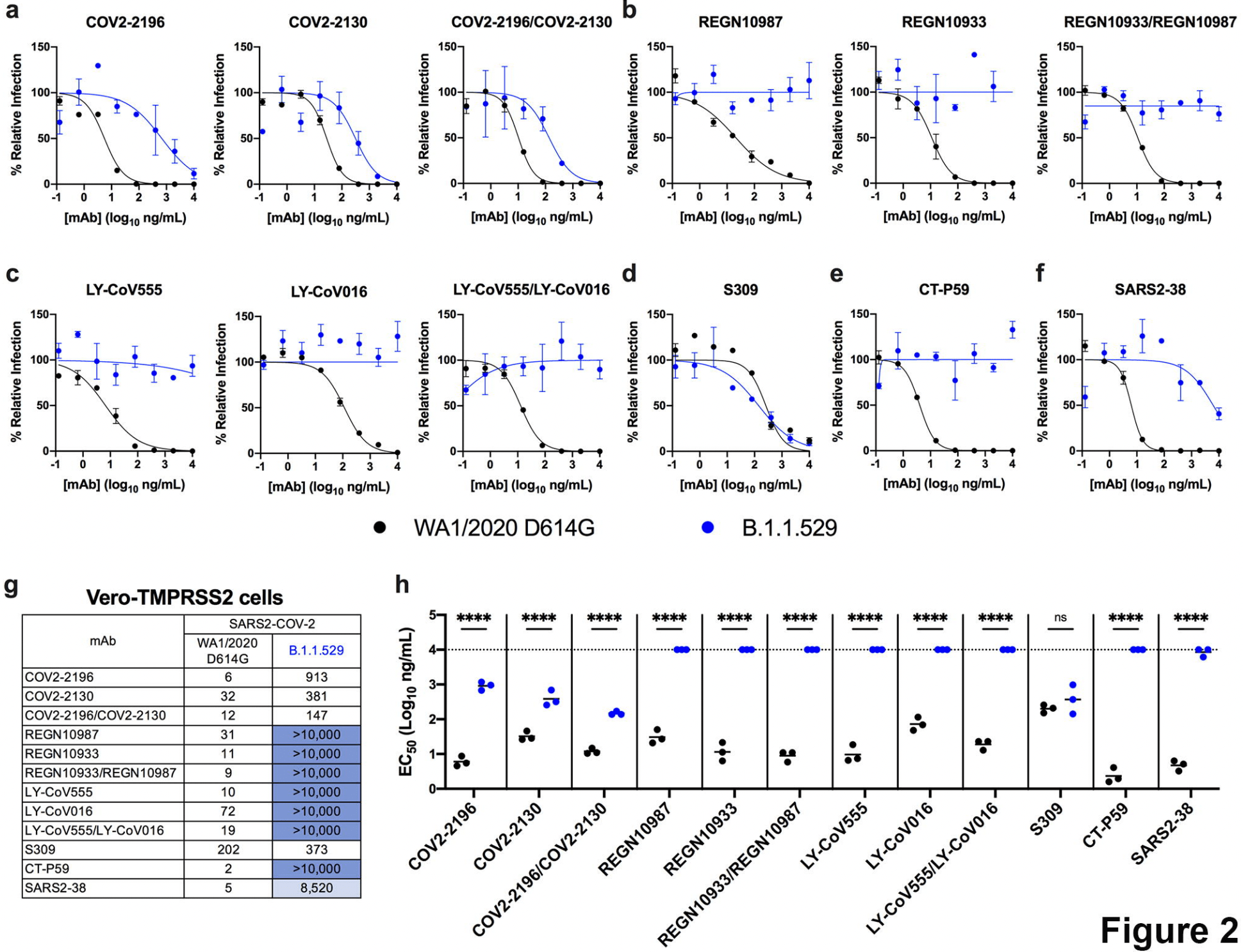
An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by several therapeutic monoclonal antibodies
et al., bioRxiv, doi:10.1101/2021.12.15.472828, Dec 2021
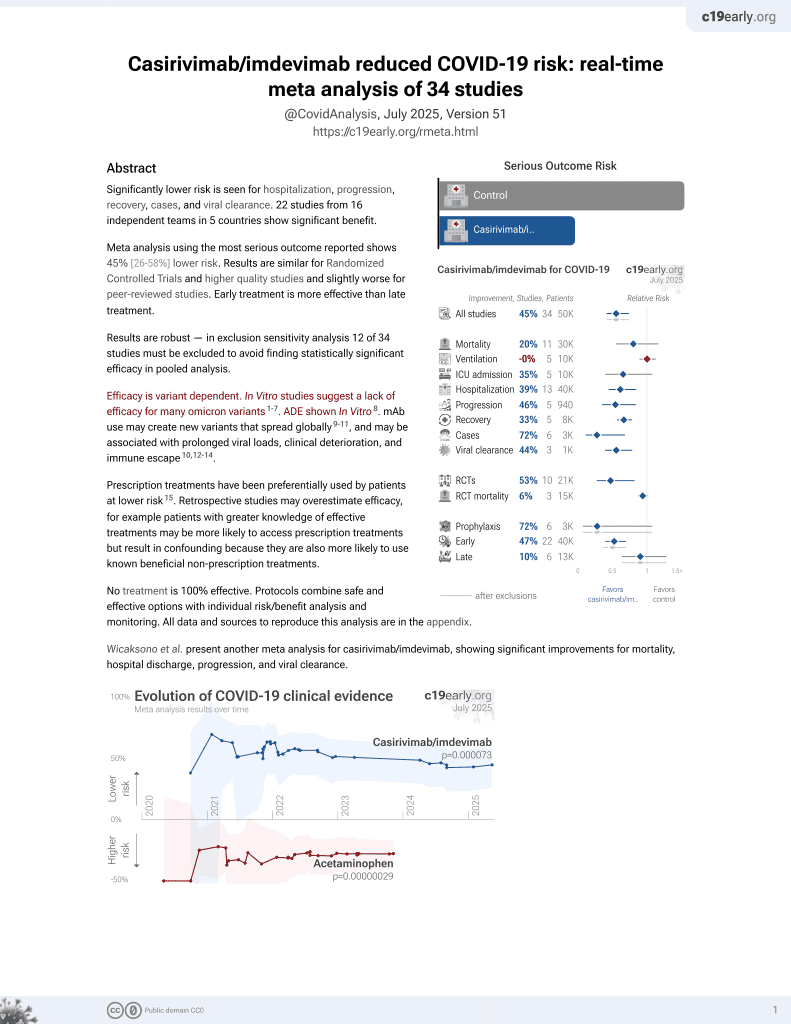
19th treatment shown to reduce risk in
March 2021, now with p = 0.000095 from 34 studies, recognized in 52 countries.
Efficacy is variant dependent.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
In vitro study (Vero-TMPRSS2 and Vero-hACE2-TMPRSS2) showing complete loss of inhibitory activity for B.1.1.529 omicron with LY-CoV555, LY-CoV016, REGN10933, REGN10987, and CT-P59, ~12-fold decrease for COV2-2196/COV2-2130, and minimal change for S309.
Efficacy is variant dependent. In Vitro research suggests a lack of efficacy for many omicron variants1-7.
Study covers casirivimab/imdevimab, bamlanivimab/etesevimab, and sotrovimab.
1.
Liu et al., Striking Antibody Evasion Manifested by the Omicron Variant of SARS-CoV-2, bioRxiv, doi:10.1101/2021.12.14.472719.
2.
Sheward et al., Variable loss of antibody potency against SARS-CoV-2 B.1.1.529 (Omicron), bioRxiv, doi:10.1101/2021.12.19.473354.
3.
VanBlargan et al., An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by several therapeutic monoclonal antibodies, bioRxiv, doi:10.1101/2021.12.15.472828.
4.
Tatham et al., Lack of Ronapreve (REGN-CoV; casirivimab and imdevimab) virological efficacy against the SARS-CoV 2 Omicron variant (B.1.1.529) in K18-hACE2 mice, bioRxiv, doi:10.1101/2022.01.23.477397.
5.
Pochtovyi et al., In Vitro Efficacy of Antivirals and Monoclonal Antibodies against SARS-CoV-2 Omicron Lineages XBB.1.9.1, XBB.1.9.3, XBB.1.5, XBB.1.16, XBB.2.4, BQ.1.1.45, CH.1.1, and CL.1, Vaccines, doi:10.3390/vaccines11101533.
VanBlargan et al., 17 Dec 2021, preprint, 10 authors.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by several therapeutic monoclonal antibodies
doi:10.1101/2021.12.15.472828
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused the global COVID-19 pandemic resulting in millions of deaths worldwide. Despite the development and deployment of highly effective antibody and vaccine countermeasures, rapidly-spreading SARS-CoV-2 variants with mutations at key antigenic sites in the spike protein jeopardize their efficacy. Indeed, the recent emergence of the highly-transmissible B.1.1.529 Omicron variant is especially concerning because of the number of mutations, deletions, and insertions in the spike protein. Here, using a panel of anti-receptor binding domain (RBD) monoclonal antibodies (mAbs) corresponding to those with emergency use authorization (EUA) or in advanced clinical development by Vir Biotechnology (S309, the parent mAbs of VIR-7381), AstraZeneca (COV2-2196 and COV2-2130, the parent mAbs of AZD8895 and AZD1061), Regeneron (REGN10933 and REGN10987), Lilly (LY-CoV555 and LY-CoV016), and Celltrion (CT-P59), we report the impact on neutralization of a prevailing, infectious B.1.1.529 Omicron isolate compared to a historical WA1/2020 D614G strain. Several highly neutralizing mAbs (LY-CoV555, LY-CoV016, REGN10933, REGN10987, and CT-P59) completely lost inhibitory activity against B.1.1.529 virus in both Vero-TMPRSS2 and Vero-hACE2-TMPRSS2 cells, whereas others were reduced (~12-fold decrease, COV2-2196 and COV2-2130 combination) or minimally affected (S309). Our results suggest that several, but not all, of the antibody products in clinical use will lose efficacy against the B.1.1.529 Omicron variant and related strains.
AUTHOR CONTRIBUTIONS L
References
Bailey, Diamond, A Crisp(r) New Perspective on SARS-CoV-2 Biology, Cell
Baum, Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies, Science
Baum, REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters, Science
Callaway, Ledford, How bad is Omicron? What scientists know so far, Nature
Callaway, Omicron likely to weaken COVID vaccine protection, Nature
Cameroni, Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift, bioRxiv
Cao, 1.529 escapes the majority of SARS-CoV-2 neutralizing antibodies of diverse epitopes, bioRxiv
Case, Neutralizing antibody and soluble ACE2 inhibition of a replicationcompetent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2, Cell Host and Microbe
Cathcart, The dual function monoclonal antibodies VIR-7831 and VIR-7832 demonstrate potent in vitro and in vivo activity against SARS-CoV-2, bioRxiv
Cele, SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection, medRxiv : the preprint server for health sciences
Chen, In vivo monoclonal antibody efficacy against SARS-CoV-2 variant strains, Nature
Chen, Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies, Nat Med
Chen, Wang, Gilby, Wei, Omicron, Infectivity, vaccine breakthrough, and antibody resistance, B
Dejnirattisai, Reduced neutralisation of SARS-COV-2 Omicron-B.1.1.529 variant by post-immunisation serum, medRxiv : the preprint server for health sciences
Ford, Machado, Janies, Predictions of the SARS-CoV-2 Omicron Variant (B.1.1.529) Spike Protein Receptor-Binding Domain Structure and Neutralizing Antibody Interactions, bioRxiv
Goddard, UCSF ChimeraX: Meeting modern challenges in visualization and analysis, Protein Sci
Golcuk, Yildiz, Gur, The Omicron Variant Increases the Interactions of SARS-CoV-2 Spike Glycoprotein with ACE2, bioRxiv
Gottlieb, Effect of Bamlanivimab as Monotherapy or in Combination With Etesevimab on Viral Load in Patients With Mild to Moderate COVID-19: A Randomized Clinical Trial, Jama
Gupta, Early Treatment for Covid-19 with SARS-CoV-2 Neutralizing Antibody Sotrovimab, N Engl J Med
Imai, Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development, Proc Natl Acad Sci U S A
Johnson, Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis, Nature
Jones, model of SARS-CoV-2 infection
Khoury, Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection, Nat Med
Kim, A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein, Nat Commun
Kim, Marks, Clemens, Looking beyond COVID-19 vaccine phase 3 trials, Nat Med
Krissinel, Henrick, Inference of macromolecular assemblies from crystalline state, J Mol Biol
Lan, Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor, Nature
Lempp, Lectins enhance SARS-CoV-2 infection and influence neutralizing antibodies, Nature
Liu, Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization, Cell Host Microbe
Pinto, Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody, Nature
Plante, Spike mutation D614G alters SARS-CoV-2 fitness, Nature
Schäfer, Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo, J Exp Med
Sempowski, Saunders, Acharya, Wiehe, Haynes, Pandemic Preparedness: Developing Vaccines and Therapeutic Antibodies For COVID-19, Cell
Shi, A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2, Nature
Starr, Greaney, Dingens, Bloom, Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016, Cell reports. Medicine
Suryadevara, Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein, Cell
Tada, Neutralization of viruses with European, South African, and United States SARS-CoV-2 variant spike proteins by convalescent sera and BNT162b2 mRNA vaccine-elicited antibodies, bioRxiv
Torjesen, Covid-19: Omicron may be more transmissible than other variants and partly resistant to existing vaccines, scientists fear, BMJ (Clinical research ed
Tortorici, Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms, Science
Vanblargan, A potently neutralizing SARS-CoV-2 antibody inhibits variants of concern by utilizing unique binding residues in a highly conserved epitope, Immunity
Wang, Antibody Resistance of SARS-CoV-2 Variants B, Nature
Wang, mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants, Nature
Wibmer, SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma
Wilhelm, Reduced Neutralization of SARS-CoV-2 Omicron Variant by Vaccine Sera and monoclonal antibodies, medRxiv : the preprint server for health sciences
Winkler, Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection, Cell
Zang, TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes, Sci Immunol
Zohar, Compromised Humoral Functional Evolution Tracks with SARS-CoV-2 Mortality, Cell
Zost, Potently neutralizing and protective human antibodies against SARS-CoV-2, Nature
Zost, Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein, Nat Med
DOI record:
{
"DOI": "10.1101/2021.12.15.472828",
"URL": "http://dx.doi.org/10.1101/2021.12.15.472828",
"abstract": "<jats:p>Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused the global COVID-19 pandemic resulting in millions of deaths worldwide. Despite the development and deployment of highly effective antibody and vaccine countermeasures, rapidly-spreading SARS-CoV-2 variants with mutations at key antigenic sites in the spike protein jeopardize their efficacy. Indeed, the recent emergence of the highly-transmissible B.1.1.529 Omicron variant is especially concerning because of the number of mutations, deletions, and insertions in the spike protein. Here, using a panel of anti-receptor binding domain (RBD) monoclonal antibodies (mAbs) corresponding to those with emergency use authorization (EUA) or in advanced clinical development by Vir Biotechnology (S309, the parent mAbs of VIR-7381), AstraZeneca (COV2-2196 and COV2-2130, the parent mAbs of AZD8895 and AZD1061), Regeneron (REGN10933 and REGN10987), Lilly (LY-CoV555 and LY-CoV016), and Celltrion (CT-P59), we report the impact on neutralization of a prevailing, infectious B.1.1.529 Omicron isolate compared to a historical WA1/2020 D614G strain. Several highly neutralizing mAbs (LY-CoV555, LY-CoV016, REGN10933, REGN10987, and CT-P59) completely lost inhibitory activity against B.1.1.529 virus in both Vero-TMPRSS2 and Vero-hACE2-TMPRSS2 cells, whereas others were reduced (~12-fold decrease, COV2-2196 and COV2-2130 combination) or minimally affected (S309). Our results suggest that several, but not all, of the antibody products in clinical use will lose efficacy against the B.1.1.529 Omicron variant and related strains.</jats:p>",
"accepted": {
"date-parts": [
[
2021,
12,
17
]
]
},
"author": [
{
"affiliation": [],
"family": "VanBlargan",
"given": "Laura A",
"sequence": "first"
},
{
"affiliation": [],
"family": "Errico",
"given": "John M",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Halfmann",
"given": "Peter",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Zost",
"given": "Seth J",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-0049-1079",
"affiliation": [],
"authenticated-orcid": false,
"family": "Crowe",
"given": "James E.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Purcell",
"given": "Lisa A",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Kawaoka",
"given": "Yoshihiro",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Corti",
"given": "Davide",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Fremont",
"given": "Daved H",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-8791-3165",
"affiliation": [],
"authenticated-orcid": false,
"family": "Diamond",
"given": "Michael",
"sequence": "additional"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2021,
12,
17
]
],
"date-time": "2021-12-17T22:40:12Z",
"timestamp": 1639780812000
},
"deposited": {
"date-parts": [
[
2021,
12,
17
]
],
"date-time": "2021-12-17T22:40:13Z",
"timestamp": 1639780813000
},
"group-title": "Microbiology",
"indexed": {
"date-parts": [
[
2021,
12,
18
]
],
"date-time": "2021-12-18T07:00:54Z",
"timestamp": 1639810854471
},
"institution": [
{
"name": "bioRxiv"
}
],
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2021,
12,
17
]
]
},
"link": [
{
"URL": "https://syndication.highwire.org/content/doi/10.1101/2021.12.15.472828",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "246",
"original-title": [],
"posted": {
"date-parts": [
[
2021,
12,
17
]
]
},
"prefix": "10.1101",
"published": {
"date-parts": [
[
2021,
12,
17
]
]
},
"publisher": "Cold Spring Harbor Laboratory",
"reference-count": 0,
"references-count": 0,
"relation": {},
"score": 1,
"short-container-title": [],
"short-title": [],
"source": "Crossref",
"subtitle": [],
"subtype": "preprint",
"title": [
"An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by several therapeutic monoclonal antibodies"
],
"type": "posted-content"
}