time": "2025-09-16T16:41:59Z",
"timestamp": 1758040919489,
"version": "3.44.0"
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2025,
9,
11
]
]
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
9,
11
]
],
"date-time": "2025-09-11T00:00:00Z",
"timestamp": 1757548800000
}
},
{
"URL": "https://creativecommons.org/licenses/by/4.0",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
9,
11
]
],
"date-time": "2025-09-11T00:00:00Z",
"timestamp": 1757548800000
}
}
],
"link": [
{
"URL": "https://link.springer.com/content/pdf/10.1007/s12602-025-10758-1.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://link.springer.com/article/10.1007/s12602-025-10758-1/fulltext.html",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://link.springer.com/content/pdf/10.1007/s12602-025-10758-1.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "297",
"original-title": [],
"prefix": "10.1007",
"published": {
"date-parts": [
[
2025,
9,
11
]
]
},
"published-online": {
"date-parts": [
[
2025,
9,
11
]
]
},
"publisher": "Springer Science and Business Media LLC",
"reference": [
{
"DOI": "10.1016/S0140-6736(20)30251-8",
"author": "R Lu",
"doi-asserted-by": "publisher",
"first-page": "565",
"journal-title": "Lancet",
"key": "10758_CR1",
"unstructured": "Lu R, Zhao X, Li J et al (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395:565–574. https://doi.org/10.1016/S0140-6736(20)30251-8",
"volume": "395",
"year": "2020"
},
{
"DOI": "10.1016/j.omtm.2022.01.015",
"author": "TA Scott",
"doi-asserted-by": "publisher",
"first-page": "355",
"journal-title": "Mol Ther",
"key": "10758_CR2",
"unstructured": "Scott TA, Supramaniam A, Idris A et al (2022) Engineered extracellular vesicles directed to the spike protein inhibit SARS-CoV-2. Mol Ther 24:355–366. https://doi.org/10.1016/j.omtm.2022.01.015",
"volume": "24",
"year": "2022"
},
{
"DOI": "10.1038/s41580-021-00418-x",
"author": "CB Jackson",
"doi-asserted-by": "publisher",
"first-page": "3",
"journal-title": "Nat Rev Mol Cell Biol",
"key": "10758_CR3",
"unstructured": "Jackson CB, Farzan M, Chen B, Choe H (2022) Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol 23:3–20. https://doi.org/10.1038/s41580-021-00418-x",
"volume": "23",
"year": "2022"
},
{
"DOI": "10.1128/JVI.01437-21",
"author": "A Acharya",
"doi-asserted-by": "publisher",
"journal-title": "J Virol",
"key": "10758_CR4",
"unstructured": "Acharya A, Pandey K, Thurman M et al (2021) Discovery and evaluation of entry inhibitors for SARS-CoV-2 and its emerging variants. J Virol 95:e01437–21. https://doi.org/10.1128/JVI.01437-21",
"volume": "95",
"year": "2021"
},
{
"DOI": "10.3389/fddsv.2022.927164",
"author": "G Valenzuela-Nieto",
"doi-asserted-by": "publisher",
"journal-title": "Front Drug Discov",
"key": "10758_CR5",
"unstructured": "Valenzuela-Nieto G, Miranda-Chacon Z, Salinas-Rebolledo C et al (2022) Nanobodies: COVID-19 and future perspectives. Front Drug Discov 2:927164. https://doi.org/10.3389/fddsv.2022.927164",
"volume": "2",
"year": "2022"
},
{
"DOI": "10.1038/s42003-022-03866-z",
"author": "MA Rossotti",
"doi-asserted-by": "publisher",
"journal-title": "Commun Biol",
"key": "10758_CR6",
"unstructured": "Rossotti MA, van Faassen H, Tran AT et al (2022) Arsenal of nanobodies shows broad-spectrum neutralization against SARS-CoV-2 variants of concern in vitro and in vivo in hamster models. Commun Biol 5:933. https://doi.org/10.1038/s42003-022-03866-z",
"volume": "5",
"year": "2022"
},
{
"DOI": "10.3389/fimmu.2024.1334829",
"author": "H De Greve",
"doi-asserted-by": "publisher",
"journal-title": "Front Immunol",
"key": "10758_CR7",
"unstructured": "De Greve H, Fioravanti A (2024) Single domain antibodies from camelids in the treatment of microbial infections. Front Immunol 15:1334829. https://doi.org/10.3389/fimmu.2024.1334829",
"volume": "15",
"year": "2024"
},
{
"DOI": "10.3390/ijms24065994",
"author": "B Jin",
"doi-asserted-by": "publisher",
"journal-title": "IJMS",
"key": "10758_CR8",
"unstructured": "Jin B, Odongo S, Radwanska M, Magez S (2023) NANOBODIES®: a review of diagnostic and therapeutic applications. IJMS 24:5994. https://doi.org/10.3390/ijms24065994",
"volume": "24",
"year": "2023"
},
{
"DOI": "10.7554/eLife.73027",
"author": "FD Mast",
"doi-asserted-by": "publisher",
"journal-title": "Elife",
"key": "10758_CR9",
"unstructured": "Mast FD, Fridy PC, Ketaren NE et al (2021) Highly synergistic combinations of nanobodies that target SARS-CoV-2 and are resistant to escape. Elife 10:e73027. https://doi.org/10.7554/eLife.73027",
"volume": "10",
"year": "2021"
},
{
"DOI": "10.1038/s41419-024-06802-7",
"author": "Q Liu",
"doi-asserted-by": "publisher",
"journal-title": "Cell Death Dis",
"key": "10758_CR10",
"unstructured": "Liu Q, Lu Y, Cai C et al (2024) A broad neutralizing nanobody against SARS-CoV-2 engineered from an approved drug. Cell Death Dis 15:458. https://doi.org/10.1038/s41419-024-06802-7",
"volume": "15",
"year": "2024"
},
{
"DOI": "10.1073/pnas.2205412119",
"author": "H Mikolajek",
"doi-asserted-by": "publisher",
"journal-title": "Proc Natl Acad Sci USA",
"key": "10758_CR11",
"unstructured": "Mikolajek H, Weckener M, Brotzakis ZF et al (2022) Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes. Proc Natl Acad Sci USA 119:e2205412119. https://doi.org/10.1073/pnas.2205412119",
"volume": "119",
"year": "2022"
},
{
"DOI": "10.1038/s41594-020-0469-6",
"author": "J Huo",
"doi-asserted-by": "publisher",
"first-page": "846",
"journal-title": "Nat Struct Mol Biol",
"key": "10758_CR12",
"unstructured": "Huo J, Le Bas A, Ruza RR et al (2020) Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat Struct Mol Biol 27:846–854. https://doi.org/10.1038/s41594-020-0469-6",
"volume": "27",
"year": "2020"
},
{
"DOI": "10.1016/j.mcp.2020.101692",
"author": "H Zare",
"doi-asserted-by": "publisher",
"journal-title": "Mol Cell Probes",
"key": "10758_CR13",
"unstructured": "Zare H, Aghamollaei H, Hosseindokht M et al (2021) Nanobodies, the potent agents to detect and treat the coronavirus infections: a systematic review. Mol Cell Probes 55:101692. https://doi.org/10.1016/j.mcp.2020.101692",
"volume": "55",
"year": "2021"
},
{
"DOI": "10.2147/IJN.S450829",
"author": "W Zhang",
"doi-asserted-by": "publisher",
"first-page": "2429",
"journal-title": "Int J Nanomedicine",
"key": "10758_CR14",
"unstructured": "Zhang W, Wang H, Wu T et al (2024) A SARS-CoV-2 nanobody displayed on the surface of human ferritin with high neutralization activity. Int J Nanomedicine 19:2429–2440. https://doi.org/10.2147/IJN.S450829",
"volume": "19",
"year": "2024"
},
{
"DOI": "10.1038/s41598-022-14263-1",
"author": "H Nguyen",
"doi-asserted-by": "publisher",
"journal-title": "Sci Rep",
"key": "10758_CR15",
"unstructured": "Nguyen H, Li MS (2022) Antibody–nanobody combination increases their neutralizing activity against SARS-CoV-2 and nanobody H11–H4 is effective against alpha, kappa and delta variants. Sci Rep 12:9701. https://doi.org/10.1038/s41598-022-14263-1",
"volume": "12",
"year": "2022"
},
{
"DOI": "10.3389/fimmu.2017.01603",
"author": "P Bannas",
"doi-asserted-by": "publisher",
"journal-title": "Front Immunol",
"key": "10758_CR16",
"unstructured": "Bannas P, Hambach J, Koch-Nolte F (2017) Nanobodies and nanobody-based human heavy chain antibodies as antitumor therapeutics. Front Immunol 8:1603. https://doi.org/10.3389/fimmu.2017.01603",
"volume": "8",
"year": "2017"
},
{
"DOI": "10.1002/cti2.1518",
"author": "H Shuwen",
"doi-asserted-by": "publisher",
"journal-title": "Clin Transl Immunol",
"key": "10758_CR17",
"unstructured": "Shuwen H, Yifei S, Xinyue W et al (2024) Advances in bacteria-based drug delivery systems for anti-tumor therapy. Clin Transl Immunol 13:e1518. https://doi.org/10.1002/cti2.1518",
"volume": "13",
"year": "2024"
},
{
"DOI": "10.1016/j.cgh.2006.03.028",
"author": "H Braat",
"doi-asserted-by": "publisher",
"first-page": "754",
"journal-title": "Clin Gastroenterol Hepatol",
"key": "10758_CR18",
"unstructured": "Braat H, Rottiers P, Hommes DW et al (2006) A phase I trial with transgenic bacteria expressing interleukin-10 in Crohn’s disease. Clin Gastroenterol Hepatol 4:754–759. https://doi.org/10.1016/j.cgh.2006.03.028",
"volume": "4",
"year": "2006"
},
{
"DOI": "10.4014/jmb.1904.04016",
"author": "PNL Vo",
"doi-asserted-by": "publisher",
"first-page": "845",
"journal-title": "J Microbiol Biotechnol",
"key": "10758_CR19",
"unstructured": "Vo PNL, Lee H-M, Na D (2019) Synthetic bacteria for therapeutics. J Microbiol Biotechnol 29:845–855. https://doi.org/10.4014/jmb.1904.04016",
"volume": "29",
"year": "2019"
},
{
"DOI": "10.1016/j.addr.2022.114364",
"author": "H Shen",
"doi-asserted-by": "publisher",
"journal-title": "Adv Drug Deliv Rev",
"key": "10758_CR20",
"unstructured": "Shen H, Aggarwal N, Wun KS et al (2022) Engineered microbial systems for advanced drug delivery. Adv Drug Deliv Rev 187:114364. https://doi.org/10.1016/j.addr.2022.114364",
"volume": "187",
"year": "2022"
},
{
"DOI": "10.1053/j.gastro.2013.09.060",
"author": "ML Hanson",
"doi-asserted-by": "publisher",
"first-page": "210",
"journal-title": "Gastroenterology",
"key": "10758_CR21",
"unstructured": "Hanson ML, Hixon JA, Li W et al (2014) Oral delivery of IL-27 recombinant bacteria attenuates immune colitis in mice. Gastroenterology 146:210-221.e13. https://doi.org/10.1053/j.gastro.2013.09.060",
"volume": "146",
"year": "2014"
},
{
"DOI": "10.1126/scitranslmed.3004212",
"doi-asserted-by": "publisher",
"key": "10758_CR22",
"unstructured": "Motta J-P, Bermúdez-Humarán LG, Deraison C et al (2012) Food-grade bacteria expressing elafin protect against inflammation and restore colon homeostasis. Sci Transl Med 4:. https://doi.org/10.1126/scitranslmed.3004212"
},
{
"DOI": "10.3389/fbioe.2022.1026248",
"author": "S Liang",
"doi-asserted-by": "publisher",
"journal-title": "Front Bioeng Biotechnol",
"key": "10758_CR23",
"unstructured": "Liang S, Wang C, Shao Y et al (2022) Recent advances in bacteria-mediated cancer therapy. Front Bioeng Biotechnol 10:1026248. https://doi.org/10.3389/fbioe.2022.1026248",
"volume": "10",
"year": "2022"
},
{
"DOI": "10.1007/s00253-020-10771-0",
"author": "NS Dosoky",
"doi-asserted-by": "publisher",
"first-page": "7657",
"journal-title": "Appl Microbiol Biotechnol",
"key": "10758_CR24",
"unstructured": "Dosoky NS, May-Zhang LS, Davies SS (2020) Engineering the gut microbiota to treat chronic diseases. Appl Microbiol Biotechnol 104:7657–7671. https://doi.org/10.1007/s00253-020-10771-0",
"volume": "104",
"year": "2020"
},
{
"DOI": "10.1038/s41591-019-0498-z",
"author": "S Chowdhury",
"doi-asserted-by": "publisher",
"first-page": "1057",
"journal-title": "Nat Med",
"key": "10758_CR25",
"unstructured": "Chowdhury S, Castro S, Coker C et al (2019) Programmable bacteria induce durable tumor regression and systemic antitumor immunity. Nat Med 25:1057–1063. https://doi.org/10.1038/s41591-019-0498-z",
"volume": "25",
"year": "2019"
},
{
"DOI": "10.1159/000100515",
"author": "I Gentschev",
"doi-asserted-by": "publisher",
"first-page": "177",
"journal-title": "Chemotherapy",
"key": "10758_CR26",
"unstructured": "Gentschev I, Spreng S, Sieber H et al (2007) Vivotif® – a ‘magic shield’ for protection against typhoid fever and delivery of heterologous antigens. Chemotherapy 53:177–180. https://doi.org/10.1159/000100515",
"volume": "53",
"year": "2007"
},
{
"DOI": "10.1158/1078-0432.CCR-23-0118",
"author": "JJ Luke",
"doi-asserted-by": "publisher",
"first-page": "2435",
"journal-title": "Clin Cancer Res",
"key": "10758_CR27",
"unstructured": "Luke JJ, Piha-Paul SA, Medina T et al (2023) Phase I study of SYNB1891, an engineered E. coli Nissle strain expressing STING agonist, with and without atezolizumab in advanced malignancies. Clin Cancer Res 29:2435–2444. https://doi.org/10.1158/1078-0432.CCR-23-0118",
"volume": "29",
"year": "2023"
},
{
"DOI": "10.1038/s41419-023-06391-x",
"author": "M Babamohamadi",
"doi-asserted-by": "publisher",
"journal-title": "Cell Death Dis",
"key": "10758_CR28",
"unstructured": "Babamohamadi M, Mohammadi N, Faryadi E et al (2024) Anti-CTLA-4 nanobody as a promising approach in cancer immunotherapy. Cell Death Dis 15:17. https://doi.org/10.1038/s41419-023-06391-x",
"volume": "15",
"year": "2024"
},
{
"DOI": "10.1126/scitranslmed.aax0876",
"author": "CR Gurbatri",
"doi-asserted-by": "publisher",
"journal-title": "Sci Transl Med",
"key": "10758_CR29",
"unstructured": "Gurbatri CR, Lia I, Vincent R et al (2020) Engineered probiotics for local tumor delivery of checkpoint blockade nanobodies. Sci Transl Med 12:eaax0876. https://doi.org/10.1126/scitranslmed.aax0876",
"volume": "12",
"year": "2020"
},
{
"DOI": "10.1038/mi.2009.116",
"author": "K Vandenbroucke",
"doi-asserted-by": "publisher",
"first-page": "49",
"journal-title": "Mucosal Immunol",
"key": "10758_CR30",
"unstructured": "Vandenbroucke K, De Haard H, Beirnaert E et al (2010) Orally administered L. lactis secreting an anti-TNF nanobody demonstrate efficacy in chronic colitis. Mucosal Immunol 3:49–56. https://doi.org/10.1038/mi.2009.116",
"volume": "3",
"year": "2010"
},
{
"DOI": "10.3389/fimmu.2017.01961",
"author": "DP Cook",
"doi-asserted-by": "publisher",
"journal-title": "Front Immunol",
"key": "10758_CR31",
"unstructured": "Cook DP, Gysemans C, Mathieu C (2018) Lactococcus lactis as a versatile vehicle for tolerogenic immunotherapy. Front Immunol 8:1961. https://doi.org/10.3389/fimmu.2017.01961",
"volume": "8",
"year": "2018"
},
{
"DOI": "10.1007/978-3-031-20537-8_5",
"doi-asserted-by": "crossref",
"key": "10758_CR32",
"unstructured": "Adekiya TA, Sumaila M, Aruleba RT, Choonara YE (2023) Mucosal targeting strategies for antiviral drug delivery. In: Shegokar R, Pathak Y (eds) Viral drug delivery systems. Springer International Publishing, Cham, pp 91–117"
},
{
"DOI": "10.1080/17425247.2023.2263363",
"author": "F Sonvico",
"doi-asserted-by": "publisher",
"first-page": "1115",
"journal-title": "Expert Opin Drug Deliv",
"key": "10758_CR33",
"unstructured": "Sonvico F, Colombo G, Quarta E et al (2023) Nasal delivery as a strategy for the prevention and treatment of COVID-19. Expert Opin Drug Deliv 20:1115–1130. https://doi.org/10.1080/17425247.2023.2263363",
"volume": "20",
"year": "2023"
},
{
"DOI": "10.3389/fmicb.2017.00846",
"author": "DM Linares",
"doi-asserted-by": "publisher",
"journal-title": "Front Microbiol",
"key": "10758_CR34",
"unstructured": "Linares DM, Gómez C, Renes E et al (2017) Lactic acid bacteria and bifidobacteria with potential to design natural biofunctional health-promoting dairy foods. Front Microbiol 8:846. https://doi.org/10.3389/fmicb.2017.00846",
"volume": "8",
"year": "2017"
},
{
"DOI": "10.3389/fbioe.2020.517166",
"author": "LM Tavares",
"doi-asserted-by": "publisher",
"journal-title": "Front Bioeng Biotechnol",
"key": "10758_CR35",
"unstructured": "Tavares LM, de Jesus LCL, da Silva TF et al (2020) Novel strategies for efficient production and delivery of live biotherapeutics and biotechnological uses of Lactococcus lactis: the lactic acid bacterium model. Front Bioeng Biotechnol 8:517166. https://doi.org/10.3389/fbioe.2020.517166",
"volume": "8",
"year": "2020"
},
{
"DOI": "10.1128/aem.61.7.2771-2774.1995",
"author": "N Klijn",
"doi-asserted-by": "publisher",
"first-page": "2771",
"issue": "7",
"journal-title": "Appl Environ Microbiol",
"key": "10758_CR36",
"unstructured": "Klijn N, Weerkamp AH, De Vos WM (1995) Genetic marking of Lactococcus lactis shows its survival in the human gastrointestinal tract. Appl Environ Microbiol 61(7):2771–2774. https://doi.org/10.1128/aem.61.7.2771-2774.1995",
"volume": "61",
"year": "1995"
},
{
"DOI": "10.1016/j.tifs.2004.03.003",
"author": "S Wessels",
"doi-asserted-by": "publisher",
"first-page": "498",
"journal-title": "Trends Food Sci Technol",
"key": "10758_CR37",
"unstructured": "Wessels S, Axelsson L, Bech Hansen E et al (2004) The lactic acid bacteria, the food chain, and their regulation. Trends Food Sci Technol 15:498–505. https://doi.org/10.1016/j.tifs.2004.03.003",
"volume": "15",
"year": "2004"
},
{
"DOI": "10.1186/s12934-016-0468-9",
"author": "C Michon",
"doi-asserted-by": "publisher",
"journal-title": "Microb Cell Fact",
"key": "10758_CR38",
"unstructured": "Michon C, Langella P, Eijsink VGH et al (2016) Display of recombinant proteins at the surface of lactic acid bacteria: strategies and applications. Microb Cell Fact 15:70. https://doi.org/10.1186/s12934-016-0468-9",
"volume": "15",
"year": "2016"
},
{
"DOI": "10.1186/1475-2859-5-22",
"author": "N Rutherford",
"doi-asserted-by": "publisher",
"journal-title": "Microb Cell Fact",
"key": "10758_CR39",
"unstructured": "Rutherford N, Mourez M (2006) Surface display of proteins by gram-negative bacterial autotransporters. Microb Cell Fact 5:22. https://doi.org/10.1186/1475-2859-5-22",
"volume": "5",
"year": "2006"
},
{
"DOI": "10.1016/j.biotechadv.2005.11.001",
"author": "XX Zhou",
"doi-asserted-by": "publisher",
"first-page": "285",
"journal-title": "Biotechnol Adv",
"key": "10758_CR40",
"unstructured": "Zhou XX, Li WF, Ma GX, Pan YJ (2006) The nisin-controlled gene expression system: construction, application and improvements. Biotechnol Adv 24:285–295. https://doi.org/10.1016/j.biotechadv.2005.11.001",
"volume": "24",
"year": "2006"
},
{
"DOI": "10.1128/AEM.01802-06",
"author": "A Kuipers",
"doi-asserted-by": "publisher",
"first-page": "7626",
"journal-title": "Appl Environ Microbiol",
"key": "10758_CR41",
"unstructured": "Kuipers A, Wierenga J, Rink R et al (2006) Sec-mediated transport of posttranslationally dehydrated peptides in Lactococcus lactis. Appl Environ Microbiol 72:7626–7633. https://doi.org/10.1128/AEM.01802-06",
"volume": "72",
"year": "2006"
},
{
"DOI": "10.1159/000106082",
"author": "E Morello",
"doi-asserted-by": "publisher",
"first-page": "48",
"journal-title": "Microb Physiol",
"key": "10758_CR42",
"unstructured": "Morello E, Bermúdez-Humarán LG, Llull D et al (2008) Lactococcus lactis, an efficient cell factory for recombinant protein production and secretion. Microb Physiol 14:48–58. https://doi.org/10.1159/000106082",
"volume": "14",
"year": "2008"
},
{
"DOI": "10.1089/fpd.2019.2727",
"author": "C Wang",
"doi-asserted-by": "publisher",
"first-page": "366",
"journal-title": "Foodborne Pathog Dis",
"key": "10758_CR43",
"unstructured": "Wang C, Zhou H, Guo F et al (2020) Oral immunization of chickens with Lactococcus lactis expressing cjaA temporarily reduces Campylobacter jejuni colonization. Foodborne Pathog Dis 17:366–372. https://doi.org/10.1089/fpd.2019.2727",
"volume": "17",
"year": "2020"
},
{
"DOI": "10.1146/annurev.biochem.77.061606.160747",
"author": "AJM Driessen",
"doi-asserted-by": "publisher",
"first-page": "643",
"journal-title": "Annu Rev Biochem",
"key": "10758_CR44",
"unstructured": "Driessen AJM, Nouwen N (2008) Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem 77:643–667. https://doi.org/10.1146/annurev.biochem.77.061606.160747",
"volume": "77",
"year": "2008"
},
{
"DOI": "10.1016/j.bbamem.2007.07.015",
"author": "P Natale",
"doi-asserted-by": "publisher",
"first-page": "1735",
"journal-title": "Biochimica et Biophysica Acta (BBA)",
"key": "10758_CR45",
"unstructured": "Natale P, Brüser T, Driessen AJM (2008) Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane—distinct translocases and mechanisms. Biochimica et Biophysica Acta (BBA) 1778:1735–1756. https://doi.org/10.1016/j.bbamem.2007.07.015",
"volume": "1778",
"year": "2008"
},
{
"DOI": "10.1111/j.1742-4658.2005.04706.x",
"author": "A Steen",
"doi-asserted-by": "publisher",
"first-page": "2854",
"journal-title": "FEBS J",
"key": "10758_CR46",
"unstructured": "Steen A, Buist G, Horsburgh GJ et al (2005) AcmA of Lactococcus lactis is an N -acetylglucosaminidase with an optimal number of LysM domains for proper functioning. FEBS J 272:2854–2868. https://doi.org/10.1111/j.1742-4658.2005.04706.x",
"volume": "272",
"year": "2005"
},
{
"DOI": "10.3389/fmicb.2019.01879",
"author": "TV Plavec",
"doi-asserted-by": "publisher",
"journal-title": "Front Microbiol",
"key": "10758_CR47",
"unstructured": "Plavec TV, Štrukelj B, Berlec A (2019) Screening for new surface anchoring domains for Lactococcus lactis. Front Microbiol 10:1879. https://doi.org/10.3389/fmicb.2019.01879",
"volume": "10",
"year": "2019"
},
{
"key": "10758_CR48",
"unstructured": "MoBiTec GmbH (2015) NICE® expression system for Lactococcus lactis. chrome-extension://efaidnbmnnnibpcajpcglclefindmkaj/https://www.mobitec.com/media/mobitec/category-preview/vector-systems/pdf/3/NICE_Expression_System-Handbook.pdf. Accessed 5 Sept 2025"
},
{
"key": "10758_CR49",
"unstructured": "QIAGEN (2003) The QIAexpressionist TM. In: Protocol 6. Determination of target protein solubility. https://www.qiagen.com/us/resources/resourcedetail?id=79ca2f7d-42fe-4d62-8676-4cfa948c9435&lang=en. Accessed 5 Sept 2025"
},
{
"DOI": "10.3389/fbioe.2022.822823",
"author": "A Zahirović",
"doi-asserted-by": "publisher",
"journal-title": "Front Bioeng Biotechnol",
"key": "10758_CR50",
"unstructured": "Zahirović A, Plavec TV, Berlec A (2022) Dual functionalized Lactococcus lactis shows tumor antigen targeting and cytokine binding in vitro. Front Bioeng Biotechnol 10:822823. https://doi.org/10.3389/fbioe.2022.822823",
"volume": "10",
"year": "2022"
},
{
"key": "10758_CR51",
"unstructured": "Held P (2018) Using phenol red to assess pH in tissue culture media. In: Agilent application note. https://www.agilent.com/cs/library/applications/phenol-red-to-assess-ph-in-tissue-culture-media-5994-3391EN-agilent.pdf. Accessed 5 Sept 2025"
},
{
"DOI": "10.1038/s41375-018-0106-0",
"author": "MC Milone",
"doi-asserted-by": "publisher",
"first-page": "1529",
"journal-title": "Leukemia",
"key": "10758_CR52",
"unstructured": "Milone MC, O’Doherty U (2018) Clinical use of lentiviral vectors. Leukemia 32:1529–1541. https://doi.org/10.1038/s41375-018-0106-0",
"volume": "32",
"year": "2018"
},
{
"DOI": "10.3390/v12091016",
"author": "A Gutierrez-Guerrero",
"doi-asserted-by": "publisher",
"issue": "9",
"journal-title": "Viruses",
"key": "10758_CR53",
"unstructured": "Gutierrez-Guerrero A, Cosset F-L, Verhoeyen E (2020) Lentiviral vector pseudotypes: precious tools to improve gene modification of hematopoietic cells for research and gene therapy. Viruses 12(9):1016. https://doi.org/10.3390/v12091016",
"volume": "12",
"year": "2020"
},
{
"DOI": "10.1038/s41596-020-0394-5",
"author": "J Nie",
"doi-asserted-by": "publisher",
"first-page": "3699",
"journal-title": "Nat Protoc",
"key": "10758_CR54",
"unstructured": "Nie J, Li Q, Wu J et al (2020) Quantification of SARS-CoV-2 neutralizing antibody by a pseudotyped virus-based assay. Nat Protoc 15:3699–3715. https://doi.org/10.1038/s41596-020-0394-5",
"volume": "15",
"year": "2020"
},
{
"DOI": "10.3390/v12050513",
"author": "KHD Crawford",
"doi-asserted-by": "publisher",
"issue": "5",
"journal-title": "Viruses",
"key": "10758_CR55",
"unstructured": "Crawford KHD, Eguia R, Dingens AS et al (2020) Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses 12(5):513. https://doi.org/10.3390/v12050513",
"volume": "12",
"year": "2020"
},
{
"DOI": "10.1016/j.procbio.2017.07.031",
"author": "Q Liu",
"doi-asserted-by": "publisher",
"first-page": "135",
"journal-title": "Process Biochem",
"key": "10758_CR56",
"unstructured": "Liu Q, Shao T, Dong Z, Bai Y (2017) Solution for promoting egl 3 gene of Trichoderma reesei high-efficiency secretory expression in Escherichia coli and Lactococcus lactis. Process Biochem 62:135–143. https://doi.org/10.1016/j.procbio.2017.07.031",
"volume": "62",
"year": "2017"
},
{
"DOI": "10.1038/s41467-020-15508-1",
"author": "MR Charbonneau",
"doi-asserted-by": "publisher",
"journal-title": "Nat Commun",
"key": "10758_CR57",
"unstructured": "Charbonneau MR, Isabella VM, Li N, Kurtz CB (2020) Developing a new class of engineered live bacterial therapeutics to treat human diseases. Nat Commun 11:1738. https://doi.org/10.1038/s41467-020-15508-1",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1016/j.it.2019.08.007",
"author": "ME Inda",
"doi-asserted-by": "publisher",
"first-page": "952",
"journal-title": "Trends Immunol",
"key": "10758_CR58",
"unstructured": "Inda ME, Broset E, Lu TK, de la Fuente-Nunez C (2019) Emerging frontiers in microbiome engineering. Trends Immunol 40:952–973. https://doi.org/10.1016/j.it.2019.08.007",
"volume": "40",
"year": "2019"
},
{
"DOI": "10.1007/s12602-017-9281-y",
"author": "W Sirichokchatchawan",
"doi-asserted-by": "publisher",
"first-page": "383",
"journal-title": "Probiotics Antimicrob Proteins",
"key": "10758_CR59",
"unstructured": "Sirichokchatchawan W, Temeeyasen G, Nilubol D, Prapasarakul N (2018) Protective effects of cell-free supernatant and live lactic acid bacteria isolated from Thai pigs against a pandemic strain of porcine epidemic diarrhea virus. Probiotics Antimicrob Proteins 10:383–390. https://doi.org/10.1007/s12602-017-9281-y",
"volume": "10",
"year": "2018"
},
{
"DOI": "10.1016/j.ijfoodmicro.2006.10.044",
"author": "T Botic",
"doi-asserted-by": "publisher",
"first-page": "227",
"journal-title": "Int J Food Microbiol",
"key": "10758_CR60",
"unstructured": "Botic T, Klingberg T, Weingartl H, Cencic A (2007) A novel eukaryotic cell culture model to study antiviral activity of potential probiotic bacteria. Int J Food Microbiol 115:227–234. https://doi.org/10.1016/j.ijfoodmicro.2006.10.044",
"volume": "115",
"year": "2007"
},
{
"DOI": "10.1371/journal.pone.0053043",
"author": "Z Wang",
"doi-asserted-by": "publisher",
"journal-title": "PLoS ONE",
"key": "10758_CR61",
"unstructured": "Wang Z, Chai W, Burwinkel M et al (2013) Inhibitory influence of Enterococcus faecium on the propagation of swine influenza A virus in vitro. PLoS ONE 8:e53043. https://doi.org/10.1371/journal.pone.0053043",
"volume": "8",
"year": "2013"
},
{
"DOI": "10.3389/fphar.2018.01208",
"author": "TA Slastnikova",
"doi-asserted-by": "publisher",
"journal-title": "Front Pharmacol",
"key": "10758_CR62",
"unstructured": "Slastnikova TA, Ulasov AV, Rosenkranz AA, Sobolev AS (2018) Targeted intracellular delivery of antibodies: the state of the art. Front Pharmacol 9:1208. https://doi.org/10.3389/fphar.2018.01208",
"volume": "9",
"year": "2018"
},
{
"DOI": "10.4014/jmb.1501.01028",
"author": "Y Song",
"doi-asserted-by": "publisher",
"first-page": "963",
"journal-title": "J Microbiol Biotechnol",
"key": "10758_CR63",
"unstructured": "Song Y, Nikoloff JM, Zhang D (2015) Improving protein production on the level of regulation of both expression and secretion pathways in Bacillus subtilis. J Microbiol Biotechnol 25:963–977. https://doi.org/10.4014/jmb.1501.01028",
"volume": "25",
"year": "2015"
},
{
"DOI": "10.1021/acs.jcim.1c00695",
"author": "M Golcuk",
"doi-asserted-by": "publisher",
"first-page": "5152",
"journal-title": "J Chem Inf Model",
"key": "10758_CR64",
"unstructured": "Golcuk M, Hacisuleyman A, Erman B et al (2021) Binding mechanism of neutralizing nanobodies targeting SARS-CoV-2 spike glycoprotein. J Chem Inf Model 61:5152–5160. https://doi.org/10.1021/acs.jcim.1c00695",
"volume": "61",
"year": "2021"
},
{
"DOI": "10.1039/D4NR02074J",
"author": "LF Cofas-Vargas",
"doi-asserted-by": "publisher",
"first-page": "18824",
"journal-title": "Nanoscale",
"key": "10758_CR65",
"unstructured": "Cofas-Vargas LF, Olivos-Ramirez GE, Chwastyk M et al (2024) Nanomechanical footprint of SARS-CoV-2 variants in complex with a potent nanobody by molecular simulations. Nanoscale 16:18824–18834. https://doi.org/10.1039/D4NR02074J",
"volume": "16",
"year": "2024"
},
{
"DOI": "10.3390/v13112214",
"author": "Q Tang",
"doi-asserted-by": "publisher",
"issue": "11",
"journal-title": "Viruses",
"key": "10758_CR66",
"unstructured": "Tang Q, Owens RJ, Naismith JH (2021) Structural biology of nanobodies against the spike protein of SARS-CoV-2. Viruses 13(11):2214. https://doi.org/10.3390/v13112214",
"volume": "13",
"year": "2021"
},
{
"DOI": "10.1186/s12934-022-01873-7",
"author": "A Zahirović",
"doi-asserted-by": "publisher",
"issue": "1",
"journal-title": "Microb Cell Fact",
"key": "10758_CR67",
"unstructured": "Zahirović A, Berlec A (2022) Targeting IL-6 by engineered Lactococcus lactis via surface-displayed affibody. Microb Cell Fact 21(1):143. https://doi.org/10.1186/s12934-022-01873-7",
"volume": "21",
"year": "2022"
},
{
"DOI": "10.1371/journal.pone.0272298",
"author": "A Frische",
"doi-asserted-by": "publisher",
"journal-title": "PLoS ONE",
"key": "10758_CR68",
"unstructured": "Frische A, Brooks PT, Gybel-Brask M et al (2022) Optimization and evaluation of a live virus SARS-CoV-2 neutralization assay. PLoS ONE 17:e0272298. https://doi.org/10.1371/journal.pone.0272298",
"volume": "17",
"year": "2022"
},
{
"DOI": "10.1038/s41467-020-18174-5",
"author": "L Hanke",
"doi-asserted-by": "publisher",
"journal-title": "Nat Commun",
"key": "10758_CR69",
"unstructured": "Hanke L, Vidakovics Perez L, Sheward DJ et al (2020) An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat Commun 11:4420. https://doi.org/10.1038/s41467-020-18174-5",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.3389/fmicb.2023.1269869",
"author": "J Wiggins",
"doi-asserted-by": "publisher",
"journal-title": "Front Microbiol",
"key": "10758_CR70",
"unstructured": "Wiggins J, Nguyen N, Wei W et al (2023) Lactic acid bacterial surface display of scytovirin inhibitors for anti-ebolavirus infection. Front Microbiol 14:1269869. https://doi.org/10.3389/fmicb.2023.1269869",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.1155/2021/6611222",
"author": "S Ahmadi Badi",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Mediators Inflamm",
"key": "10758_CR71",
"unstructured": "Ahmadi Badi S, Tarashi S, Fateh A et al (2021) From the role of microbiota in gut-lung axis to SARS-CoV-2 pathogenesis. Mediators Inflamm 2021:1–12. https://doi.org/10.1155/2021/6611222",
"volume": "2021",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2020.05.042",
"author": "YJ Hou",
"doi-asserted-by": "publisher",
"first-page": "429",
"journal-title": "Cell",
"key": "10758_CR72",
"unstructured": "Hou YJ, Okuda K, Edwards CE et al (2020) SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell 182:429-446.e14. https://doi.org/10.1016/j.cell.2020.05.042",
"volume": "182",
"year": "2020"
},
{
"DOI": "10.1128/JVI.02292-10",
"author": "L Liu",
"doi-asserted-by": "publisher",
"first-page": "4025",
"journal-title": "J Virol",
"key": "10758_CR73",
"unstructured": "Liu L, Wei Q, Alvarez X et al (2011) Epithelial cells lining salivary gland ducts are early target cells of severe acute respiratory syndrome coronavirus infection in the upper respiratory tracts of rhesus macaques. J Virol 85:4025–4030. https://doi.org/10.1128/JVI.02292-10",
"volume": "85",
"year": "2011"
},
{
"DOI": "10.1016/j.virol.2020.10.002",
"author": "R Bhattacharya",
"doi-asserted-by": "publisher",
"first-page": "107",
"journal-title": "Virology",
"key": "10758_CR74",
"unstructured": "Bhattacharya R, Gupta AM, Mitra S et al (2021) A natural food preservative peptide nisin can interact with the SARS-CoV-2 spike protein receptor human ACE2. Virology 552:107–111. https://doi.org/10.1016/j.virol.2020.10.002",
"volume": "552",
"year": "2021"
},
{
"DOI": "10.1007/s00125-023-06014-2",
"author": "C Mathieu",
"doi-asserted-by": "publisher",
"first-page": "27",
"journal-title": "Diabetologia",
"key": "10758_CR75",
"unstructured": "Mathieu C, Wiedeman A, Cerosaletti K et al (2024) A first-in-human, open-label phase 1b and a randomised, double-blind phase 2a clinical trial in recent-onset type 1 diabetes with AG019 as monotherapy and in combination with teplizumab. Diabetologia 67:27–41. https://doi.org/10.1007/s00125-023-06014-2",
"volume": "67",
"year": "2024"
},
{
"DOI": "10.3389/fcimb.2020.00440",
"author": "LM Endam",
"doi-asserted-by": "publisher",
"journal-title": "Front Cell Infect Microbiol",
"key": "10758_CR76",
"unstructured": "Endam LM, Alromaih S, Gonzalez E et al (2020) Intranasal application of Lactococcus lactis W136 is safe in chronic rhinosinusitis patients with previous sinus surgery. Front Cell Infect Microbiol 10:440. https://doi.org/10.3389/fcimb.2020.00440",
"volume": "10",
"year": "2020"
},
{
"DOI": "10.1002/alr.22521",
"author": "D Cho",
"doi-asserted-by": "publisher",
"first-page": "444",
"journal-title": "Int Forum Allergy Rhinol",
"key": "10758_CR77",
"unstructured": "Cho D, Skinner D, Lim DJ et al (2020) The impact of Lactococcus lactis (probiotic nasal rinse) co-culture on growth of patient-derived strains of Pseudomonas aeruginosa. Int Forum Allergy Rhinol 10:444–449. https://doi.org/10.1002/alr.22521",
"volume": "10",
"year": "2020"
},
{
"DOI": "10.1002/cncr.28365",
"author": "SA Limaye",
"doi-asserted-by": "publisher",
"first-page": "4268",
"journal-title": "Cancer",
"key": "10758_CR78",
"unstructured": "Limaye SA, Haddad RI, Cilli F et al (2013) Phase 1b, multicenter, single blinded, placebo-controlled, sequential dose escalation study to assess the safety and tolerability of topically applied AG013 in subjects with locally advanced head and neck cancer receiving induction chemotherapy. Cancer 119:4268–4276. https://doi.org/10.1002/cncr.28365",
"volume": "119",
"year": "2013"
},
{
"DOI": "10.1186/s12934-015-0198-4",
"author": "LG Bermúdez-Humarán",
"doi-asserted-by": "publisher",
"journal-title": "Microb Cell Fact",
"key": "10758_CR79",
"unstructured": "Bermúdez-Humarán LG, Motta J-P, Aubry C et al (2015) Serine protease inhibitors protect better than IL-10 and TGF-β anti-inflammatory cytokines against mouse colitis when delivered by recombinant lactococci. Microb Cell Fact 14:26. https://doi.org/10.1186/s12934-015-0198-4",
"volume": "14",
"year": "2015"
},
{
"DOI": "10.1128/IAI.66.7.3183-3189.1998",
"author": "L Steidler",
"doi-asserted-by": "publisher",
"first-page": "3183",
"journal-title": "Infect Immun",
"key": "10758_CR80",
"unstructured": "Steidler L, Robinson K, Chamberlain L et al (1998) Mucosal delivery of murine interleukin-2 (IL-2) and IL-6 by recombinant strains of Lactococcus lactis coexpressing antigen and cytokine. Infect Immun 66:3183–3189. https://doi.org/10.1128/IAI.66.7.3183-3189.1998",
"volume": "66",
"year": "1998"
},
{
"DOI": "10.1016/j.oraloncology.2010.04.008",
"author": "S Caluwaerts",
"doi-asserted-by": "publisher",
"first-page": "564",
"journal-title": "Oral Oncol",
"key": "10758_CR81",
"unstructured": "Caluwaerts S, Vandenbroucke K, Steidler L et al (2010) AG013, a mouth rinse formulation of Lactococcus lactis secreting human trefoil factor 1, provides a safe and efficacious therapeutic tool for treating oral mucositis. Oral Oncol 46:564–570. https://doi.org/10.1016/j.oraloncology.2010.04.008",
"volume": "46",
"year": "2010"
},
{
"DOI": "10.3390/antiox9111049",
"author": "K Yumoto",
"doi-asserted-by": "publisher",
"journal-title": "Antioxidants",
"key": "10758_CR82",
"unstructured": "Yumoto K, Sato T, Nakashima K et al (2020) Nasally administered Lactococcus lactis secreting heme oxygenase-1 attenuates murine emphysema. Antioxidants 9:1049. https://doi.org/10.3390/antiox9111049",
"volume": "9",
"year": "2020"
},
{
"DOI": "10.3390/agriculture11030186",
"author": "E Okello",
"doi-asserted-by": "publisher",
"journal-title": "Agriculture",
"key": "10758_CR83",
"unstructured": "Okello E, Moonens K, Erume J, De Greve H (2021) Orally fed recombinant Lactococcus lactis displaying surface anti-fimbrial nanobodies protects piglets against Escherichia coli causing post-weaning diarrhea. Agriculture 11:186. https://doi.org/10.3390/agriculture11030186",
"volume": "11",
"year": "2021"
},
{
"DOI": "10.1158/1535-7163.MCT-19-0375",
"author": "AH Mohseni",
"doi-asserted-by": "publisher",
"first-page": "717",
"journal-title": "Mol Cancer Ther",
"key": "10758_CR84",
"unstructured": "Mohseni AH, Taghinezhad-S S, Keyvani H (2020) The first clinical use of a recombinant Lactococcus lactis expressing human papillomavirus type 16 E7 oncogene oral vaccine: a phase I safety and immunogenicity trial in healthy women volunteers. Mol Cancer Ther 19:717–727. https://doi.org/10.1158/1535-7163.MCT-19-0375",
"volume": "19",
"year": "2020"
},
{
"DOI": "10.1007/s12602-023-10155-6",
"author": "H Zhang",
"doi-asserted-by": "publisher",
"first-page": "2150",
"journal-title": "Probiotics Antimicrob Proteins",
"key": "10758_CR85",
"unstructured": "Zhang H, Dong M, Xu H et al (2024) Recombinant Lactococcus lactis expressing human LL-37 prevents deaths from viral infections in piglets and chicken. Probiotics Antimicrob Proteins 16:2150–2160. https://doi.org/10.1007/s12602-023-10155-6",
"volume": "16",
"year": "2024"
},
{
"DOI": "10.1186/s12951-024-02904-8",
"author": "M Yang",
"doi-asserted-by": "publisher",
"journal-title": "J Nanobiotechnol",
"key": "10758_CR86",
"unstructured": "Yang M, Gu K, Xu Q et al (2024) Recombinant Lactococcus lactis secreting FliC protein nanobodies for resistance against Salmonella enteritidis invasion in the intestinal tract. J Nanobiotechnol 22:629. https://doi.org/10.1186/s12951-024-02904-8",
"volume": "22",
"year": "2024"
},
{
"DOI": "10.3389/fbioe.2021.794742",
"author": "KV Teymennet-Ramírez",
"doi-asserted-by": "publisher",
"journal-title": "Front Bioeng Biotechnol",
"key": "10758_CR87",
"unstructured": "Teymennet-Ramírez KV, Martínez-Morales F, Trejo-Hernández MR (2022) Yeast surface display system: strategies for improvement and biotechnological applications. Front Bioeng Biotechnol 9:794742. https://doi.org/10.3389/fbioe.2021.794742",
"volume": "9",
"year": "2022"
},
{
"DOI": "10.1186/1471-2164-10-425",
"author": "G Mathiesen",
"doi-asserted-by": "publisher",
"journal-title": "BMC Genomics",
"key": "10758_CR88",
"unstructured": "Mathiesen G, Sveen A, Brurberg M et al (2009) Genome-wide analysis of signal peptide functionality in Lactobacillus plantarum WCFS1. BMC Genomics 10:425. https://doi.org/10.1186/1471-2164-10-425",
"volume": "10",
"year": "2009"
},
{
"DOI": "10.1186/s12951-024-02573-7",
"author": "Y Yang",
"doi-asserted-by": "publisher",
"journal-title": "J Nanobiotechnol",
"key": "10758_CR89",
"unstructured": "Yang Y, Li F, Du L (2024) Therapeutic nanobodies against SARS-CoV-2 and other pathogenic human coronaviruses. J Nanobiotechnol 22:304. https://doi.org/10.1186/s12951-024-02573-7",
"volume": "22",
"year": "2024"
},
{
"DOI": "10.1007/s11274-024-04157-x",
"author": "M Yoda",
"doi-asserted-by": "publisher",
"journal-title": "World J Microbiol Biotechnol",
"key": "10758_CR90",
"unstructured": "Yoda M, Takase S, Suzuki K et al (2024) Development of engineered IL-36γ-hypersecreting Lactococcus lactis to improve the intestinal environment. World J Microbiol Biotechnol 40:363. https://doi.org/10.1007/s11274-024-04157-x",
"volume": "40",
"year": "2024"
},
{
"DOI": "10.1038/s12276-020-00507-5",
"author": "F Namai",
"doi-asserted-by": "publisher",
"first-page": "1627",
"journal-title": "Exp Mol Med",
"key": "10758_CR91",
"unstructured": "Namai F, Shigemori S, Ogita T et al (2020) Microbial therapeutics for acute colitis based on genetically modified Lactococcus lactis hypersecreting IL-1Ra in mice. Exp Mol Med 52:1627–1636. https://doi.org/10.1038/s12276-020-00507-5",
"volume": "52",
"year": "2020"
},
{
"DOI": "10.1186/s12934-024-02321-4",
"author": "F Zhang",
"doi-asserted-by": "publisher",
"journal-title": "Microb Cell Fact",
"key": "10758_CR92",
"unstructured": "Zhang F, Ni L, Zhang Z et al (2024) Recombinant L. lactis vaccine LL-plSAM-WAE targeting four virulence factors provides mucosal immunity against H. pylori infection. Microb Cell Fact 23:61. https://doi.org/10.1186/s12934-024-02321-4",
"volume": "23",
"year": "2024"
},
{
"DOI": "10.1128/aem.62.10.3662-3667.1996",
"author": "PG De Ruyter",
"doi-asserted-by": "publisher",
"first-page": "3662",
"journal-title": "Appl Environ Microbiol",
"key": "10758_CR93",
"unstructured": "De Ruyter PG, Kuipers OP, De Vos WM (1996) Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol 62:3662–3667. https://doi.org/10.1128/aem.62.10.3662-3667.1996",
"volume": "62",
"year": "1996"
}
],
"reference-count": 93,
"references-count": 93,
"relation": {},
"resource": {
"primary": {
"URL": "https://link.springer.com/10.1007/s12602-025-10758-1"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Construction of Synthetic Probiotic Bacteria for In Situ Delivery of Anti-SARS-CoV-2 Nanobodies",
"type": "journal-article",
"update-policy": "https://doi.org/10.1007/springer_crossmark_policy"
}