SARS-CoV-2 infectivity can be modulated through bacterial grooming of the glycocalyx
et al., mBio, doi:10.1128/mbio.04015-24, Apr 2025
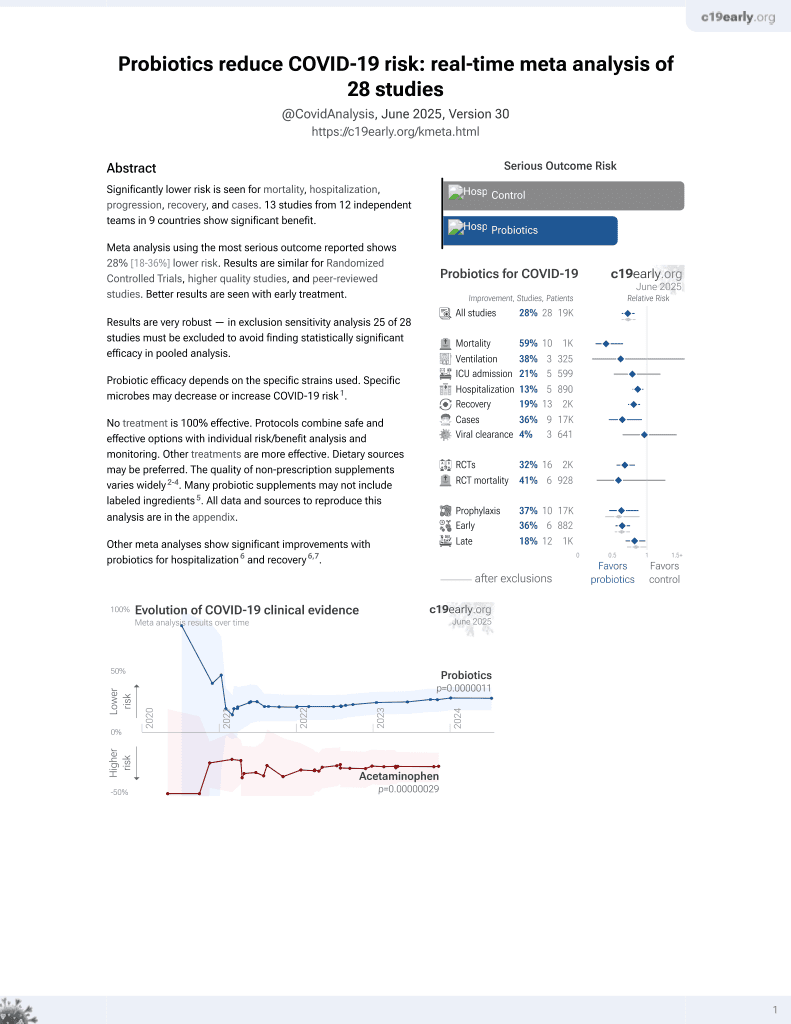
Probiotics for COVID-19
20th treatment shown to reduce risk in
March 2021, now with p = 0.00000044 from 29 studies.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
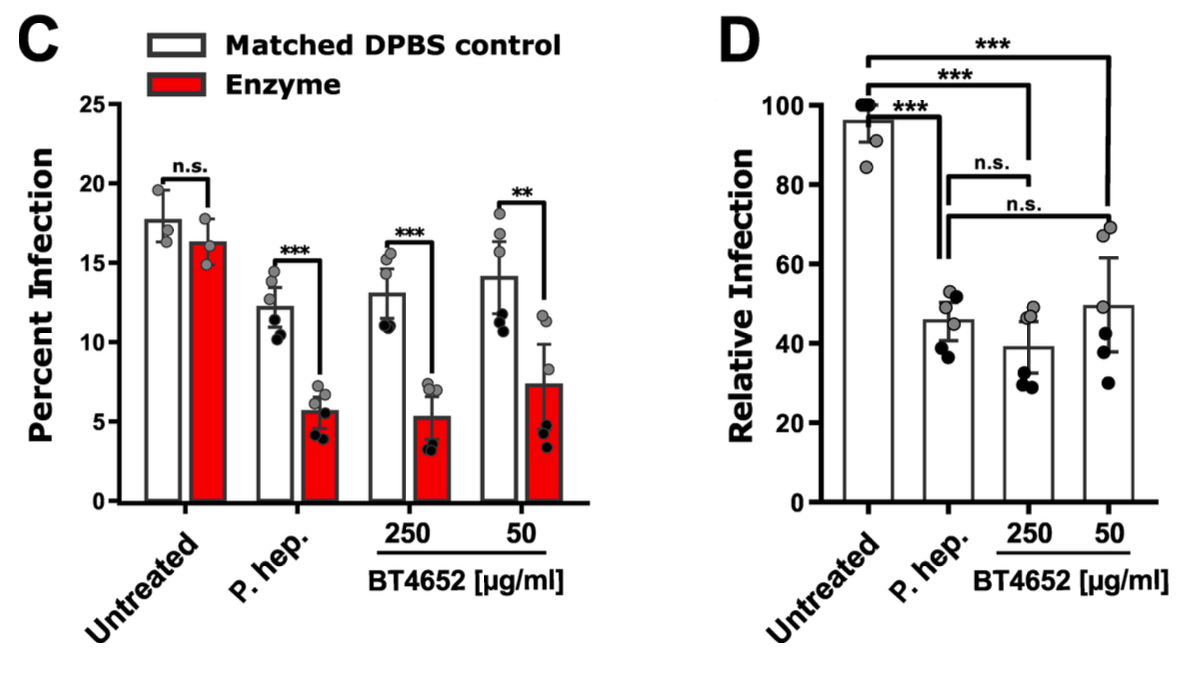
In vitro study showing that certain gut bacteria can modulate SARS-CoV-2 infection by degrading heparan sulfate (HS) on cell surfaces. Authors found that the abundance of HS-modifying bacteria inversely correlates with age, sex, and COVID-19 severity, suggesting a potential protective role. Through metabolic task analysis of large microbiome datasets, they identified bacteria with high HS-catabolic capacity, particularly Bacteroides species. In experiments, cell-free supernatants from B. ovatus and B. thetaiotaomicron reduced cell-surface HS by 60% and decreased SARS-CoV-2 spike protein binding 20-30 fold. Purified bacterial HS lyases (especially BT4652) reduced authentic SARS-CoV-2 infection of Vero E6 cells by ~2.5-fold. Additionally, engineered probiotic E. coli Nissle 1917 expressing B. thetaiotaomicron HS-modifying enzymes effectively blocked SARS-CoV-2 binding to human cells. These findings suggest that bacterial "grooming" of the glycocalyx may influence viral susceptibility and offer potential for novel therapeutic approaches.
Bacteroides species may inhibit SARS-CoV-2 infection by degrading heparan sulfate on cell surfaces, preventing initial viral attachment. Engineered E. coli Nissle 1917 expressing bacterial heparin lyases could serve as a probiotic therapy to prevent SARS-CoV-2 binding and entry. Bacterial HS-modifying enzymes like BT4652 may offer novel therapeutic strategies by modifying the glycocalyx to block SARS-CoV-2 cell entry.
Probiotic efficacy depends on the specific strains used. Specific microbes may decrease or increase COVID-19 risk1.
2 preclinical studies support the efficacy of probiotics for COVID-19:
1.
Li et al., Large-scale genetic correlation studies explore the causal relationship and potential mechanism between gut microbiota and COVID-19-associated risks, BMC Microbiology, doi:10.1186/s12866-024-03423-0.
Martino et al., 9 Apr 2025, USA, peer-reviewed, 65 authors.
Contact: robknight@ucsd.edu.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
SARS-CoV-2 infectivity can be modulated through bacterial grooming of the glycocalyx
mBio, doi:10.1128/mbio.04015-24
The gastrointestinal (GI) tract is a site of replication of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and GI symptoms are often reported by patients. SARS-CoV-2 cell entry depends upon heparan sulfate (HS) proteoglycans, which commensal bacteria that bathe the human mucosa are known to modify. To explore human gut HS-modifying bacterial abundances and how their presence may impact SARS-CoV-2 infection, we developed a task-based analysis of proteoglycan degradation on large-scale shotgun metagenomic data. We observed that gut bacteria with high predicted catabolic capacity for HS differ by age and sex, factors associated with coronavirus disease 2019 (COVID-19) severity, and directly by disease severity during/after infection, but do not vary between subjects with COVID-19 comorbidities or by diet. Gut commensal bacterial HS-modifying enzymes reduce spike protein binding and infection of authentic SARS-CoV-2, suggesting that bacterial grooming of the GI mucosa may impact viral susceptibility. IMPORTANCE Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus responsible for coronavirus disease 2019, can infect the gastrointestinal (GI) tract, and individuals who exhibit GI symptoms often have more severe disease. The GI tract's glycocalyx, a component of the mucosa covering the large intestine, plays a key role in viral entry by binding SARS-CoV-2's spike protein via heparan sulfate (HS). Here, using metabolic task analysis of multiple large microbiome sequencing data sets of the human gut microbiome, we identify a key commensal human intestinal bacteria capable of grooming glycocalyx HS and modulating SARS-CoV-2 infectivity in vitro. Moreover, we engineered the common probiotic Escherichia coli Nissle 1917 (EcN) to effectively block SARS-CoV-2 binding and infection of human cell cultures. Understanding these microbial interactions could lead to better risk assessments and novel therapies targeting viral entry mechanisms.
AUTHOR AFFILIATIONS
ADDITIONAL FILES The following material is available online.
Supplemental Material Supplemental Material (mBio04015-24-s0001.pdf). Supplemental figures and tables.
References
Ali, Sweeney, In pursuit of microbiome-based therapies for acute respiratory failure, Am J Respir Crit Care Med, doi:10.1164/rccm.202008-3146ED
Altschul, Gish, Miller, Myers, Lipman, Basic local alignment search tool, J Mol Biol, doi:10.1093/bioinformatics/btu170
Bankevich, Nurk, Antipov, Gurevich, Dvorkin et al., SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing, J Comput Biol, doi:10.1089/cmb.2012.0021
Beck, Schloss, Venkataraman, Twigg H 3rd, Jablonski et al., Multicenter comparison of lung and oral microbiomes of HIVinfected and HIV-uninfected Individuals, Am J Respir Crit Care Med, doi:10.1164/rccm.202008-3146ED
Belda-Ferre, Knight, Nathan, National Institutes of Health (NIH) 1P30DK120515, Lewis HHS | National Institutes of Health (NIH) R
Benjdia, Leprince, Sandström, Vaudry, Berteau, Mechanistic investigations of anaerobic sulfatase-maturating enzyme: direct Cbeta H-atom abstraction catalyzed by a radical AdoMet enzyme, J Am Chem Soc, doi:10.1021/ja901571p
Benjdia, Martens, Gordon, Berteau, Sulfatases and a radical S-adenosyl-L-methionine (AdoMet) enzyme are key for mucosal foraging and fitness of the prominent human gut symbiont, Bacteroides thetaiotaomicron, J Biol Chem, doi:10.3389/fnut.2022.934568
Bolger, Lohse, Usadel, Trimmomatic: a flexible trimmer for Illumina sequence data, Bioinformatics, doi:10.1093/bioinformatics/btu170
Borodulin, Tolonen, Jousilahti, Jula, Juolevi et al., Cohort profile: the national FINRISK study, Int J Epidemiol, doi:10.3389/fnut.2022.934568
Brownlee, Havler, Dettmar, Allen, Pearson, Colonic mucus: secretion and turnover in relation to dietary fibre intake, Proc Nutr Soc, doi:10.1079/pns2003206
Budden, Gellatly, Wood, Cooper, Morrison et al., Emerging pathogenic links between microbiota and the gut-lung axis, Nat Rev Microbiol, doi:10.1038/nrmicro.2016.142
Cagno, Tseligka, Jones, Tapparel, Heparan sulfate proteoglycans and viral attachment: true receptors or adaptation bias?, Viruses, doi:10.1038/nature18847
Cao, What do we know so far about gastrointestinal and liver injuries induced by SARS-CoV-2 virus?, Gastroenterol Res, doi:10.1136/gutjnl-2021-324280
Cartmell, Lowe, Baslé, Firbank, Ndeh et al., How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans, Proc Natl Acad Sci U S A, doi:10.1073/pnas.1704367114
Cheung, Goh, Lim, Tien, Lim et al., Residual SARS-CoV-2 viral antigens detected in GI and hepatic tissues from five recovered patients with COVID-19, Gut, doi:10.1136/gutjnl-2021-324280
Chu, Chan, Wang, Yuen, Chai et al., SARS-Cov-2 induces a more robust innate immune response and replicates less efficiently than SARS-CoV in the human intestines: an ex vivo study with implications on pathogenesis of COVID-19, Cell Mol Gastroenterol Hepatol, doi:10.1136/gutjnl-2021-324280
Clausen, Sandoval, Spliid, Pihl, Perrett et al., SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2, Cell, doi:10.1016/j.sbi.2022.102439
De La Cuesta-Zuluaga, Kelley, Chen, Escobar, Mueller et al., Ageand sex-dependent patterns of gut microbial diversity in human adults, mSystems, doi:10.3389/fnut.2022.934568
Desai, Seekatz, Koropatkin, Kamada, Hickey et al., A dietary fiberdeprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility, Cell, doi:10.3389/fnut.2022.934568
Didion, Martin, Collins, Atropos: specific, sensitive, and speedy trimming of sequencing reads, PeerJ, doi:10.1093/bioinformatics/btu170
Earle, Billings, Sigal, Lichtman, Hansson et al., Quantitative imaging of gut microbiota spatial organization, Cell Host Microbe, doi:10.3389/fnut.2022.934568
Eilam, Zarecki, Oberhardt, Ursell, Kupiec et al., Glycan degradation (GlyDeR) analysis predicts mammalian gut microbiota abundance and host diet-specific adaptations, MBio, doi:10.1128/mBio.01526-14
Elena, James, Lacey, None
Elshazli, Kline, Elgaml, Aboutaleb, Salim et al., Gastroenterology manifestations and COVID-19 outcomes: a meta-analysis of 25,252 cohorts among the first and second waves, J Med Virol, doi:10.1136/gutjnl-2021-324280
Esko, Selleck, Order out of chaos: assembly of ligand binding sites in heparan sulfate, Annu Rev Biochem, doi:10.1146/annurev.biochem.71.110601.135458
Galliher, Cooney, Langer, Linhardt, Heparinase production by Flavobacterium heparinum, Appl Environ Microbiol, doi:10.1128/aem.41.2.360-365.1981
Grondin, Tamura, Déjean, Abbott, Brumer, Polysaccharide utilization loci: fueling microbial communities, J Bacteriol, doi:10.1128/JB.00860-16
Guo, Tao, Flavell, Zhu, Potential intestinal infection and faecal-oral transmission of SARS-CoV-2, Nat Rev Gastroenterol Hepatol, doi:10.1136/gutjnl-2021-324280
Han, Duan, Yang, Nilsson-Payant, Wang et al., Identification of SARS-CoV-2 inhibitors using lung and colonic organoids, Nature New Biol, doi:10.1038/s41586-020-2901-9
Hayashi, Wagatsuma, Nojima, Yamakawa, Ichimiya et al., The characteristics of gastrointestinal symptoms in patients with severe COVID-19: a systematic review and meta-analysis, J Gastroenterol, doi:10.1136/gutjnl-2021-324280
Hedemann, Theil, Knudsen, The thickness of the intestinal mucous layer in the colon of rats fed various sources of nondigestible carbohydrates is positively correlated with the pool of SCFA but negatively correlated with the proportion of butyric acid in digesta, Br J Nutr, doi:10.1017/S0007114508143549
Hillmann, Al-Ghalith, Shields-Cutler, Zhu, Gohl et al., Evaluating the information content of shallow shotgun metagenomics, mSystems, doi:10.1093/bioinformatics/btu170
Hobbs, Pluvinage, Boraston, Glycan-metabolizing enzymes in microbe-host interactions: the Streptococcus pneumoniae paradigm, FEBS Lett, doi:10.1002/1873-3468.13045
Inoyue, None
Jeffrey, Alfred Benzon Foundation, The Alfred Benzon Foundation) Thomas Mandel Clausen Academy of Finland 321351 and 354447 Teemu Niiranen Emil Aaltosen Säätiö
Jeffrey, Novo, Foundation, None, doi:10.13039/501100009708
Jennifer, Benbow, Spielfogel Cheryl, Anderson Maria, James, None
Jennifer, Benbow, Spielfogel Cheryl, Anderson, Elena et al., National Institutes of Health (NIH)
Jennifer, Benbow, Spielfogel Cheryl, Anderson, Elena et al., National Institutes of Health (NIH) P
Jennifer, Benbow, Spielfogel Cheryl, Anderson, Elena et al., National Institutes of Health (NIH) P
Jennifer, Benbow, Spielfogel Cheryl, Anderson, None, Research Article mBio
Jin, Ananthanarayanan, Brown, Rager, Bram et al., SARS CoV-2 detected in neonatal stool remote from maternal COVID-19 during pregnancy, Pediatr Res, doi:10.1136/gutjnl-2021-324280
Johansson, Numerical python: scientific computing and data science applications with Numpy, doi:10.1093/bioinformatics/btu170
Johansson, Phillipson, Petersson, Velcich, Holm et al., The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria, Proc Natl Acad Sci U S A, doi:10.1073/pnas.0803124105
Kanehisa, Furumichi, Tanabe, Sato, Morishima, KEGG: new perspectives on genomes, pathways, diseases and drugs, Nucleic Acids Res, doi:10.1093/nar/gkw1092
Kearns, Sandoval, Casalino, Clausen, Rosenfeld et al., Spike-heparan sulfate interactions in SARS-CoV-2 infection, Curr Opin Struct Biol, doi:10.1016/j.sbi.2022.102439
Koropatkin, Cameron, Martens, How glycan metabolism shapes the human gut microbiota, Nat Rev Microbiol, doi:10.1038/nrmicro2746
Lagesen, Hallin, Rødland, Staerfeldt, Rognes et al., RNAmmer: consistent and rapid annotation of ribosomal RNA genes, Nucleic Acids Res, doi:10.1093/bioinformatics/btu170
Langdon, Performance of genetic programming optimised Bowtie2 on genome comparison and analytic testing (GCAT) bench marks, BioData Min, doi:10.1186/s13040-014-0034-0
Langmead, Salzberg, Fast gapped-read alignment with Bowtie 2, Nat Methods, doi:10.1093/bioinformatics/btu170
Lee, Kim, Park, OrthoANI: an improved algorithm and software for calculating average nucleotide identity, Int J Syst Evol Microbiol, doi:10.1099/ijsem.0.000760
Lewis, None
Linhardt, Galliher, Cooney, Polysaccharide lyases, Appl Biochem Biotechnol, doi:10.1007/BF02798420
Lloyd-Price, Mahurkar, Rahnavard, Crabtree, Orvis et al., Erratum: strains, functions and dynamics in the expanded human microbiome project, Nature New Biol, doi:10.1038/nature24485
Lombard, Ramulu, Drula, Coutinho, Henrissat, The carbohydrate-active enzymes database (CAZy) in 2013, Nucleic Acids Res, doi:10.1093/nar/gkt1178
Machado, Andrejev, Tramontano, Patil, Fast automated reconstruction of genome-scale metabolic models for microbial species and communities, Nucleic Acids Res, doi:10.1093/nar/gky537
Magnúsdóttir, Heinken, Kutt, Ravcheev, Bauer et al., Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota, Nat Biotechnol, doi:10.1038/nbt.3703
Magoč, Salzberg, FLASH: fast length adjustment of short reads to improve genome assemblies, Bioinformatics, doi:10.1093/bioinformatics/btr507
Marotz, Belda-Ferre, Ali, Das, Huang et al., SARS-CoV-2 detection status associates with bacterial community composition in patients and the hospital environment, Microbiome, doi:10.3389/fnut.2022.934568
Marotz, Salido, Zhu, Mcdonald, Taylor et al., None
Martino, Dilmore, Burcham, Metcalf, Jeste et al., Microbiota succession throughout life from the cradle to the grave, Nat Rev Microbiol, doi:10.1164/rccm.202008-3146ED
Mcdonald, Hyde, Debelius, Morton, Gonzalez et al., American gut: an open platform for citizen science microbiome research, mSystems, doi:10.1038/nature18847
Mcdonald, Knight, University of California
Mehta, Mcauley, Brown, Sanchez, Tattersall et al., COVID-19: consider cytokine storm syndromes and immunosuppression, Lancet, doi:10.1136/gutjnl-2021-324280
Natarajan, Zlitni, Brooks, Vance, Dahlen et al., Gastrointestinal symptoms and fecal shedding of SARS-CoV-2 RNA suggest prolonged gastrointestinal infection, Med, doi:10.1136/gutjnl-2021-324280
Ndeh, Baslé, Strahl, Yates, Mcclurgg et al., Metabolism of multiple glycosaminoglycans by Bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus, Nat Commun, doi:10.1038/s41467-020-14509-4
Neurath, Überla, Ng, Gut as viral reservoir: lessons from gut viromes, HIV and COVID-19, Gut, doi:10.1136/gutjnl-2021-324280
Pruitt, Tatusova, Maglott, NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins, Nucleic Acids Res, doi:10.1093/bioinformatics/btu170
Redd, Zhou, Hathorn, Mccarty, Bazarbashi et al., Prevalence and characteristics of gastroin testinal symptoms in patients with severe acute respiratory syndrome coronavirus 2 infection in the united states: a multicenter cohort study, Gastroenterology, doi:10.3389/fnut.2022.934568
Reed, Famili, Thiele, Palsson, Towards multidimensional genome annotation, Nat Rev Genet, doi:10.1038/nrg1769
Richelle, Chiang, Kuo, Ne, Increasing consensus of context-specific metabolic models by integrating data-inferred cell functions, PLoS Comput Biol, doi:10.1093/bioinformatics/btu170
Richelle, Joshi, Lewis, Assessing key decisions for transcriptomic data integration in biochemical networks, PLoS Comput Biol, doi:10.1371/journal.pcbi.1007185
Richelle, Kellman, Wenzel, Chiang, Reagan et al., Model-based assessment of mammalian cell metabolic functionalities using omics data, Cell Rep Methods, doi:10.3389/fnut.2022.934568
Robb, Hobbs, Woodiga, Shapiro-Ward, Suits et al., Molecular characteri zation of N-glycan degradation and transport in Streptococcus pneumoniae and its contribution to virulence, PLoS Pathog, doi:10.1371/journal.ppat.1006090
Salomaa, National Institutes of Health (NIH) R01ES027595 Mohit Jain ANID Becas Chile Doctorado, Erick Armingol Research Article mBio
Salosensaari, Laitinen, Havulinna, Meric, Cheng et al., Taxonomic signatures of long-term mortality risk in human gut microbiota, Epidemiology, doi:10.3389/fnut.2022.934568
Sanders, Nurk, Salido, Minich, Xu et al., Optimizing sequencing protocols for leaderboard metagenomics by combining long and short reads, Genome Biol, doi:10.1186/s13059-019-1834-9
Thaiss, Zmora, Levy, Elinav, The microbiome and innate immunity, Nature New Biol, doi:10.1038/nature18847
Ulmer, Vilén, Namburi, Benjdia, Beneteau et al., Characterization of glycosaminoglycan (GAG) sulfatases from the human gut symbiont bacteroides thetaiotaomicron reveals the first GAG-specific bacterial endosulfatase, J Biol Chem, doi:10.1074/jbc.M114.573303
Vajargah, Zargarzadeh, Ebrahimzadeh, Mousavi, Mobasheran et al., Association of fruits, vegetables, and fiber intake with COVID-19 severity and symptoms in hospitalized patients: a cross-sectional study, Front Nutr, doi:10.3389/fnut.2022.934568
Vespa, Pugliese, Colapietro, Aghemo, Stay (GI) Healthy: COVID-19 and gastrointestinal manifestations, Tech Innov Gastrointest Endosc, doi:10.1136/gutjnl-2021-324280
Villapol, Gastrointestinal symptoms associated with COVID-19: impact on the gut microbiome, Transl Res, doi:10.1136/gutjnl-2021-324280
Weiss, Esko, Tor, Targeting heparin and heparan sulfate protein interactions, Org Biomol Chem, doi:10.1039/C7OB01058C
Williamson, Walker, Bhaskaran, Bacon, Bates et al., OpenSAFELY: factors associated with COVID-19 death in 17 million patients, Nature New Biol, doi:10.3389/fnut.2022.934568
Woo, Shafiq, Fitzek, Dottermusch, Altmeppen et al., Vagus nerve inflammation contributes to dysautonomia in COVID-19, Acta Neuropathol, doi:10.1136/gutjnl-2021-324280
Yin, Mao, Yang, Chen, Mao et al., dbCAN: a web resource for automated carbohydrate-active enzyme annotation, Nucleic Acids Res, doi:10.1093/nar/gks479
Zaramela, Martino, Alisson-Silva, Rees, Diaz et al., Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates, Nat Microbiol, doi:10.3389/fnut.2022.934568
Zhang, Kang, Gong, Xu, Wang et al., Digestive system is a potential route of COVID-19: an analysis of single-cell coexpression pattern of key proteins in viral entry process, Gut, doi:10.1136/gutjnl-2020-320953
Zhang, Yohe, Huang, Entwistle, Wu et al., dbCAN2: a meta server for automated carbohydrate-active enzyme annotation, Nucleic Acids Res, doi:10.1093/nar/gky418
Zhu, Mai, Pfeiffer, Janssen, Asnicar et al., Phylogenomics of 10,575 genomes reveals evolutionary proximity between domains bacteria and archaea, Nat Commun, doi:10.3389/fnut.2022.934568
Zhu, Vázquez, Shi, Niina, Laxmi et al., None, National Institutes of Health (NIH) P
Zmora, Suez, Elinav, You are what you eat: diet, health and the gut microbiota, Nat Rev Gastroenterol Hepatol, doi:10.1038/s41575-018-0061-2
DOI record:
{
"DOI": "10.1128/mbio.04015-24",
"ISSN": [
"2150-7511"
],
"URL": "http://dx.doi.org/10.1128/mbio.04015-24",
"abstract": "<jats:title>ABSTRACT</jats:title>\n <jats:sec>\n <jats:title/>\n <jats:p>The gastrointestinal (GI) tract is a site of replication of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and GI symptoms are often reported by patients. SARS-CoV-2 cell entry depends upon heparan sulfate (HS) proteoglycans, which commensal bacteria that bathe the human mucosa are known to modify. To explore human gut HS-modifying bacterial abundances and how their presence may impact SARS-CoV-2 infection, we developed a task-based analysis of proteoglycan degradation on large-scale shotgun metagenomic data. We observed that gut bacteria with high predicted catabolic capacity for HS differ by age and sex, factors associated with coronavirus disease 2019 (COVID-19) severity, and directly by disease severity during/after infection, but do not vary between subjects with COVID-19 comorbidities or by diet. Gut commensal bacterial HS-modifying enzymes reduce spike protein binding and infection of authentic SARS-CoV-2, suggesting that bacterial grooming of the GI mucosa may impact viral susceptibility.</jats:p>\n <jats:sec>\n <jats:title>IMPORTANCE</jats:title>\n <jats:p>\n Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus responsible for coronavirus disease 2019, can infect the gastrointestinal (GI) tract, and individuals who exhibit GI symptoms often have more severe disease. The GI tract’s glycocalyx, a component of the mucosa covering the large intestine, plays a key role in viral entry by binding SARS-CoV-2’s spike protein via heparan sulfate (HS). Here, using metabolic task analysis of multiple large microbiome sequencing data sets of the human gut microbiome, we identify a key commensal human intestinal bacteria capable of grooming glycocalyx HS and modulating SARS-CoV-2 infectivity\n <jats:italic>in vitro</jats:italic>\n . Moreover, we engineered the common probiotic\n <jats:italic>Escherichia coli</jats:italic>\n Nissle 1917 (EcN) to effectively block SARS-CoV-2 binding and infection of human cell cultures. Understanding these microbial interactions could lead to better risk assessments and novel therapies targeting viral entry mechanisms.\n </jats:p>\n </jats:sec>\n </jats:sec>",
"alternative-id": [
"10.1128/mbio.04015-24"
],
"assertion": [
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Received",
"name": "received",
"order": 0,
"value": "2024-12-23"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "2025-01-30"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Published",
"name": "published",
"order": 3,
"value": "2025-02-25"
}
],
"author": [
{
"ORCID": "https://orcid.org/0000-0001-9334-1258",
"affiliation": [
{
"name": "Department of Pediatrics, University of California San Diego School of Medicine",
"place": [
"La Jolla, USA"
]
},
{
"name": "Bioinformatics and Systems Biology Program, University of California San Diego",
"place": [
"La Jolla, USA"
]
},
{
"name": "Center for Microbiome Innovation, University of California San Diego",
"place": [
"La Jolla, USA"
]
}
],
"authenticated-orcid": false,
"family": "Martino",
"given": "Cameron",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Department of Pediatrics, University of California San Diego School of Medicine",
"place": [
"La Jolla, USA"
]
},
{
"name": "Bioinformatics and Systems Biology Program, University of California San Diego",
"place": [
"La Jolla, USA"
]
}
],
"family": "Kellman",
"given": "Benjamin P.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Cellular and Molecular Medicine, University of California San Diego",
"place": [
"La Jolla, USA"
]
}
],
"family": "Sandoval",
"given": "Daniel R.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Cellular and Molecular Medicine, University of California San Diego",
"place": [
"La Jolla, USA"
]
},
{
"name": "Copenhagen Center for Glycomics, Department of Molecular and Cellular Medicine, Faculty of Health and Medical Sciences, University of Copenhagen",
"place": [
"Copenhagen, Denmark"
]
}
],
"family": "Clausen",
"given": "Thomas Mandel",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Bioengineering, University of California San Diego",
"place": [
"La Jolla, USA"
]
}
],
"family": "Cooper",
"given": "Robert",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Université Paris-Saclay, INRAE, AgroParisTech, Micalis Institute, ChemSyBio",
"place": [
"78350, Jouy-en-Josas, France"
]
}
],
"family": "Benjdia",
"given": "Alhosna",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Université Paris-Saclay, INRAE, AgroParisTech, Micalis Institute, ChemSyBio",
"place": [
"78350, Jouy-en-Josas, France"
]
},
{
"name": "Sorbonne Université, Faculty of Sciences and Engineering, IBPS, UMR 8263 CNRS-SU, ERL INSERM U1345, Development, Adaptation and Ageing",
"place": [
"F-75252 Paris, France"
]
}
],
"family": "Soualmia",
"given": "Feryel",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Medicine, University of California San Diego",
"place": [
"La Jolla, USA"
]
}
],
"family": "Clark",
"given": "Alex E.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Medicine, University of California San Diego",
"place": [
"La Jolla, USA"
]
}
],
"family": "Garretson",
"given": "Aaron F.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Pediatrics, University of California San Diego School of Medicine",
"place": [
"La Jolla, USA"
]
}
],
"family": "Marotz",
"given": "Clarisse A.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Center for Microbiome Innovation, University of California San Diego",
"place": [
"La Jolla, USA"
]
}
],
"family": "Song",
"given": "Se Jin",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Center for Microbiome Innovation, University of California San Diego",
"place": [
"La Jolla, USA"
]
}
],
"family": "Wandro",
"given": "Stephen",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Pediatrics, University of California San Diego School of Medicine",
"place": [
"La Jolla, USA"
]
},
{
"name": "Department of Biochemistry, Ribeirão Preto Medical School, University of São Paulo",
"place": [
"Ribeirão Preto, Brazil"
]
}
],
"family": "Zaramela",
"given": "Livia S.",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-0631-2817",
"affiliation": [
{
"name": "Department of Pediatrics, University of California San Diego School of Medicine",
"place": [
"La Jolla, USA"
]
},
{
"name": "Center for Microbiome Innovation, University of California San Diego",
"place": [
"La Jolla, USA"
]
},
{
"name": "Department of Bioengineering, University of California San Diego",
"place": [
"La Jolla, USA"
]
}
],
"authenticated-orcid": true,
"family": "Salido",
"given": "Rodolfo A.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Pediatrics, University of California San Diego School of Medicine",
"place": [
"La Jolla, USA"
]
},
{
"name": "School of Life Sciences, Arizona State University",
"place": [
"Tempe, USA"
]
}
],
"family": "Zhu",
"given": "Qiyun",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Pediatrics, University of California San Diego School of Medicine",
"place": [
"La Jolla, USA"
]
},
{
"name": "Bioinformatics and Systems Biology Program, University of California San Diego",
"place": [
"La Jolla, USA"
]
}
],
"family": "Armingol",
"given": "Erick",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Center for Microbiome Innovation, University of California San Diego",
"place": [
"La Jolla, USA"
]
},
{
"name": "Jacobs School of Engineering, University of California San Diego",
"place": [
"La Jolla, USA"
]
}
],
"family": "Vázquez-Baeza",
"given": "Yoshiki",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Pediatrics, University of California San Diego School of Medicine",
"place": [
"La Jolla, USA"
]
}
],
"family": "McDonald",
"given": "Daniel",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Pediatrics, University of California San Diego School of Medicine",
"place": [
"La Jolla, USA"
]
},
{
"name": "Bioinformatics and Systems Biology Program, University of California San Diego",
"place": [
"La Jolla, USA"
]
}
],
"family": "Sorrentino",
"given": "James T.",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-4957-5417",
"affiliation": [
{
"name": "Biomedical Sciences Graduate Program, University of California San Diego",
"place": [
"La Jolla, USA"
]
}
],
"authenticated-orcid": true,
"family": "Taylor",
"given": "Bryn",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Pediatrics, University of California San Diego School of Medicine",
"place": [
"La Jolla, USA"
]
}
],
"family": "Belda-Ferre",
"given": "Pedro",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-6733-2059",
"affiliation": [
{
"name": "Department of Pediatrics, University of California San Diego School of Medicine",
"place": [
"La Jolla, USA"
]
},
{
"name": "Scripps Institution of Oceanography, University of California San Diego",
"place": [
"La Jolla, USA"
]
}
],
"authenticated-orcid": true,
"family": "Das",
"given": "Promi",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Pediatrics, University of California San Diego School of Medicine",
"place": [
"La Jolla, USA"
]
}
],
"family": "Ali",
"given": "Farhana",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Pediatrics, University of California San Diego School of Medicine",
"place": [
"La Jolla, USA"
]
},
{
"name": "Department of Bioengineering, University of California San Diego",
"place": [
"La Jolla, USA"
]
},
{
"name": "Merck & Co., Inc.",
"place": [
"Rahway, USA"
]
}
],
"family": "Liang",
"given": "Chenguang",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Bioengineering, University of California San Diego",
"place": [
"La Jolla, USA"
]
},
{
"name": "Department of Biological & Medical Informatics, University of California San Francisco, School of Pharmacy",
"place": [
"San Francisco, USA"
]
}
],
"family": "Zhang",
"given": "Yujie",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Surgery, Division of Surgical Outcomes and Precision Medicine Research, Medical School, University of Minnesota",
"place": [
"Minneapolis, USA"
]
},
{
"name": "National Institutes of Health, National Cancer Institute, Center for Cancer Research, Animal Models and Retroviral Vaccine Section",
"place": [
"Bethesda, USA"
]
}
],
"family": "Schifanella",
"given": "Luca",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Infectious diseases, Luigi Sacco Hospital, Milan and Department of Biomedical and Clinical Sciences (DIBIC), University of Milan",
"place": [
"Milan, Italy"
]
}
],
"family": "Covizzi",
"given": "Alice",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Infectious diseases, Luigi Sacco Hospital, Milan and Department of Biomedical and Clinical Sciences (DIBIC), University of Milan",
"place": [
"Milan, Italy"
]
}
],
"family": "Lai",
"given": "Alessia",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Infectious diseases, Luigi Sacco Hospital, Milan and Department of Biomedical and Clinical Sciences (DIBIC), University of Milan",
"place": [
"Milan, Italy"
]
}
],
"family": "Riva",
"given": "Agostino",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Surgery, Division of Surgical Outcomes and Precision Medicine Research, Medical School, University of Minnesota",
"place": [
"Minneapolis, USA"
]
}
],
"family": "Basting",
"given": "Christopher",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Surgery, Division of Surgical Outcomes and Precision Medicine Research, Medical School, University of Minnesota",
"place": [
"Minneapolis, USA"
]
}
],
"family": "Broedlow",
"given": "Courtney Ann",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Public Health and Welfare, Finnish Institute for Health and Welfare",
"place": [
"Helsinki and Turku, Finland"
]
},
{
"name": "Institute for Molecular Medicine Finland, FIMM - HiLIFE",
"place": [
"Helsinki, Finland"
]
}
],
"family": "Havulinna",
"given": "Aki S.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Public Health and Welfare, Finnish Institute for Health and Welfare",
"place": [
"Helsinki and Turku, Finland"
]
}
],
"family": "Jousilahti",
"given": "Pekka",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-4556-8444",
"affiliation": [
{
"name": "Department of Pediatrics, University of California San Diego School of Medicine",
"place": [
"La Jolla, USA"
]
}
],
"authenticated-orcid": true,
"family": "Estaki",
"given": "Mehrbod",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Pediatrics, University of California San Diego School of Medicine",
"place": [
"La Jolla, USA"
]
},
{
"name": "Sano Centre for Computational Medicine",
"place": [
"Krakow, Poland"
]
}
],
"family": "Kosciolek",
"given": "Tomasz",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Laureate Institute for Brain Research",
"place": [
"Tulsa, USA"
]
}
],
"family": "Kuplicki",
"given": "Rayus",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Laureate Institute for Brain Research",
"place": [
"Tulsa, USA"
]
}
],
"family": "Victor",
"given": "Teresa A.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Laureate Institute for Brain Research",
"place": [
"Tulsa, USA"
]
}
],
"family": "Paulus",
"given": "Martin P.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Division of Health Analytics, Department of Computational and Quantitative Medicine, City of Hope",
"place": [
"Duarte, USA"
]
}
],
"family": "Savage",
"given": "Kristen E.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Division of Health Analytics, Department of Computational and Quantitative Medicine, City of Hope",
"place": [
"Duarte, USA"
]
},
{
"name": "UC Health Data Warehouse, University of California Irvine",
"place": [
"Irvine, USA"
]
}
],
"family": "Benbow",
"given": "Jennifer L.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Division of Health Analytics, Department of Computational and Quantitative Medicine, City of Hope",
"place": [
"Duarte, USA"
]
}
],
"family": "Spielfogel",
"given": "Emma S.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Herbert Wertheim School of Public Health and Human Longevity Science, University of California San Diego",
"place": [
"La Jolla, USA"
]
}
],
"family": "Anderson",
"given": "Cheryl A. M.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Herbert Wertheim School of Public Health and Human Longevity Science, University of California San Diego",
"place": [
"La Jolla, USA"
]
}
],
"family": "Martinez",
"given": "Maria Elena",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Division of Health Analytics, Department of Computational and Quantitative Medicine, City of Hope",
"place": [
"Duarte, USA"
]
}
],
"family": "Lacey",
"given": "James V.",
"sequence": "additional",
"suffix": "Jr"
},
{
"affiliation": [
{
"name": "Department of Pediatrics, University of California San Diego School of Medicine",
"place": [
"La Jolla, USA"
]
},
{
"name": "Center for Microbiome Innovation, University of California San Diego",
"place": [
"La Jolla, USA"
]
},
{
"name": "Faculty of Dentistry, The University of Hong Kong",
"place": [
"Hong Kong, China"
]
}
],
"family": "Huang",
"given": "Shi",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-8663-1019",
"affiliation": [
{
"name": "IBM T. J. Watson Research Center",
"place": [
"Yorktown Heights, USA"
]
}
],
"authenticated-orcid": true,
"family": "Haiminen",
"given": "Niina",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "IBM T. J. Watson Research Center",
"place": [
"Yorktown Heights, USA"
]
}
],
"family": "Parida",
"given": "Laxmi",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "AI and Cognitive Software, IBM Research-Almaden",
"place": [
"San Jose, USA"
]
}
],
"family": "Kim",
"given": "Ho-Cheol",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Pediatrics, University of California San Diego School of Medicine",
"place": [
"La Jolla, USA"
]
},
{
"name": "Center for Microbiome Innovation, University of California San Diego",
"place": [
"La Jolla, USA"
]
},
{
"name": "Scripps Institution of Oceanography, University of California San Diego",
"place": [
"La Jolla, USA"
]
}
],
"family": "Gilbert",
"given": "Jack A.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Division of Pulmonary, Critical Care and Sleep Medicine, Department of Medicine, University of California San Diego",
"place": [
"La Jolla, USA"
]
}
],
"family": "Sweeney",
"given": "Daniel A.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Pediatrics, University of California San Diego School of Medicine",
"place": [
"La Jolla, USA"
]
},
{
"name": "Scripps Institution of Oceanography, University of California San Diego",
"place": [
"La Jolla, USA"
]
}
],
"family": "Allard",
"given": "Sarah M.",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-5655-8300",
"affiliation": [
{
"name": "Center for Microbiome Innovation, University of California San Diego",
"place": [
"La Jolla, USA"
]
},
{
"name": "International Biomedical Research Alliance",
"place": [
"Bethesda, USA"
]
}
],
"authenticated-orcid": false,
"family": "Swafford",
"given": "Austin D.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Division of Cardiology, Brigham and Women’s Hospital",
"place": [
"Boston, USA"
]
},
{
"name": "Cedars-Sinai Medical Center",
"place": [
"Los Angeles, USA"
]
}
],
"family": "Cheng",
"given": "Susan",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Health Data Research UK Cambridge, Wellcome Genome Campus and University of Cambridge",
"place": [
"Cambridge, United Kingdom"
]
},
{
"name": "Cambridge Baker Systems Genomics Initiative, Baker Heart and Diabetes Institute",
"place": [
"Melbourne, Australia"
]
},
{
"name": "Cambridge Baker Systems Genomics Initiative, Department of Public Health and Primary Care, University of Cambridge",
"place": [
"Cambridge, United Kingdom"
]
}
],
"family": "Inouye",
"given": "Michael",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Public Health and Welfare, Finnish Institute for Health and Welfare",
"place": [
"Helsinki and Turku, Finland"
]
},
{
"name": "Division of Medicine, Turku University Hospital and University of Turku",
"place": [
"Turku, Finland"
]
}
],
"family": "Niiranen",
"given": "Teemu",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Pharmacology, University of California, San Diego",
"place": [
"La Jolla, USA"
]
}
],
"family": "Jain",
"given": "Mohit",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Public Health and Welfare, Finnish Institute for Health and Welfare",
"place": [
"Helsinki and Turku, Finland"
]
}
],
"family": "Salomaa",
"given": "Veikko",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-8062-3296",
"affiliation": [
{
"name": "Department of Pediatrics, University of California San Diego School of Medicine",
"place": [
"La Jolla, USA"
]
},
{
"name": "Center for Microbiome Innovation, University of California San Diego",
"place": [
"La Jolla, USA"
]
},
{
"name": "Department of Bioengineering, University of California San Diego",
"place": [
"La Jolla, USA"
]
}
],
"authenticated-orcid": true,
"family": "Zengler",
"given": "Karsten",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-2968-5480",
"affiliation": [
{
"name": "Department of Surgery, Division of Surgical Outcomes and Precision Medicine Research, Medical School, University of Minnesota",
"place": [
"Minneapolis, USA"
]
}
],
"authenticated-orcid": true,
"family": "Klatt",
"given": "Nichole R.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Bioengineering, University of California San Diego",
"place": [
"La Jolla, USA"
]
},
{
"name": "Molecular Biology Section, Division of Biological Science, University of California San Diego",
"place": [
"La Jolla, USA"
]
}
],
"family": "Hasty",
"given": "Jeff",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Université Paris-Saclay, INRAE, AgroParisTech, Micalis Institute, ChemSyBio",
"place": [
"78350, Jouy-en-Josas, France"
]
}
],
"family": "Berteau",
"given": "Olivier",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Medicine, University of California San Diego",
"place": [
"La Jolla, USA"
]
}
],
"family": "Carlin",
"given": "Aaron F.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Cellular and Molecular Medicine, University of California San Diego",
"place": [
"La Jolla, USA"
]
},
{
"name": "Glycobiology Research and Training Center, University of California San Diego",
"place": [
"La Jolla, USA"
]
}
],
"family": "Esko",
"given": "Jeffrey D.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Pediatrics, University of California San Diego School of Medicine",
"place": [
"La Jolla, USA"
]
},
{
"name": "Center for Microbiome Innovation, University of California San Diego",
"place": [
"La Jolla, USA"
]
},
{
"name": "Department of Bioengineering, University of California San Diego",
"place": [
"La Jolla, USA"
]
},
{
"name": "Jacobs School of Engineering, University of California San Diego",
"place": [
"La Jolla, USA"
]
},
{
"name": "Center for Molecular Medicine, Complex Carbohydrate Research Center, and Dept of Biochemistry and Molecular Biology, University of Georgia",
"place": [
"Athens, USA"
]
},
{
"name": "Department of Biotechnology and Biomedicine, Technical University of Denmark",
"place": [
"Lyngby, Denmark"
]
}
],
"family": "Lewis",
"given": "Nathan E.",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-0975-9019",
"affiliation": [
{
"name": "Department of Pediatrics, University of California San Diego School of Medicine",
"place": [
"La Jolla, USA"
]
},
{
"name": "Center for Microbiome Innovation, University of California San Diego",
"place": [
"La Jolla, USA"
]
},
{
"name": "Department of Bioengineering, University of California San Diego",
"place": [
"La Jolla, USA"
]
},
{
"name": "Department of Computer Science and Engineering, University of California San Diego",
"place": [
"La Jolla, USA"
]
}
],
"authenticated-orcid": true,
"family": "Knight",
"given": "Rob",
"sequence": "additional"
}
],
"container-title": "mBio",
"container-title-short": "mBio",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"journals.asm.org"
]
},
"created": {
"date-parts": [
[
2025,
2,
25
]
],
"date-time": "2025-02-25T14:08:27Z",
"timestamp": 1740492507000
},
"deposited": {
"date-parts": [
[
2025,
4,
9
]
],
"date-time": "2025-04-09T13:06:35Z",
"timestamp": 1744203995000
},
"editor": [
{
"affiliation": [],
"family": "Miller",
"given": "Samuel I.",
"sequence": "additional"
}
],
"funder": [
{
"award": [
"AI Horizons Network"
],
"name": "IBM | IBM Research"
},
{
"award": [
"P01 HL131474"
],
"name": "HHS | National Institutes of Health"
},
{
"DOI": "10.13039/501100009708",
"award": [
"NNF20SA0066621"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100009708",
"id-type": "DOI"
}
],
"name": "Novo Nordisk Foundation"
},
{
"award": [
"2031989"
],
"name": "National Science Foundation"
},
{
"DOI": "10.13039/501100007469",
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/501100007469",
"id-type": "DOI"
}
],
"name": "Alfred Benzon Foundation"
},
{
"DOI": "10.13039/501100002341",
"award": [
"321351 and 354447"
],
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/501100002341",
"id-type": "DOI"
}
],
"name": "Academy of Finland"
},
{
"DOI": "10.13039/501100004756",
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/501100004756",
"id-type": "DOI"
}
],
"name": "Emil Aaltosen Säätiö"
},
{
"DOI": "10.13039/501100005633",
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/501100005633",
"id-type": "DOI"
}
],
"name": "Sydäntutkimussäätiö"
},
{
"award": [
"R01ES027595"
],
"name": "HHS | National Institutes of Health"
},
{
"award": [
"2018-72190270"
],
"name": "ANID Becas Chile Doctorado"
},
{
"DOI": "10.13039/501100002341",
"award": [
"321356"
],
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/501100002341",
"id-type": "DOI"
}
],
"name": "Academy of Finland"
},
{
"award": [
"N/A"
],
"name": "UC San Diego Center for Microbiome Innovation"
},
{
"DOI": "10.13039/501100002341",
"award": [
"335525"
],
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/501100002341",
"id-type": "DOI"
}
],
"name": "Academy of Finland"
},
{
"award": [
"101046041"
],
"name": "Union's Horizon Europe Research and Innovation 459 Actions"
},
{
"DOI": "10.13039/501100001665",
"award": [
"ANR-17-CE11-0014"
],
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/501100001665",
"id-type": "DOI"
}
],
"name": "French National Research Agency"
},
{
"DOI": "10.13039/501100001665",
"award": [
"ANR-20-CE44-0005"
],
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/501100001665",
"id-type": "DOI"
}
],
"name": "French National Research Agency"
},
{
"award": [
"R01GM069811"
],
"name": "HHS | National Institutes of Health"
},
{
"name": "Munz Chair of Cardiovascular Prediction and Prevention"
},
{
"name": "NIHR | NIHR Cambridge Biomedical Research Centre"
},
{
"DOI": "10.13039/501100000274",
"award": [
"RG/13/13/30194"
],
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/501100000274",
"id-type": "DOI"
}
],
"name": "British Heart Foundation"
},
{
"DOI": "10.13039/501100000274",
"award": [
"RG/18/13/33946"
],
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/501100000274",
"id-type": "DOI"
}
],
"name": "British Heart Foundation"
},
{
"award": [
"BRC-1215-20014"
],
"name": "NIHR | NIHR Cambridge Biomedical Research Centre"
},
{
"award": [
"1DP1AT010885"
],
"name": "HHS | National Institutes of Health"
},
{
"DOI": "10.13039/501100023699",
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/501100023699",
"id-type": "DOI"
}
],
"name": "Health Data Research UK"
},
{
"award": [
"U01-CA199277"
],
"name": "HHS | National Institutes of Health"
},
{
"award": [
"P30-CA033572"
],
"name": "HHS | National Institutes of Health"
},
{
"award": [
"P30-CA023100"
],
"name": "HHS | National Institutes of Health"
},
{
"award": [
"UM1-CA164917"
],
"name": "HHS | National Institutes of Health"
},
{
"award": [
"R01-CA077398"
],
"name": "HHS | National Institutes of Health"
},
{
"award": [
"2038509"
],
"name": "National Science Foundation"
},
{
"award": [
"R00RG2503"
],
"name": "UC | University of California, San Diego"
},
{
"DOI": "10.13039/100014894",
"award": [
"3022"
],
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/100014894",
"id-type": "DOI"
}
],
"name": "Emerald Foundation"
},
{
"award": [
"1P30DK120515"
],
"name": "HHS | National Institutes of Health"
},
{
"award": [
"R35GM119850"
],
"name": "HHS | National Institutes of Health"
},
{
"award": [
"UH2AI153029"
],
"name": "HHS | National Institutes of Health"
}
],
"indexed": {
"date-parts": [
[
2025,
4,
10
]
],
"date-time": "2025-04-10T04:19:00Z",
"timestamp": 1744258740512,
"version": "3.40.4"
},
"is-referenced-by-count": 0,
"issue": "4",
"issued": {
"date-parts": [
[
2025,
4,
9
]
]
},
"journal-issue": {
"issue": "4",
"published-print": {
"date-parts": [
[
2025,
4,
9
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
4,
9
]
],
"date-time": "2025-04-09T00:00:00Z",
"timestamp": 1744156800000
}
},
{
"URL": "https://journals.asm.org/non-commercial-tdm-license",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
4,
9
]
],
"date-time": "2025-04-09T00:00:00Z",
"timestamp": 1744156800000
}
}
],
"link": [
{
"URL": "https://journals.asm.org/doi/pdf/10.1128/mbio.04015-24",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://journals.asm.org/doi/pdf/10.1128/mbio.04015-24",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "235",
"original-title": [],
"prefix": "10.1128",
"published": {
"date-parts": [
[
2025,
4,
9
]
]
},
"published-print": {
"date-parts": [
[
2025,
4,
9
]
]
},
"publisher": "American Society for Microbiology",
"reference": [
{
"DOI": "10.1038/s41586-020-2901-9",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_2_2"
},
{
"DOI": "10.1002/jmv.26836",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_3_2"
},
{
"DOI": "10.1016/j.tige.2021.01.006",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_4_2"
},
{
"DOI": "10.1016/j.trsl.2020.08.004",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_5_2"
},
{
"DOI": "10.14740/gr1350",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_6_2"
},
{
"DOI": "10.1007/s00535-021-01778-z",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_7_2"
},
{
"DOI": "10.1038/s41575-021-00416-6",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_8_2"
},
{
"DOI": "10.1016/j.jcmgh.2020.09.017",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_9_2"
},
{
"DOI": "10.1136/gutjnl-2021-324622",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_10_2"
},
{
"DOI": "10.1016/S0140-6736(20)30628-0",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_11_2"
},
{
"DOI": "10.1016/j.medj.2022.04.001",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_12_2"
},
{
"DOI": "10.1007/s00401-023-02612-x",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_13_2"
},
{
"DOI": "10.1038/s41390-022-02266-7",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_14_2"
},
{
"DOI": "10.1136/gutjnl-2021-324280",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_15_2"
},
{
"DOI": "10.1016/j.cell.2020.09.033",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_16_2"
},
{
"DOI": "10.1016/j.sbi.2022.102439",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_17_2"
},
{
"DOI": "10.1136/gutjnl-2020-320953",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_18_2"
},
{
"DOI": "10.1146/annurev.biochem.71.110601.135458",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_19_2"
},
{
"DOI": "10.3390/v11070596",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_20_2"
},
{
"DOI": "10.1128/mSystems.00031-18",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_21_2"
},
{
"DOI": "10.1038/nature18847",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_22_2"
},
{
"DOI": "10.1038/nature24485",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_23_2"
},
{
"DOI": "10.1073/pnas.0803124105",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_24_2"
},
{
"DOI": "10.1007/BF02798420",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_25_2"
},
{
"DOI": "10.1128/JB.00860-16",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_26_2"
},
{
"DOI": "10.1073/pnas.1704367114",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_27_2"
},
{
"DOI": "10.1038/s41467-020-14509-4",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_28_2"
},
{
"DOI": "10.1074/jbc.M114.573303",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_29_2"
},
{
"DOI": "10.1128/aem.41.2.360-365.1981",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_30_2"
},
{
"DOI": "10.1039/C7OB01058C",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_31_2"
},
{
"DOI": "10.1016/j.chom.2015.09.002",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_32_2"
},
{
"DOI": "10.1016/j.crmeth.2021.100040",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_33_2"
},
{
"DOI": "10.1093/ije/dyx239",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_34_2"
},
{
"DOI": "10.1101/2019.12.30.19015842",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_35_2"
},
{
"DOI": "10.1053/j.gastro.2020.04.045",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_36_2"
},
{
"DOI": "10.1038/s41586-020-2521-4",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_37_2"
},
{
"DOI": "10.1038/s41467-019-13443-4",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_38_2"
},
{
"DOI": "10.1038/s41564-019-0564-9",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_39_2"
},
{
"DOI": "10.1016/j.cell.2016.10.043",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_40_2"
},
{
"DOI": "10.1128/mSystems.00261-19",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_41_2"
},
{
"DOI": "10.1186/s40168-021-01083-0",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_42_2"
},
{
"DOI": "10.1074/jbc.M111.228841",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_43_2"
},
{
"DOI": "10.1017/S0007114508143549",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_44_2"
},
{
"DOI": "10.1079/pns2003206",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_45_2"
},
{
"DOI": "10.3389/fnut.2022.934568",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_46_2"
},
{
"DOI": "10.1038/s41575-018-0061-2",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_47_2"
},
{
"DOI": "10.1038/s41579-022-00768-z",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_48_2"
},
{
"DOI": "10.1038/nature11234",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_49_2"
},
{
"DOI": "10.1164/rccm.201501-0128OC",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_50_2"
},
{
"DOI": "10.1164/rccm.202008-3146ED",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_51_2"
},
{
"DOI": "10.1038/nrmicro.2016.142",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_52_2"
},
{
"DOI": "10.1038/nrg1769",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_53_2"
},
{
"DOI": "10.1038/nbt.3703",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_54_2"
},
{
"DOI": "10.1093/nar/gky537",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_55_2"
},
{
"DOI": "10.1128/mBio.01526-14",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_56_2"
},
{
"DOI": "10.1038/nrmicro2746",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_57_2"
},
{
"DOI": "10.1093/nar/gkw1092",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_58_2"
},
{
"DOI": "10.1002/1873-3468.13045",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_59_2"
},
{
"DOI": "10.1371/journal.ppat.1006090",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_60_2"
},
{
"DOI": "10.1093/nar/gkt1178",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_61_2"
},
{
"DOI": "10.1093/nar/gks479",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_62_2"
},
{
"DOI": "10.1093/nar/gky418",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_63_2"
},
{
"DOI": "10.1371/journal.pcbi.1007185",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_64_2"
},
{
"DOI": "10.1371/journal.pcbi.1006867",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_65_2"
},
{
"DOI": "10.7717/peerj.3720",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_66_2"
},
{
"DOI": "10.1038/nmeth.1923",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_67_2"
},
{
"DOI": "10.1128/mSystems.00069-18",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_68_2"
},
{
"DOI": "10.1093/nar/gkl842",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_69_2"
},
{
"author": "Johansson R",
"key": "e_1_3_6_70_2",
"unstructured": "Johansson R. 2018. Numerical python: scientific computing and data science applications with Numpy, SciPy and Matplotlib. Apress.",
"volume-title": "Numerical python: scientific computing and data science applications with Numpy, SciPy and Matplotlib",
"year": "2018"
},
{
"DOI": "10.1093/nar/gkm160",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_71_2"
},
{
"DOI": "10.1016/S0022-2836(05)80360-2",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_72_2"
},
{
"DOI": "10.1186/s13059-019-1834-9",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_73_2"
},
{
"DOI": "10.1093/bioinformatics/btu170",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_74_2"
},
{
"DOI": "10.1186/s13040-014-0034-0",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_75_2"
},
{
"DOI": "10.1093/bioinformatics/btr507",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_76_2"
},
{
"DOI": "10.1089/cmb.2012.0021",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_77_2"
},
{
"DOI": "10.1099/ijsem.0.000760",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_78_2"
},
{
"DOI": "10.1021/ja901571p",
"doi-asserted-by": "publisher",
"key": "e_1_3_6_79_2"
}
],
"reference-count": 78,
"references-count": 78,
"relation": {},
"resource": {
"primary": {
"URL": "https://journals.asm.org/doi/10.1128/mbio.04015-24"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "SARS-CoV-2 infectivity can be modulated through bacterial grooming of the glycocalyx",
"type": "journal-article",
"update-policy": "https://doi.org/10.1128/asmj-crossmark-policy-page",
"volume": "16"
}