In Vitro Efficacy of Antivirals and Monoclonal Antibodies against SARS-CoV-2 Omicron Lineages XBB.1.9.1, XBB.1.9.3, XBB.1.5, XBB.1.16, XBB.2.4, BQ.1.1.45, CH.1.1, and CL.1
et al., Vaccines, doi:10.3390/vaccines11101533, Sep 2023
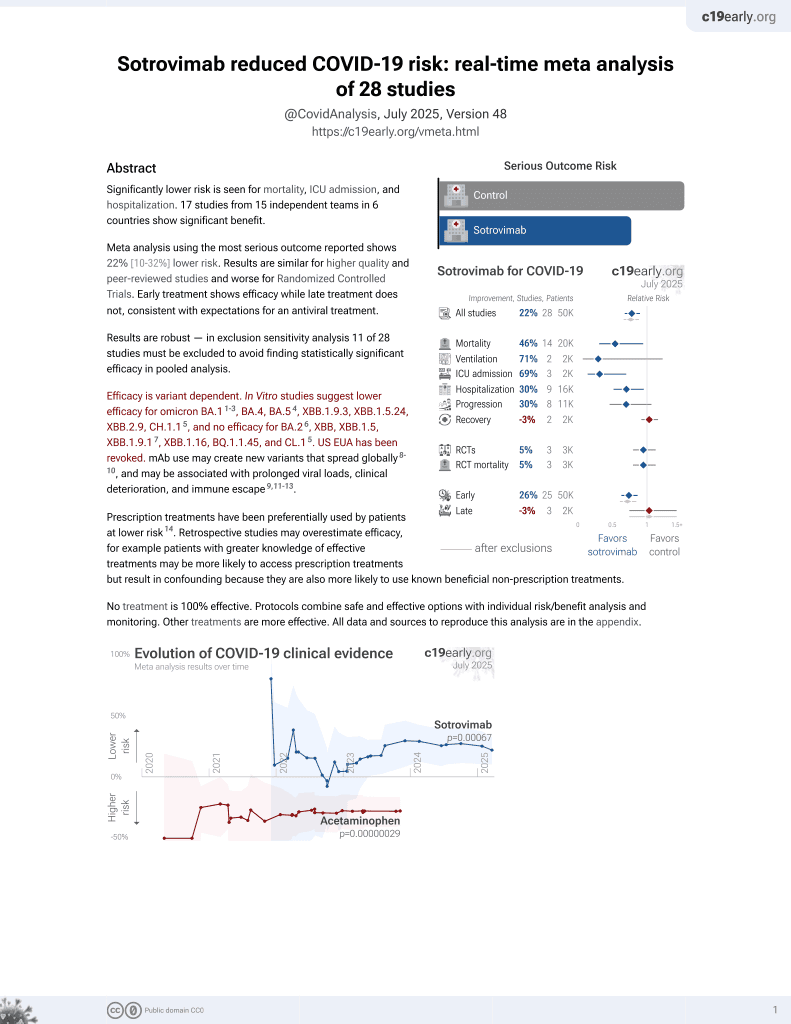
Sotrovimab for COVID-19
45th treatment shown to reduce risk in
August 2022, now with p = 0.00048 from 29 studies, recognized in 42 countries.
Efficacy is variant dependent.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
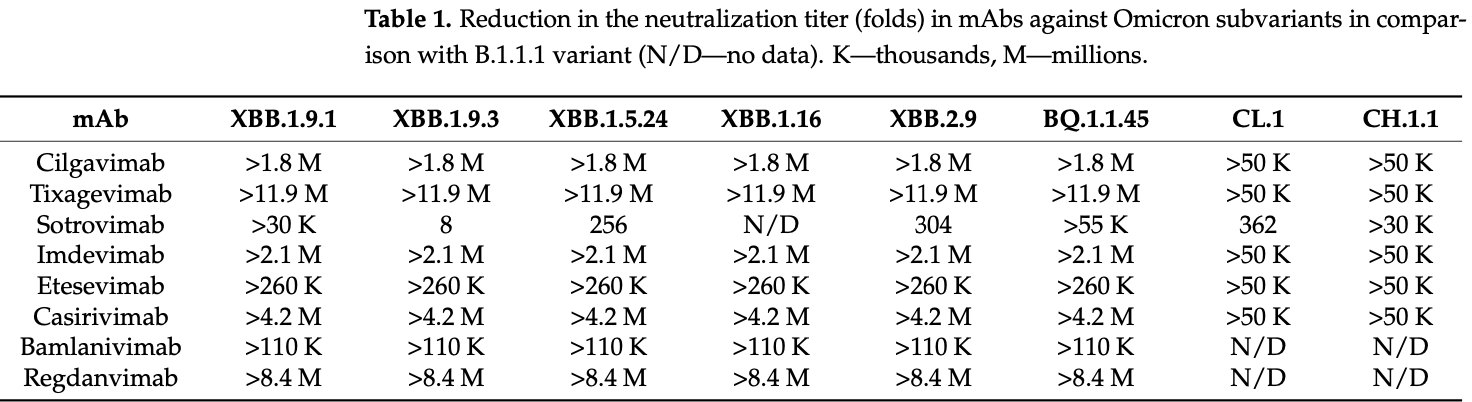
In vitro study showing sharply reduced neutralization of SARS-CoV-2 variants XBB.1.9.1, XBB.1.9.3, XBB.1.5, XBB.1.16, XBB.2.4, BQ.1.1.45, CH.1.1, and CL.1 with monoclonal antibodies cilgavimab, tixagevimab, imdevimab, etsevimab, casirivimab, bamlanivimab, and regdanvimab. Only sotrovimab retained measurable, though reduced neutralizing activity.
Efficacy was retained for remdesivir, molnupiravir, and nirmatrelvir, however IC50 values for remdesivir were ~3 times higher for XBB.1.* variants compared with B.1.1, BQ.1.1.45, CL.1, CH.1.1, and XBB.2.9, and the IC50 for nirmatrelvir for XBB.2.9 was ~2 times higher compared with previous variants.
Efficacy is variant dependent. In Vitro studies predict lower efficacy for BA.11-3, BA.4, BA.54, XBB.1.9.3, XBB.1.5.24, XBB.2.9, CH.1.15, and no efficacy for BA.26, XBB, XBB.1.5, ХВВ.1.9.17, XBB.1.16, BQ.1.1.45, and CL.15. US EUA has been revoked.
Study covers paxlovid, remdesivir, tixagevimab/cilgavimab, casirivimab/imdevimab, bamlanivimab/etesevimab, regdanvimab, and sotrovimab.
1.
Liu et al., Striking Antibody Evasion Manifested by the Omicron Variant of SARS-CoV-2, bioRxiv, doi:10.1101/2021.12.14.472719.
2.
Sheward et al., Variable loss of antibody potency against SARS-CoV-2 B.1.1.529 (Omicron), bioRxiv, doi:10.1101/2021.12.19.473354.
3.
VanBlargan et al., An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by several therapeutic monoclonal antibodies, bioRxiv, doi:10.1101/2021.12.15.472828.
4.
Haars et al., Prevalence of SARS-CoV-2 Omicron Sublineages and Spike Protein Mutations Conferring Resistance against Monoclonal Antibodies in a Swedish Cohort during 2022–2023, Microorganisms, doi:10.3390/microorganisms11102417.
5.
Pochtovyi et al., In Vitro Efficacy of Antivirals and Monoclonal Antibodies against SARS-CoV-2 Omicron Lineages XBB.1.9.1, XBB.1.9.3, XBB.1.5, XBB.1.16, XBB.2.4, BQ.1.1.45, CH.1.1, and CL.1, Vaccines, doi:10.3390/vaccines11101533.
Pochtovyi et al., 28 Sep 2023, peer-reviewed, 16 authors, study period September 2022 - May 2023.
Contact: a.pochtovyy@gamaleya.org (corresponding author), kustovad70@gmail.com, wowaniada@yandex.ru.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
In Vitro Efficacy of Antivirals and Monoclonal Antibodies against SARS-CoV-2 Omicron Lineages XBB.1.9.1, XBB.1.9.3, XBB.1.5, XBB.1.16, XBB.2.4, BQ.1.1.45, CH.1.1, and CL.1
Vaccines, doi:10.3390/vaccines11101533
The spread of COVID-19 continues, expressed by periodic wave-like increases in morbidity and mortality. The reason for the periodic increases in morbidity is the emergence and spread of novel genetic variants of SARS-CoV-2. A decrease in the efficacy of monoclonal antibodies (mAbs) has been reported, especially against Omicron subvariants. There have been reports of a decrease in the efficacy of specific antiviral drugs as a result of mutations in the genes of non-structural proteins. This indicates the urgent need for practical healthcare to constantly monitor pathogen variability and its effect on the efficacy of preventive and therapeutic drugs. As part of this study, we report the results of the continuous monitoring of COVID-19 in Moscow using genetic and virological methods. As a result of this monitoring, we determined the dominant genetic variants and identified the variants that are most widespread, not only in Moscow, but also in other countries. A collection of viruses from more than 500 SARS-CoV-2 isolates has been obtained and characterized. The genetic lines XBB.1.9.1, XBB.1.9.3, XBB.1.5, XBB.1.16, XBB.2.4, BQ.1.1.45, CH.1.1, and CL.1, representing the greatest concern, were identified among the dominant variants. We studied the in vitro efficacy of mAbs Tixagevimab + Cilgavimab (Evusheld), Sotrovimab, Regdanvimab, Casirivimab + Imdevimab (Ronapreve), and Bebtelovimab, as well as the specific antiviral drugs Remdesivir, Molnupiravir, and Nirmatrelvir, against these genetic lines. At the current stage of the COVID-19 pandemic, the use of mAbs developed against early SARS-CoV-2 variants has little prospect. Specific antiviral drugs retain their activity, but further monitoring is needed to assess the risk of their efficacy being reduced and adjust recommendations for their use.
Conflicts of Interest: The authors declare no conflict of interest.
References
Addetia, Piccoli, Case, Park, Beltramello et al., Effector Function and Immune Imprinting of Omicron Variants, Nature, doi:10.1038/s41586-023-06487-6
Akimkin, Popova, Khafizov, Dubodelov, Ugleva et al., COVID-19: Evolution of the Pandemic in Russia. Report II: Dynamics of the Circulation of SARS-CoV-2 Genetic Variants, J. Mikrobiol. Epidemiol. Immunobiol, doi:10.36233/0372-9311-295
Arora, Kempf, Nehlmeier, Schulz, Jäck et al., Omicron Sublineage BQ.1.1 Resistance to Monoclonal Antibodies, Lancet Infect. Dis, doi:10.1016/S1473-3099(22)00733-2
Baden, El Sahly, Essink, Kotloff, Frey et al., Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine, N. Engl. J. Med, doi:10.1056/NEJMoa2035389
Dubey, Choudhary, Kumar, Tomar, Emerging SARS-CoV-2 Variants: Genetic Variability and Clinical Implications, Curr. Microbiol, doi:10.1007/s00284-021-02724-1
Focosi, Maggi, Mcconnell, Casadevall, Very Low Levels of Remdesivir Resistance in SARS-COV-2 Genomes after 18 Months of Massive Usage during the COVID19 Pandemic: A GISAID Exploratory Analysis, Antivir. Res, doi:10.1016/j.antiviral.2022.105247
Focosi, Spezia, Capria, Gueli, Mcconnell et al., Rise of the BQ.1.1.37 SARS-CoV-2 Sublineage, Italy, Diagnostics, doi:10.3390/diagnostics13051000
Fong, Rockett, Agius, Chandra, Johnson-Mckinnon et al., In Silico Detection of Drug Resistance Conferring Mutations in Subpopulations of SARS-CoV-2 Genomes, BMC Infect. Dis, doi:10.1186/s12879-023-08236-6
Garrison, Marth, Haplotype-Based Variant Detection from Short-Read Sequencing, doi:10.48550/arXiv.1207.3907
Godkov, Ogarkova, Gushchin, Kleymenov, Mazunina et al., Revaccination in Age-Risk Groups with Sputnik V Is Immunologically Effective and Depends on the Initial Neutralizing SARS-CoV-2 IgG Antibodies Level, Vaccines, doi:10.3390/vaccines11010090
Goodell, COVID-19 and Finance: Agendas for Future Research, Financ. Res. Lett, doi:10.1016/j.frl.2020.101512
Greasley, Noell, Plotnikova, Ferre, Liu et al., Structural Basis for the in Vitro Efficacy of Nirmatrelvir against SARS-CoV-2 Variants, J. Biol. Chem, doi:10.1016/j.jbc.2022.101972
Gushchin, Pochtovyi, Kustova, Ogarkova, Tarnovetskii et al., Dynamics of SARS-CoV-2 Major Genetic Lineages in Moscow in the Context of Vaccine Prophylaxis, Int. J. Mol. Sci, doi:10.3390/ijms232314670
Iketani, Mohri, Culbertson, Hong, Duan et al., Multiple Pathways for SARS-CoV-2 Resistance to Nirmatrelvir, Nature, doi:10.1038/s41586-022-05514-2
Imai, Ito, Kiso, Yamayoshi, Uraki et al., Efficacy of Antiviral Agents against Omicron Subvariants BQ.1.1 and XBB, N. Engl. J. Med, doi:10.1056/NEJMc2214302
Jochmans, Liu, Donckers, Stoycheva, Boland et al., The Substitutions L50F, E166A, and L167F in SARS-CoV-2 3CLpro Are Selected by a Protease Inhibitor In Vitro and Confer Resistance to Nirmatrelvir, mBio, doi:10.1128/mbio.02815-22
Klink, Safina, Nabieva, Shvyrev, Garushyants et al., The Rise and Spread of the SARS-CoV-2 AY.122 Lineage in Russia, Virus Evol, doi:10.1093/ve/veac017
Kumar, Malla, Dubey, With Corona Outbreak: Nature Started Hitting the Reset Button Globally, Front. Public Health, doi:10.3389/fpubh.2020.569353
Kurhade, Zou, Xia, Liu, Chang et al., Low Neutralization of SARS-CoV-2 Omicron BA.2.75.2, BQ.1.1 and XBB.1 by Parental mRNA Vaccine or a BA.5 Bivalent Booster, Nat. Med, doi:10.1038/s41591-022-02162-x
Lan, Ge, Yu, Shan, Zhou et al., Structure of the SARS-CoV-2 Spike Receptor-Binding Domain Bound to the ACE2 Receptor, Nature, doi:10.1038/s41586-020-2180-5
Lau, Cheng, Leung, Lee, Hachim et al., Real-World COVID-19 Vaccine Effectiveness against the Omicron BA.2 Variant in a SARS-CoV-2 Infection-Naive Population, Nat. Med, doi:10.1038/s41591-023-02219-5
Li, Durbin, Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform, Bioinformatics, doi:10.1093/bioinformatics/btp324
Li, Handsaker, Wysoker, Fennell, Ruan et al., The Sequence Alignment/Map Format and SAMtools, Bioinformatics, doi:10.1093/bioinformatics/btp352
Logunov, Dolzhikova, Shcheblyakov, Tukhvatulin, Zubkova et al., Safety and Efficacy of an rAd26 and rAd5 Vector-Based Heterologous Prime-Boost COVID-19 Vaccine: An Interim Analysis of a Randomised Controlled Phase 3 Trial in Russia, Lancet, doi:10.1016/S0140-6736(21)00234-8
Martin, Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads, EMBnet. J, doi:10.14806/ej.17.1.200
Miteva, Kitanova, Batselova, Lazova, Chervenkov et al., The End or a New Era of Development of SARS-CoV-2 Virus: Genetic Variants Responsible for Severe COVID-19 and Clinical Efficacy of the Most Commonly Used Vaccines in Clinical Practice, Vaccines, doi:10.3390/vaccines11071181
Moeller, Passow, Harki, Aihara, SARS-CoV-2 nsp14 Exoribonuclease Removes the Natural Antiviral 3 -Deoxy-3 ,4 -Didehydro-Cytidine Nucleotide from RNA, Viruses, doi:10.3390/v14081790
Pinto, Park, Beltramello, Walls, Tortorici et al., Cross-Neutralization of SARS-CoV-2 by a Human Monoclonal SARS-CoV Antibody, Nature, doi:10.1038/s41586-020-2349-y
Planas, Bruel, Staropoli, Guivel-Benhassine, Porrot et al., Resistance of Omicron Subvariants BA.2.75.2, BA.4.6, and BQ.1.1 to Neutralizing Antibodies, Nat. Commun, doi:10.1038/s41467-023-36561-6
Polack, Thomas, Kitchin, Absalon, Gurtman et al., Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine, N. Engl. J. Med, doi:10.1056/NEJMoa2034577
Quinlan, Hall, Bedtools, A Flexible Suite of Utilities for Comparing Genomic Features, Bioinformatics, doi:10.1093/bioinformatics/btq033
Reed, Muench, A Simple Method of estimating fifty per cent endpoints, Am. J. Epidemiol, doi:10.1093/oxfordjournals.aje.a118408
Rognes, Flouri, Nichols, Quince, Mahé et al., A Versatile Open Source Tool for Metagenomics, PeerJ, doi:10.7717/peerj.2584
Shah, Woo, Omicron: A Heavily Mutated SARS-CoV-2 Variant Exhibits Stronger Binding to ACE2 and Potently Escapes Approved COVID-19 Therapeutic Antibodies, Front. Immunol, doi:10.3389/fimmu.2021.830527
Siniavin, Streltsova, Nikiforova, Kudryavtsev, Grinkina et al., Snake Venom Phospholipase A2s Exhibit Strong Virucidal Activity against SARS-CoV-2 and Inhibit the Viral Spike Glycoprotein Interaction with ACE2, Cell. Mol. Life Sci, doi:10.1007/s00018-021-03985-6
Stevens, Pruijssers, Lee, Gordon, Tchesnokov et al., Mutations in the SARS-CoV-2 RNA-Dependent RNA Polymerase Confer Resistance to Remdesivir by Distinct Mechanisms, Sci. Transl. Med, doi:10.1126/scitranslmed.abo0718
Uraki, Ito, Kiso, Yamayoshi, Iwatsuki-Horimoto et al., Antiviral and Bivalent Vaccine Efficacy against an Omicron XBB.1.5 Isolate, Lancet Infect. Dis, doi:10.1016/S1473-3099(23)00070-1
Voysey, Clemens, Madhi, Weckx, Folegatti et al., Safety and Efficacy of the ChAdOx1 nCoV-19 Vaccine (AZD1222) against SARS-CoV-2: An Interim Analysis of Four Randomised Controlled Trials in Brazil, South Africa, and the UK, Lancet, doi:10.1016/S0140-6736(20)32661-1
Wang, Guo, Iketani, Nair, Li et al., Antibody Evasion by SARS-CoV-2 Omicron Subvariants BA, BA.4 and BA.5, doi:10.1038/s41586-022-05053-w
Xie, Edwards, Adam, Leung, Tsang et al., Resurgence of Omicron BA.2 in SARS-CoV-2 Infection-Naive Hong Kong, Nat. Commun, doi:10.1038/s41467-023-38201-5
Yamasoba, Uriu, Plianchaisuk, Kosugi, Pan et al., Virological Characteristics of the SARS-CoV-2 Omicron XBB.1.16 Variant, Lancet Infect. Dis, doi:10.1016/S1473-3099(23)00278-5
Yazawa, Yamazaki, Saga, Itamochi, Inasaki et al., Evaluation of SARS-CoV-2 Isolation in Cell Culture from Nasal/nasopharyngeal Swabs or Saliva Specimens of Patients with COVID-19, Sci. Rep, doi:10.1038/s41598-023-35915-w
DOI record:
{
"DOI": "10.3390/vaccines11101533",
"ISSN": [
"2076-393X"
],
"URL": "http://dx.doi.org/10.3390/vaccines11101533",
"abstract": "<jats:p>The spread of COVID-19 continues, expressed by periodic wave-like increases in morbidity and mortality. The reason for the periodic increases in morbidity is the emergence and spread of novel genetic variants of SARS-CoV-2. A decrease in the efficacy of monoclonal antibodies (mAbs) has been reported, especially against Omicron subvariants. There have been reports of a decrease in the efficacy of specific antiviral drugs as a result of mutations in the genes of non-structural proteins. This indicates the urgent need for practical healthcare to constantly monitor pathogen variability and its effect on the efficacy of preventive and therapeutic drugs. As part of this study, we report the results of the continuous monitoring of COVID-19 in Moscow using genetic and virological methods. As a result of this monitoring, we determined the dominant genetic variants and identified the variants that are most widespread, not only in Moscow, but also in other countries. A collection of viruses from more than 500 SARS-CoV-2 isolates has been obtained and characterized. The genetic lines XBB.1.9.1, XBB.1.9.3, XBB.1.5, XBB.1.16, XBB.2.4, BQ.1.1.45, CH.1.1, and CL.1, representing the greatest concern, were identified among the dominant variants. We studied the in vitro efficacy of mAbs Tixagevimab + Cilgavimab (Evusheld), Sotrovimab, Regdanvimab, Casirivimab + Imdevimab (Ronapreve), and Bebtelovimab, as well as the specific antiviral drugs Remdesivir, Molnupiravir, and Nirmatrelvir, against these genetic lines. At the current stage of the COVID-19 pandemic, the use of mAbs developed against early SARS-CoV-2 variants has little prospect. Specific antiviral drugs retain their activity, but further monitoring is needed to assess the risk of their efficacy being reduced and adjust recommendations for their use.</jats:p>",
"alternative-id": [
"vaccines11101533"
],
"author": [
{
"ORCID": "http://orcid.org/0000-0003-1107-9351",
"affiliation": [
{
"name": "Federal State Budget Institution “National Research Centre for Epidemiology and Microbiology Named after Honorary Academician N. F. Gamaleya” of the Ministry of Health of the Russian Federation, 123098 Moscow, Russia"
},
{
"name": "Department of Virology, Biological Faculty, Lomonosov Moscow State University, 119991 Moscow, Russia"
}
],
"authenticated-orcid": false,
"family": "Pochtovyi",
"given": "Andrei A.",
"sequence": "first"
},
{
"ORCID": "http://orcid.org/0000-0002-8382-275X",
"affiliation": [
{
"name": "Federal State Budget Institution “National Research Centre for Epidemiology and Microbiology Named after Honorary Academician N. F. Gamaleya” of the Ministry of Health of the Russian Federation, 123098 Moscow, Russia"
},
{
"name": "Department of Virology, Biological Faculty, Lomonosov Moscow State University, 119991 Moscow, Russia"
}
],
"authenticated-orcid": false,
"family": "Kustova",
"given": "Daria D.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-7576-2059",
"affiliation": [
{
"name": "Federal State Budget Institution “National Research Centre for Epidemiology and Microbiology Named after Honorary Academician N. F. Gamaleya” of the Ministry of Health of the Russian Federation, 123098 Moscow, Russia"
},
{
"name": "Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, 117997 Moscow, Russia"
}
],
"authenticated-orcid": false,
"family": "Siniavin",
"given": "Andrei E.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Federal State Budget Institution “National Research Centre for Epidemiology and Microbiology Named after Honorary Academician N. F. Gamaleya” of the Ministry of Health of the Russian Federation, 123098 Moscow, Russia"
}
],
"family": "Dolzhikova",
"given": "Inna V.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Federal State Budget Institution “National Research Centre for Epidemiology and Microbiology Named after Honorary Academician N. F. Gamaleya” of the Ministry of Health of the Russian Federation, 123098 Moscow, Russia"
}
],
"family": "Shidlovskaya",
"given": "Elena V.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Moscow Healthcare Department, 127006 Moscow, Russia"
}
],
"family": "Shpakova",
"given": "Olga G.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Federal State Budget Institution “National Research Centre for Epidemiology and Microbiology Named after Honorary Academician N. F. Gamaleya” of the Ministry of Health of the Russian Federation, 123098 Moscow, Russia"
}
],
"family": "Vasilchenko",
"given": "Lyudmila A.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Federal State Budget Institution “National Research Centre for Epidemiology and Microbiology Named after Honorary Academician N. F. Gamaleya” of the Ministry of Health of the Russian Federation, 123098 Moscow, Russia"
}
],
"family": "Glavatskaya",
"given": "Arina A.",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-7399-7628",
"affiliation": [
{
"name": "Federal State Budget Institution “National Research Centre for Epidemiology and Microbiology Named after Honorary Academician N. F. Gamaleya” of the Ministry of Health of the Russian Federation, 123098 Moscow, Russia"
}
],
"authenticated-orcid": false,
"family": "Kuznetsova",
"given": "Nadezhda A.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Federal State Budget Institution “National Research Centre for Epidemiology and Microbiology Named after Honorary Academician N. F. Gamaleya” of the Ministry of Health of the Russian Federation, 123098 Moscow, Russia"
}
],
"family": "Iliukhina",
"given": "Anna A.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Federal State Budget Institution “National Research Centre for Epidemiology and Microbiology Named after Honorary Academician N. F. Gamaleya” of the Ministry of Health of the Russian Federation, 123098 Moscow, Russia"
}
],
"family": "Shelkov",
"given": "Artem Y.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Federal State Budget Institution “National Research Centre for Epidemiology and Microbiology Named after Honorary Academician N. F. Gamaleya” of the Ministry of Health of the Russian Federation, 123098 Moscow, Russia"
}
],
"family": "Grinkevich",
"given": "Olesia M.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Moscow Healthcare Department, 127006 Moscow, Russia"
}
],
"family": "Komarov",
"given": "Andrei G.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Federal State Budget Institution “National Research Centre for Epidemiology and Microbiology Named after Honorary Academician N. F. Gamaleya” of the Ministry of Health of the Russian Federation, 123098 Moscow, Russia"
}
],
"family": "Logunov",
"given": "Denis Y.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Federal State Budget Institution “National Research Centre for Epidemiology and Microbiology Named after Honorary Academician N. F. Gamaleya” of the Ministry of Health of the Russian Federation, 123098 Moscow, Russia"
},
{
"name": "Department of Virology, Biological Faculty, Lomonosov Moscow State University, 119991 Moscow, Russia"
}
],
"family": "Gushchin",
"given": "Vladimir A.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Federal State Budget Institution “National Research Centre for Epidemiology and Microbiology Named after Honorary Academician N. F. Gamaleya” of the Ministry of Health of the Russian Federation, 123098 Moscow, Russia"
},
{
"name": "Department of Infectiology and Virology, Federal State Autonomous Educational Institution of Higher Education I.M. Sechenov, First Moscow State Medical University of the Ministry of Health of the Russian Federation (Sechenov University), 119435 Moscow, Russia"
}
],
"family": "Gintsburg",
"given": "Alexander L.",
"sequence": "additional"
}
],
"container-title": "Vaccines",
"container-title-short": "Vaccines",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2023,
9,
28
]
],
"date-time": "2023-09-28T05:42:12Z",
"timestamp": 1695879732000
},
"deposited": {
"date-parts": [
[
2023,
9,
28
]
],
"date-time": "2023-09-28T07:15:20Z",
"timestamp": 1695885320000
},
"funder": [
{
"DOI": "10.13039/501100017638",
"award": [
"123031400022-0"
],
"doi-asserted-by": "publisher",
"name": "Ministry of Health of the Russian Federation"
}
],
"indexed": {
"date-parts": [
[
2023,
9,
29
]
],
"date-time": "2023-09-29T08:46:55Z",
"timestamp": 1695977215482
},
"is-referenced-by-count": 0,
"issue": "10",
"issued": {
"date-parts": [
[
2023,
9,
28
]
]
},
"journal-issue": {
"issue": "10",
"published-online": {
"date-parts": [
[
2023,
10
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2023,
9,
28
]
],
"date-time": "2023-09-28T00:00:00Z",
"timestamp": 1695859200000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/2076-393X/11/10/1533/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "1533",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2023,
9,
28
]
]
},
"published-online": {
"date-parts": [
[
2023,
9,
28
]
]
},
"publisher": "MDPI AG",
"reference": [
{
"key": "ref_1",
"unstructured": "(2023, August 01). WHO Coronavirus (COVID-19). Available online: https://covid19.who.int/."
},
{
"DOI": "10.1056/NEJMoa2034577",
"article-title": "Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine",
"author": "Polack",
"doi-asserted-by": "crossref",
"first-page": "2603",
"journal-title": "N. Engl. J. Med.",
"key": "ref_2",
"volume": "383",
"year": "2020"
},
{
"DOI": "10.1056/NEJMoa2035389",
"article-title": "Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine",
"author": "Baden",
"doi-asserted-by": "crossref",
"first-page": "403",
"journal-title": "N. Engl. J. Med.",
"key": "ref_3",
"volume": "384",
"year": "2021"
},
{
"DOI": "10.1016/S0140-6736(21)00234-8",
"article-title": "Safety and Efficacy of an rAd26 and rAd5 Vector-Based Heterologous Prime-Boost COVID-19 Vaccine: An Interim Analysis of a Randomised Controlled Phase 3 Trial in Russia",
"author": "Logunov",
"doi-asserted-by": "crossref",
"first-page": "671",
"journal-title": "Lancet",
"key": "ref_4",
"volume": "397",
"year": "2021"
},
{
"DOI": "10.1016/S0140-6736(20)32661-1",
"article-title": "Safety and Efficacy of the ChAdOx1 nCoV-19 Vaccine (AZD1222) against SARS-CoV-2: An Interim Analysis of Four Randomised Controlled Trials in Brazil, South Africa, and the UK",
"author": "Voysey",
"doi-asserted-by": "crossref",
"first-page": "99",
"journal-title": "Lancet",
"key": "ref_5",
"volume": "397",
"year": "2021"
},
{
"key": "ref_6",
"unstructured": "European Medicines Agency (2023, August 01). COVID-19 Medicines. Available online: https://www.ema.europa.eu/en/human-regulatory/overview/public-health-threats/coronavirus-disease-covid-19/covid-19-medicines."
},
{
"key": "ref_7",
"unstructured": "(2023, August 01). Emergency Use Authorization (EUA) for the Treatment of COVID-19. Available online: https://www.covid19.lilly.com/bebtelovimab."
},
{
"DOI": "10.1007/s00284-021-02724-1",
"doi-asserted-by": "crossref",
"key": "ref_8",
"unstructured": "Dubey, A., Choudhary, S., Kumar, P., and Tomar, S. (2022). Emerging SARS-CoV-2 Variants: Genetic Variability and Clinical Implications. Curr. Microbiol., 79."
},
{
"DOI": "10.1038/s41591-023-02219-5",
"article-title": "Real-World COVID-19 Vaccine Effectiveness against the Omicron BA.2 Variant in a SARS-CoV-2 Infection-Naive Population",
"author": "Lau",
"doi-asserted-by": "crossref",
"first-page": "348",
"journal-title": "Nat. Med.",
"key": "ref_9",
"volume": "29",
"year": "2023"
},
{
"DOI": "10.3390/vaccines11010090",
"doi-asserted-by": "crossref",
"key": "ref_10",
"unstructured": "Godkov, M.A., Ogarkova, D.A., Gushchin, V.A., Kleymenov, D.A., Mazunina, E.P., Bykonia, E.N., Pochtovyi, A.A., Shustov, V.V., Shcheblyakov, D.V., and Komarov, A.G. (2022). Revaccination in Age-Risk Groups with Sputnik V Is Immunologically Effective and Depends on the Initial Neutralizing SARS-CoV-2 IgG Antibodies Level. Vaccines, 11."
},
{
"DOI": "10.3390/vaccines11071181",
"doi-asserted-by": "crossref",
"key": "ref_11",
"unstructured": "Miteva, D., Kitanova, M., Batselova, H., Lazova, S., Chervenkov, L., Peshevska-Sekulovska, M., Sekulovski, M., Gulinac, M., Vasilev, G.V., and Tomov, L. (2023). The End or a New Era of Development of SARS-CoV-2 Virus: Genetic Variants Responsible for Severe COVID-19 and Clinical Efficacy of the Most Commonly Used Vaccines in Clinical Practice. Vaccines, 11."
},
{
"DOI": "10.1038/s41591-022-02162-x",
"article-title": "Low Neutralization of SARS-CoV-2 Omicron BA.2.75.2, BQ.1.1 and XBB.1 by Parental mRNA Vaccine or a BA.5 Bivalent Booster",
"author": "Kurhade",
"doi-asserted-by": "crossref",
"first-page": "344",
"journal-title": "Nat. Med.",
"key": "ref_12",
"volume": "29",
"year": "2023"
},
{
"DOI": "10.1038/s41586-023-06487-6",
"article-title": "Neutralization, Effector Function and Immune Imprinting of Omicron Variants",
"author": "Addetia",
"doi-asserted-by": "crossref",
"first-page": "592",
"journal-title": "Nature",
"key": "ref_13",
"volume": "621",
"year": "2023"
},
{
"DOI": "10.1038/s41598-023-35915-w",
"article-title": "Evaluation of SARS-CoV-2 Isolation in Cell Culture from Nasal/nasopharyngeal Swabs or Saliva Specimens of Patients with COVID-19",
"author": "Yazawa",
"doi-asserted-by": "crossref",
"first-page": "8893",
"journal-title": "Sci. Rep.",
"key": "ref_14",
"volume": "13",
"year": "2023"
},
{
"DOI": "10.1093/oxfordjournals.aje.a118408",
"article-title": "A Simple Method of estimating fifty per cent endpoints",
"author": "Reed",
"doi-asserted-by": "crossref",
"first-page": "493",
"journal-title": "Am. J. Epidemiol.",
"key": "ref_15",
"volume": "27",
"year": "1938"
},
{
"DOI": "10.3390/ijms232314670",
"doi-asserted-by": "crossref",
"key": "ref_16",
"unstructured": "Gushchin, V.A., Pochtovyi, A.A., Kustova, D.D., Ogarkova, D.A., Tarnovetskii, I.Y., Belyaeva, E.D., Divisenko, E.V., Vasilchenko, L.A., Shidlovskaya, E.V., and Kuznetsova, N.A. (2022). Dynamics of SARS-CoV-2 Major Genetic Lineages in Moscow in the Context of Vaccine Prophylaxis. Int. J. Mol. Sci., 23."
},
{
"DOI": "10.1093/ve/veac017",
"article-title": "The Rise and Spread of the SARS-CoV-2 AY.122 Lineage in Russia",
"author": "Klink",
"doi-asserted-by": "crossref",
"first-page": "veac017",
"journal-title": "Virus Evol.",
"key": "ref_17",
"volume": "8",
"year": "2022"
},
{
"DOI": "10.14806/ej.17.1.200",
"article-title": "Cutadapt Removes Adapter Sequences from High- Throughput Sequencing Reads",
"author": "Martin",
"doi-asserted-by": "crossref",
"first-page": "10",
"journal-title": "EMBnet. J.",
"key": "ref_18",
"volume": "17",
"year": "2011"
},
{
"DOI": "10.7717/peerj.2584",
"article-title": "VSEARCH: A Versatile Open Source Tool for Metagenomics",
"author": "Rognes",
"doi-asserted-by": "crossref",
"first-page": "e2584",
"journal-title": "PeerJ",
"key": "ref_19",
"volume": "2016",
"year": "2016"
},
{
"DOI": "10.1093/bioinformatics/btp324",
"article-title": "Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "1754",
"journal-title": "Bioinformatics",
"key": "ref_20",
"volume": "25",
"year": "2009"
},
{
"key": "ref_21",
"unstructured": "Garrison, E., and Marth, G. (2012). Haplotype-Based Variant Detection from Short-Read Sequencing. arXiv."
},
{
"DOI": "10.1093/bioinformatics/btp352",
"article-title": "The Sequence Alignment/Map Format and SAMtools",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "2078",
"journal-title": "Bioinformatics",
"key": "ref_22",
"volume": "25",
"year": "2009"
},
{
"DOI": "10.1093/bioinformatics/btq033",
"article-title": "BEDTools: A Flexible Suite of Utilities for Comparing Genomic Features",
"author": "Quinlan",
"doi-asserted-by": "crossref",
"first-page": "841",
"journal-title": "Bioinformatics",
"key": "ref_23",
"volume": "26",
"year": "2010"
},
{
"key": "ref_24",
"unstructured": "(2023, August 01). ARTIC: A Bioinformatics Pipeline for Working with Virus Sequencing Data Sequenced with Nanopore. Available online: https://artic.network/."
},
{
"key": "ref_25",
"unstructured": "Official Website U.S. Food & Drug (2023, August 01). Coronavirus (COVID-19). Drugs, Available online: https://www.fda.gov/drugs/emergency-preparedness-drugs/coronavirus-covid-19-drugs."
},
{
"key": "ref_26",
"unstructured": "(2023, August 01). Temporary Guidelines «Prevention, Diagnosis and Treatment of New Coronavirus Infection (COVID-19)», Version 17, Available online: https://static-0.minzdrav.gov.ru/system/attachments/attaches/000/061/252/original/%D0%92%D0%9C%D0%A0_COVID-19_V17.pdf."
},
{
"DOI": "10.1007/s00018-021-03985-6",
"article-title": "Snake Venom Phospholipase A2s Exhibit Strong Virucidal Activity against SARS-CoV-2 and Inhibit the Viral Spike Glycoprotein Interaction with ACE2",
"author": "Siniavin",
"doi-asserted-by": "crossref",
"first-page": "7777",
"journal-title": "Cell. Mol. Life Sci.",
"key": "ref_27",
"volume": "78",
"year": "2021"
},
{
"DOI": "10.1056/NEJMc2214302",
"article-title": "Efficacy of Antiviral Agents against Omicron Subvariants BQ.1.1 and XBB",
"author": "Imai",
"doi-asserted-by": "crossref",
"first-page": "89",
"journal-title": "N. Engl. J. Med.",
"key": "ref_28",
"volume": "388",
"year": "2023"
},
{
"DOI": "10.1016/S1473-3099(22)00733-2",
"article-title": "Omicron Sublineage BQ.1.1 Resistance to Monoclonal Antibodies",
"author": "Arora",
"doi-asserted-by": "crossref",
"first-page": "22",
"journal-title": "Lancet Infect. Dis.",
"key": "ref_29",
"volume": "23",
"year": "2023"
},
{
"DOI": "10.3389/fpubh.2020.569353",
"article-title": "With Corona Outbreak: Nature Started Hitting the Reset Button Globally",
"author": "Kumar",
"doi-asserted-by": "crossref",
"first-page": "569353",
"journal-title": "Front. Public Health",
"key": "ref_30",
"volume": "8",
"year": "2020"
},
{
"DOI": "10.1016/j.frl.2020.101512",
"article-title": "COVID-19 and Finance: Agendas for Future Research",
"author": "Goodell",
"doi-asserted-by": "crossref",
"first-page": "101512",
"journal-title": "Financ. Res. Lett.",
"key": "ref_31",
"volume": "35",
"year": "2020"
},
{
"DOI": "10.36233/0372-9311-295",
"article-title": "COVID-19: Evolution of the Pandemic in Russia. Report II: Dynamics of the Circulation of SARS-CoV-2 Genetic Variants",
"author": "Akimkin",
"doi-asserted-by": "crossref",
"first-page": "381",
"journal-title": "J. Mikrobiol. Epidemiol. Immunobiol.",
"key": "ref_32",
"volume": "99",
"year": "2022"
},
{
"DOI": "10.1038/s41467-023-38201-5",
"article-title": "Resurgence of Omicron BA.2 in SARS-CoV-2 Infection-Naive Hong Kong",
"author": "Xie",
"doi-asserted-by": "crossref",
"first-page": "2422",
"journal-title": "Nat. Commun.",
"key": "ref_33",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.1016/S1473-3099(23)00278-5",
"article-title": "Virological Characteristics of the SARS-CoV-2 Omicron XBB.1.16 Variant",
"author": "Yamasoba",
"doi-asserted-by": "crossref",
"first-page": "655",
"journal-title": "Lancet Infect. Dis.",
"key": "ref_34",
"volume": "23",
"year": "2023"
},
{
"DOI": "10.1038/s41467-023-36561-6",
"article-title": "Resistance of Omicron Subvariants BA.2.75.2, BA.4.6, and BQ.1.1 to Neutralizing Antibodies",
"author": "Planas",
"doi-asserted-by": "crossref",
"first-page": "824",
"journal-title": "Nat. Commun.",
"key": "ref_35",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.3390/diagnostics13051000",
"doi-asserted-by": "crossref",
"key": "ref_36",
"unstructured": "Focosi, D., Spezia, P.G., Capria, A.-L., Gueli, F., McConnell, S., Novazzi, F., and Pistello, M. (2023). Rise of the BQ.1.1.37 SARS-CoV-2 Sublineage, Italy. Diagnostics, 13."
},
{
"DOI": "10.1038/s41586-022-05053-w",
"article-title": "Antibody Evasion by SARS-CoV-2 Omicron Subvariants BA.2.12.1, BA.4 and BA.5",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "603",
"journal-title": "Nature",
"key": "ref_37",
"volume": "608",
"year": "2022"
},
{
"DOI": "10.3389/fimmu.2021.830527",
"article-title": "Omicron: A Heavily Mutated SARS-CoV-2 Variant Exhibits Stronger Binding to ACE2 and Potently Escapes Approved COVID-19 Therapeutic Antibodies",
"author": "Shah",
"doi-asserted-by": "crossref",
"first-page": "830527",
"journal-title": "Front. Immunol.",
"key": "ref_38",
"volume": "12",
"year": "2022"
},
{
"DOI": "10.1038/s41586-020-2180-5",
"article-title": "Structure of the SARS-CoV-2 Spike Receptor-Binding Domain Bound to the ACE2 Receptor",
"author": "Lan",
"doi-asserted-by": "crossref",
"first-page": "215",
"journal-title": "Nature",
"key": "ref_39",
"volume": "581",
"year": "2020"
},
{
"DOI": "10.1038/s41586-020-2349-y",
"article-title": "Cross-Neutralization of SARS-CoV-2 by a Human Monoclonal SARS-CoV Antibody",
"author": "Pinto",
"doi-asserted-by": "crossref",
"first-page": "290",
"journal-title": "Nature",
"key": "ref_40",
"volume": "583",
"year": "2020"
},
{
"DOI": "10.1038/s41586-022-05514-2",
"article-title": "Multiple Pathways for SARS-CoV-2 Resistance to Nirmatrelvir",
"author": "Iketani",
"doi-asserted-by": "crossref",
"first-page": "558",
"journal-title": "Nature",
"key": "ref_41",
"volume": "613",
"year": "2023"
},
{
"DOI": "10.1128/mbio.02815-22",
"doi-asserted-by": "crossref",
"key": "ref_42",
"unstructured": "Jochmans, D., Liu, C., Donckers, K., Stoycheva, A., Boland, S., Stevens, S.K., De Vita, C., Vanmechelen, B., Maes, P., and Trüeb, B. (2023). The Substitutions L50F, E166A, and L167F in SARS-CoV-2 3CLpro Are Selected by a Protease Inhibitor In Vitro and Confer Resistance to Nirmatrelvir. mBio, 14."
},
{
"DOI": "10.1101/2022.01.17.476556",
"doi-asserted-by": "crossref",
"key": "ref_43",
"unstructured": "Greasley, S.E., Noell, S., Plotnikova, O., Ferre, R., Liu, W., Bolanos, B., Fennell, K., Nicki, J., Craig, T., and Zhu, Y. (2022). Structural Basis for the in Vitro Efficacy of Nirmatrelvir against SARS-CoV-2 Variants. J. Biol. Chem., 298."
},
{
"key": "ref_44",
"unstructured": "Official Website Chemical & Engineering News (2023, August 01). Pfizer Unveils Its Oral SARS-CoV-2 Inhibitor. Available online: https://cen.acs.org/acs-news/acs-meeting-news/Pfizer-unveils-oral-SARS-CoV/99/i13."
},
{
"DOI": "10.3390/v14081790",
"doi-asserted-by": "crossref",
"key": "ref_45",
"unstructured": "Moeller, N.H., Passow, K.T., Harki, D.A., and Aihara, H. (2022). SARS-CoV-2 nsp14 Exoribonuclease Removes the Natural Antiviral 3′-Deoxy-3′,4′-Didehydro-Cytidine Nucleotide from RNA. Viruses, 14."
},
{
"DOI": "10.1016/j.antiviral.2022.105247",
"article-title": "Very Low Levels of Remdesivir Resistance in SARS-COV-2 Genomes after 18 Months of Massive Usage during the COVID19 Pandemic: A GISAID Exploratory Analysis",
"author": "Focosi",
"doi-asserted-by": "crossref",
"first-page": "105247",
"journal-title": "Antivir. Res.",
"key": "ref_46",
"volume": "198",
"year": "2022"
},
{
"DOI": "10.1186/s12879-023-08236-6",
"doi-asserted-by": "crossref",
"key": "ref_47",
"unstructured": "Fong, W., Rockett, R.J., Agius, J.E., Chandra, S., Johnson-Mckinnon, J., Sim, E., Lam, C., Arnott, A., Gall, M., and Draper, J. (2023). SABRes: In Silico Detection of Drug Resistance Conferring Mutations in Subpopulations of SARS-CoV-2 Genomes. BMC Infect. Dis., 23."
},
{
"DOI": "10.1126/scitranslmed.abo0718",
"article-title": "Mutations in the SARS-CoV-2 RNA-Dependent RNA Polymerase Confer Resistance to Remdesivir by Distinct Mechanisms",
"author": "Stevens",
"doi-asserted-by": "crossref",
"first-page": "eabo0718",
"journal-title": "Sci. Transl. Med.",
"key": "ref_48",
"volume": "14",
"year": "2022"
},
{
"DOI": "10.1016/S1473-3099(23)00070-1",
"article-title": "Antiviral and Bivalent Vaccine Efficacy against an Omicron XBB.1.5 Isolate",
"author": "Uraki",
"doi-asserted-by": "crossref",
"first-page": "402",
"journal-title": "Lancet Infect. Dis.",
"key": "ref_49",
"volume": "23",
"year": "2023"
}
],
"reference-count": 49,
"references-count": 49,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.mdpi.com/2076-393X/11/10/1533"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"Pharmacology (medical)",
"Infectious Diseases",
"Drug Discovery",
"Pharmacology",
"Immunology"
],
"subtitle": [],
"title": "In Vitro Efficacy of Antivirals and Monoclonal Antibodies against SARS-CoV-2 Omicron Lineages XBB.1.9.1, XBB.1.9.3, XBB.1.5, XBB.1.16, XBB.2.4, BQ.1.1.45, CH.1.1, and CL.1",
"type": "journal-article",
"volume": "11"
}
pochtovyi