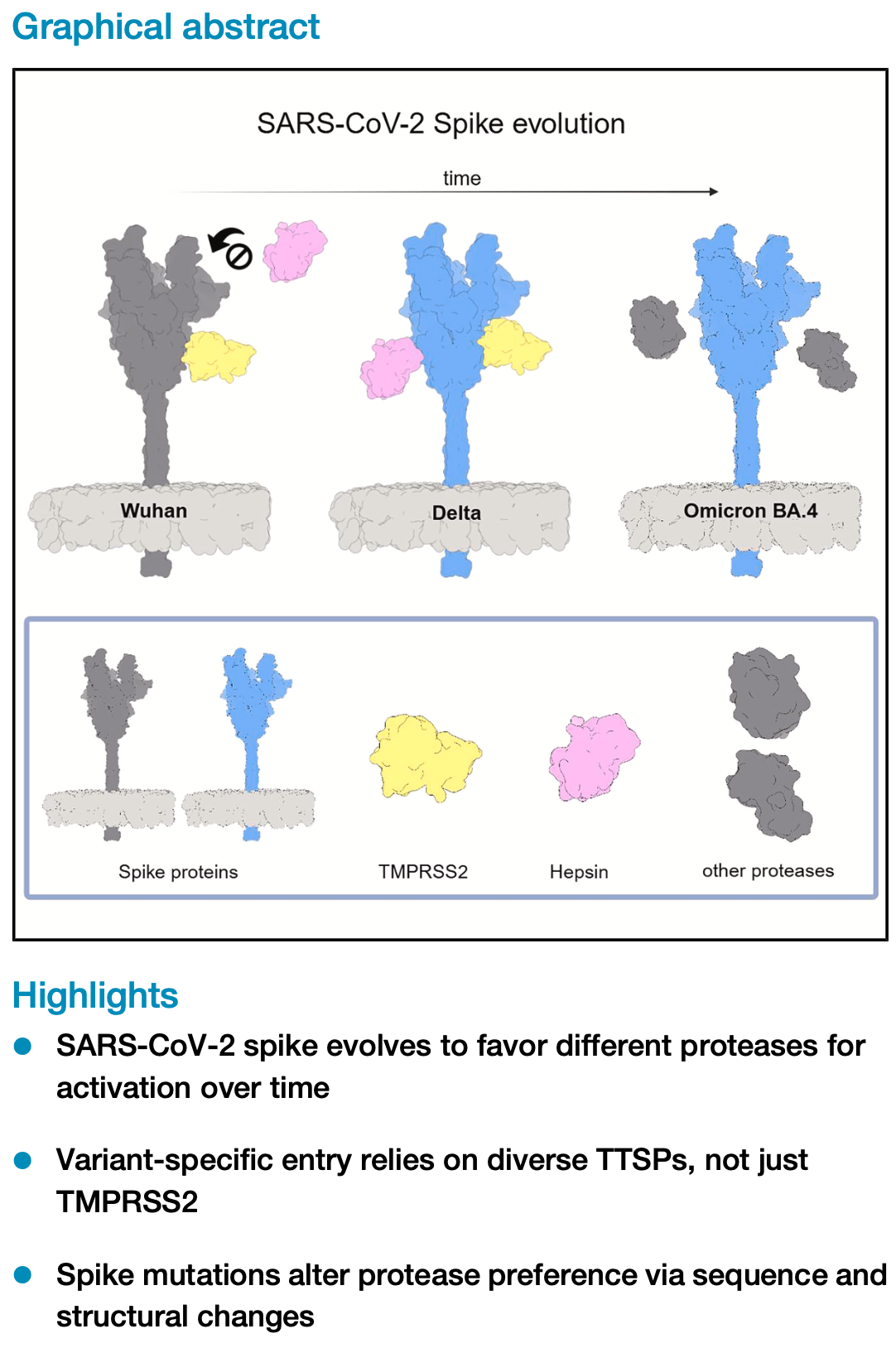
Evolution of the SARS-CoV-2 spike protein in utilizing host transmembrane serine proteases
et al., iScience, doi:10.1016/j.isci.2025.113318, Sep 2025
In vitro study showing variant-dependent differences in SARS-CoV-2 spike protein activation by host transmembrane serine proteases (TTSPs). Authors find that all SARS-CoV-2 variants (Wuhan, Alpha, Beta, Gamma, Delta, and Omicron) require TTSPs for entry in human airway epithelial cells, with camostat (TTSP inhibitor) blocking replication while cathepsin inhibitor E64D was ineffective. In A549 cells, different variants showed distinct protease preferences: Delta variant preferentially used hepsin and TMPRSS2, while Omicron BA.1 primarily relied on TMPRSS2. Authors demonstrate that spike protein mutations affect not only ACE2 recognition but also protease utilization for viral entry, suggesting this evolution influences viral transmission and tissue tropism.
11 preclinical studies support the efficacy of camostat for COVID-19:
1.
Manikyam et al., INP-Guided Network Pharmacology Discloses Multi-Target Therapeutic Strategy Against Cytokine and IgE Storms in the SARS-CoV-2 NB.1.8.1 Variant, Research Square, doi:10.21203/rs.3.rs-6819274/v1.
2.
González-Paz et al., Biophysical Analysis of Potential Inhibitors of SARS-CoV-2 Cell Recognition and Their Effect on Viral Dynamics in Different Cell Types: A Computational Prediction from In Vitro Experimental Data, ACS Omega, doi:10.1021/acsomega.3c06968.
3.
Umar et al., Inhibitory potentials of ivermectin, nafamostat, and camostat on spike protein and some nonstructural proteins of SARS-CoV-2: Virtual screening approach, Jurnal Teknologi Laboratorium, doi:10.29238/teknolabjournal.v11i1.344.
4.
Unal et al., Favipiravir, umifenovir and camostat mesylate: a comparative study against SARS-CoV-2, bioRxiv, doi:10.1101/2022.01.11.475889.
5.
Sgrignani et al., Computational Identification of a Putative Allosteric Binding Pocket in TMPRSS2, Frontiers in Molecular Biosciences, doi:10.3389/fmolb.2021.666626.
6.
Milewska et al., Evolution of the SARS-CoV-2 spike protein in utilizing host transmembrane serine proteases, iScience, doi:10.1016/j.isci.2025.113318.
7.
Schultz et al., Pyrimidine inhibitors synergize with nucleoside analogues to block SARS-CoV-2, Nature, doi:10.1038/s41586-022-04482-x.
Milewska et al., 30 Sep 2025, peer-reviewed, 4 authors.
Contact: apoma@ippt.pan.pl (corresponding author), apoma@ippt.pan.pl (corresponding author), k.pyrc@uj.edu.pl.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Evolution of the SARS-CoV-2 spike protein in utilizing host transmembrane serine proteases
iScience, doi:10.1016/j.isci.2025.113318
SARS-CoV-2 entry into host cells depends on proteolytic activation of the spike protein by host proteases, a process shaped by spike mutations that influence viral specificity and infectivity. Using human airway epithelial models, this study investigated how different SARS-CoV-2 variants interact with host serine proteases. The Delta variant exhibited enhanced and stable binding to Hepsin through stronger ionic and hydrophobic interactions, promoting efficient spike activation and cell entry. In contrast, Omicron BA.1 showed weaker Hepsin binding and relied more on TMPRSS2 or cathepsins, depending on the cellular context. These findings reveal how variant-specific differences in protease usage are linked to spike protein mutations and cleavage site evolution. By illuminating the dynamic interplay between viral adaptation and host protease specificity, this work provides insights into mechanisms that influence viral transmission and immune evasion, with implications for developing targeted antiviral strategies and understanding the evolution of emerging SARS-CoV-2 variants.
AUTHOR CONTRIBUTIONS A.M., experimental design, carrying out experiments, data analysis, figure preparation, and manuscript writing. L.F.C.-V., carrying out experiments, data analysis, and figure preparation. A.B.P. and K.P., experimental design, obtaining funding, and manuscript writing.
DECLARATION OF INTERESTS The authors declare no conflict of interest.
STAR★METHODS Detailed methods are provided in the online version of this paper and include the following: , penicillin (100 U/mL; Thermo Fisher Scientific), and streptomycin (100 μg/mL; Thermo Fisher Scientific). A549 ACE2/TMPRSS2 cells were supplemented with blasticidin S (10 μg/mL, Sigma-Aldrich), and puromycin (0.5 μg/mL, Sigma-Aldrich) to maintain the ACE2+TMPRSS2+ population. Cells were cultured at 37 • C, 5% CO2, and 95% humidity. All cell lines were tested every two weeks for mycoplasma contamination either by in-house DAPI staining or LookOut® Mycoplasma PCR Detection Kit (Sigma-Aldrich).
Human epithelium (HAE) cultures Human airway epithelial cells (Epithelix SAS, Archamps, France) were maintained in BEGM medium. Before the test, cells were trypsinised and transferred to a permeable transwell insert supports (f = 6.5 mm). Cell differentiation was stimulated by medium additives. The removal of the media from the apical side was performed after the cells reached confluence. Cells were cultured for 3 to 5 weeks to form well-differentiated, pseudostratified mucociliary epithelia, as previously described.
..
References
Antalis, Bugge, Wu, Membrane-anchored serine proteases in health and disease, Prog. Mol. Biol. Transl. Sci, doi:10.1016/B978-0-12-385504-6.00001-4
Baggen, Jacquemyn, Persoons, Vanstreels, Pye et al., TMEM106B is a receptor mediating ACE2-independent SARS-CoV-2 cell entry, Cell, doi:10.1016/j.cell.2023.06.005
Barnes, Infection of liver hepatocytes with SARS-CoV-2, Nat. Metab, doi:10.1038/s42255-022-00554-4
Bayati, Kumar, Francis, Mcpherson, SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis, J. Biol. Chem, doi:10.1016/j.jbc.2021.100306
Beaulieu, Gravel, Cloutier, Marois, Colombo et al., Matriptase proteolytically activates influenza virus and promotes multicycle replication in the human airway epithelium, J. Virol, doi:10.1128/JVI.03005-12
Benlarbi, Laroche, Fink, Fu, Mulloy et al., Identification and differential usage of a host metalloproteinase entry pathway by SARS-CoV-2 Delta and Omicron, iScience, doi:10.1016/j.isci.2022.105316
Blee, He, Yang, Ye, Yan et al., Controls Luminal Epithelial Lineage and Antiandrogen Sensitivity, Clin. Cancer Res, doi:10.1158/1078-0432.CCR-18-0653
Bosch, Bartelink, Rottier, Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide, J. Virol, doi:10.1128/JVI.00415-08
Cantuti-Castelvetri, Ojha, Pedro, Djannatian, Franz et al., Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity, Science, doi:10.1126/science.abd2985
Case, Aktulga, Belfon, Cerutti, Cisneros et al., AmberTools, J. Chem. Inf. Model, doi:10.1021/acs.jcim.3c01153
Cerutti, Duke, Darden, Lybrand, Staggered Mesh Ewald: An extension of the Smooth Particle-Mesh Ewald method adding great versatility, J. Chem. Theory Comput, doi:10.1021/ct9001015
Chan, Huang, Hu, Chai, Shi et al., Altered host protease determinants for SARS-CoV-2 Omicron, Sci. Adv, doi:10.1126/sciadv.add3867
Chandran, Sullivan, Felbor, Whelan, Cunningham, Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection, Science, doi:10.1126/science.1110656
Clausen, Sandoval, Spliid, Pihl, Perrett et al., SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2, Cell, doi:10.1016/j.cell.2020.09.033
Dominguez, Boelens, Bonvin, HADDOCK: a protein-protein docking approach based on biochemical or biophysical information, J. Am. Chem. Soc, doi:10.1021/ja026939x
Eastman, Swails, Chodera, Mcgibbon, Zhao et al., OpenMM 7: Rapid development of high performance algorithms for molecular dynamics, PLoS Comput. Biol, doi:10.1371/journal.pcbi.1005659
Ebert, Deussing, Peters, Dermody, Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells, J. Biol. Chem, doi:10.1074/jbc.M201107200
Essalmani, Jain, Susan-Resiga, Andre ´o, Evagelidis et al., Distinctive Roles of Furin and TMPRSS2 in SARS-CoV-2 Infectivity, J. Virol, doi:10.1128/jvi.00128-22
Fuentes-Prior, Priming of SARS-CoV-2 S protein by several membrane-bound serine proteinases could explain enhanced viral infectivity and systemic COVID-19 infection, J. Biol. Chem, doi:10.1074/jbc.REV120.015980
Glowacka, Bertram, Mu ¨ller, Allen, Soilleux et al., Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response, J. Virol, doi:10.1128/JVI.02232-10
Herter, Piper, Aaron, Gabriele, Cutler et al., Hepatocyte growth factor is a preferred in vitro substrate for human hepsin, a membraneanchored serine protease implicated in prostate and ovarian cancers, Biochem. J, doi:10.1042/BJ20041955
Hoffmann, Kleine-Weber, Schroeder, Kru ¨ger, Herrler et al., SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor, Cell, doi:10.1016/j.cell.2020.02.052
Hoffmann, Mo ¨sbauer, Hofmann-Winkler, Kaul, Kleine-Weber et al., Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2, Nature, doi:10.1038/s41586-020-2575-3
Hopkins, Le Grand, Walker, Roitberg, Long-Time-Step Molecular Dynamics through Hydrogen Mass Repartitioning, J. Chem. Theory Comput, doi:10.1021/ct5010406
Hou, Okuda, Edwards, Martinez, Asakura et al., SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract, Cell, doi:10.1016/j.cell.2020.05.042
Inoue, Tanaka, Tanaka, Inoue, Morita et al., Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted, J. Virol, doi:10.1128/JVI.00253-07
Islam, Dhawan, Nafady, Emran, Mitra et al., Understanding the omicron variant (B.1.1.529) of SARS-CoV-2: Mutational impacts, concerns, and the possible solutions, Ann. Med. Surg, doi:10.1016/j.amsu.2022.103737
Iwata-Yoshikawa, Okamura, Shimizu, Hasegawa, Takeda et al., TMPRSS2 Contributes to Virus Spread and Immunopathology in the Airways of Murine Models after Coronavirus Infection, J. Virol, doi:10.1128/JVI.01815-18
Izadi, Anandakrishnan, Onufriev, Building Water Models: A Different Approach, J. Phys. Chem. Lett, doi:10.1021/jz501780a
Jean-Paul, Ciccotti, Herman, Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes, J. Comput. Phys, doi:10.1016/0021-9991(77)90098-5
Jung, Kmiec, Koepke, Zech, Jacob et al., Omicron: What Makes the Latest SARS-CoV-2 Variant of Concern So Concerning?, J. Virol, doi:10.1128/jvi.02077-21
Kakizaki, Hashimoto, Nagata, Yamamoto, Okura et al., The respective roles of TMPRSS2 and cathepsins for SARS-CoV-2 infection in human respiratory organoids, J. Virol, doi:10.1128/jvi.01853-24
Kawase, Shirato, Van Der Hoek, Taguchi, Matsuyama, Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry, J. Virol, doi:10.1128/JVI.00094-12
Kim, Kang, Kim, Park, Heo et al., The host protease KLK5 primes and activates spike proteins to promote human betacoronavirus replication and lung inflammation, Sci. Signal, doi:10.1126/scisignal.adn3785
Laporte, Naesens, Airway proteases: an emerging drug target for influenza and other respiratory virus infections, Curr. Opin. Virol, doi:10.1016/j.coviro.2017.03.018
Li, Wang, Sun, Wu, Hepsin: a multifunctional transmembrane serine protease in pathobiology, FEBS J, doi:10.1111/febs.15663
Limburg, Harbig, Bestle, Stein, Moulton et al., TMPRSS2 Is the Major Activating Protease of Influenza A Virus in Primary Human Airway Cells and Influenza B Virus in Human Type II Pneumocytes, J. Virol, doi:10.1128/JVI.00649-19
Lu, Zhao, Li, Niu, Yang et al., Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding, Lancet, doi:10.1016/S0140-6736(20)30251-8
Luostari, Hartikainen, Tengstro ¨m, Palvimo, Kataja et al., Type II transmembrane serine protease gene variants associate with breast cancer, PLoS One, doi:10.1371/journal.pone.0102519
Magnen, Gueugnon, Guillon, Baranek, Thibault et al., Kallikrein-Related Peptidase 5 Contributes to H3N2 Influenza Virus Infection in Human Lungs, J. Virol, doi:10.1128/JVI.00421-17
Matsuyama, Nagata, Shirato, Kawase, Takeda et al., Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2, J. Virol, doi:10.1128/JVI.01542-10
Meng, Abdullahi, Ferreira, Goonawardane, Saito et al., Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity, Nature, doi:10.1038/s41586-022-04474-x
Meng, Goddard, Pettersen, Couch, Pearson et al., UCSF ChimeraX: Tools for structure building and analysis, Protein Sci, doi:10.1002/pro.4792
Milewska, Falkowski, Kulczycka, Bielecka, Naskalska et al., Kallikrein 13 serves as a priming protease during infection by the human coronavirus HKU1, Sci. Signal, doi:10.1126/scisignal.aba9902
Milewska, Kula-Pacurar, Wadas, Suder, Szczepanski et al., Replication of Severe Acute Respiratory Syndrome Coronavirus 2 in Human Respiratory Epithelium, J. Virol, doi:10.1128/JVI.00957-20
Moreira, Chwastyk, Baker, Guzman, Poma, All atom simulations snapshots and contact maps analysis scripts for SARS-CoV-2002 and SARS-CoV-2 spike proteins with and without ACE2 enzyme (Zenodo), doi:10.5281/zenodo.3817446
Moreira, Chwastyk, Baker, Guzman, Poma, Quantitative determination of mechanical stability in the novel coronavirus spike protein, Nanoscale, doi:10.1039/d0nr03969a
Myerburg, Mckenna, Luke, Frizzell, Kleyman et al., Prostasin expression is regulated by airway surface liquid volume and is increased in cystic fibrosis, Am. J. Physiol. Lung Cell. Mol. Physiol, doi:10.1152/ajplung.00437.2007
Mykytyn, Breugem, Geurts, Beumer, Schipper et al., SARS-CoV-2 Omicron entry is type II transmembrane serine protease-mediated in human airway and intestinal organoid models, J. Virol, doi:10.1128/jvi.00851-23
Oberst, Singh, Ozdemirli, Dickson, Johnson et al., Characterization of matriptase expression in normal human tissues, J. Histochem. Cytochem, doi:10.1177/002215540305100805
Ou, Mou, Zhang, Ojha, Choe et al., Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2, PLoS Pathog, doi:10.1371/journal.ppat.1009212
Poma, Cieplak, Theodorakis, Combining the MARTINI and Structure-Based Coarse-Grained Approaches for the Molecular Dynamics Studies of Conformational Transitions in Proteins, J. Chem. Theor. Comput, doi:10.1021/acs.jctc.6b00986
Ray, Minh Tran, Santos Natividade, Moreira, Simpson et al., Single-Molecule Investigation of the Binding Interface Stability of SARS-CoV-2 Variants with ACE2, ACS Nanosci. Au, doi:10.1021/acsnanoscienceau.3c00060
Sali, Blundell, Comparative protein modelling by satisfaction of spatial restraints, J. Mol. Biol, doi:10.1006/jmbi.1993.1626
Salomon-Ferrer, Go ¨tz, Poole, Le Grand, Walker, Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald, J. Chem. Theory Comput, doi:10.1021/ct400314y
Shen, Mao, Wu, Tanaka, Zhang, TMPRSS2: A potential target for treatment of influenza virus and coronavirus infections, Biochimie, doi:10.1016/j.biochi.2017.07.016
Shi, Li, Lai, Johnson, Yewdell et al., Omicron Spike confers enhanced infectivity and interferon resistance to SARS-CoV-2 in human nasal tissue, Nat. Commun, doi:10.1038/s41467-024-45075-8
Simmonett, Brooks, A compression strategy for particle mesh Ewald theory, J. Chem. Phys, doi:10.1063/5.0040966
Simmons, Gosalia, Rennekamp, Reeves, Diamond et al., Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry, Proc. Natl. Acad. Sci. USA, doi:10.1073/pnas.0505577102
Sindhikara, Kim, Voter, Roitberg, Bad Seeds Sprout Perilous Dynamics: Stochastic Thermostat Induced Trajectory Synchronization in Biomolecules, J. Chem. Theory Comput, doi:10.1021/ct800573m
Sungnak, Huang, Be ´cavin, Berg, Queen et al., SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes, Nat. Med, doi:10.1038/s41591-020-0868-6
Szabo, Bugge, Type II transmembrane serine proteases in development and disease, Int. J. Biochem. Cell Biol, doi:10.1016/j.biocel.2007.11.013
Szlachcic, Dabrowska, Milewska, Ziojla, Blaszczyk et al., SARS-CoV-2 infects an, iScience, doi:10.1016/j.isci.2022.104594
Van Zundert, Rodrigues, Trellet, Schmitz, Kastritis et al., The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes, J. Mol. Biol, doi:10.1016/j.jmb.2015.09.014
Wang, Liu, Zhang, Wang, Hong et al., Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies, Nat. Commun, doi:10.1038/s41467-022-28528-w
Wang, Yang, Liu, Xu, Shao et al., Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry, Nat. Commun, doi:10.1038/s41467-023-42527-5
Wanner, Andrieux, Badia-I-Mompel, Edler, Pfefferle et al., Molecular consequences of SARS-CoV-2 liver tropism, Nat. Metab, doi:10.1038/s42255-022-00552-6
Wennerstro ¨m, Nervall, Bjelic, Bjørn, Molecular dynamics simulations of water and biomolecules with a Monte Carlo constant pressure algorithm, Chem. Phys. Lett, doi:10.1016/j.cplett.2003.12.039
Willett, Grove, Maclean, Wilkie, De Lorenzo et al., SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway, Nat. Microbiol, doi:10.1038/s41564-022-01143-7
Wolter, Jassat, Walaza, Welch, Moultrie et al., Early assessment of the clinical severity of the SARS-CoV-2 omicron variant in South Africa: a data linkage study, Lancet, doi:10.1016/S0140-6736(22)00017-4
Xu, Wu, Cao, Gu, Liu et al., Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins, Cell Res, doi:10.1038/s41422-022-00672-4
Zhao, Lu, Peng, Chen, Meng et al., SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells, Emerg. Microbes Infect, doi:10.1080/22221751.2021.2023329
Zhao, Zhao, Wang, Zhou, Ma et al., Single-Cell RNA Expression Profiling of ACE2, the Receptor of SARS-CoV-2, Am. J. Respir. Crit. Care Med, doi:10.1164/rccm.202001-0179LE
Zhu, Feng, Hu, Wang, Yu et al., A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry, Nat. Commun, doi:10.1038/s41467-021-21213-4
DOI record:
{
"DOI": "10.1016/j.isci.2025.113318",
"ISSN": [
"2589-0042"
],
"URL": "http://dx.doi.org/10.1016/j.isci.2025.113318",
"alternative-id": [
"S2589004225015792"
],
"article-number": "113318",
"assertion": [
{
"label": "This article is maintained by",
"name": "publisher",
"value": "Elsevier"
},
{
"label": "Article Title",
"name": "articletitle",
"value": "Evolution of the SARS-CoV-2 spike protein in utilizing host transmembrane serine proteases"
},
{
"label": "Journal Title",
"name": "journaltitle",
"value": "iScience"
},
{
"label": "CrossRef DOI link to publisher maintained version",
"name": "articlelink",
"value": "https://doi.org/10.1016/j.isci.2025.113318"
},
{
"label": "Content Type",
"name": "content_type",
"value": "article"
},
{
"label": "Copyright",
"name": "copyright",
"value": "© 2025 The Authors. Published by Elsevier Inc."
}
],
"author": [
{
"ORCID": "https://orcid.org/0000-0003-0895-8697",
"affiliation": [],
"authenticated-orcid": false,
"family": "Milewska",
"given": "Aleksandra",
"sequence": "first"
},
{
"affiliation": [],
"family": "Cofas-Vargas",
"given": "Luis Fernando",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Poma",
"given": "Adolfo B.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Pyrć",
"given": "Krzysztof",
"sequence": "additional"
}
],
"container-title": "iScience",
"container-title-short": "iScience",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"cell.com",
"elsevier.com",
"sciencedirect.com"
]
},
"created": {
"date-parts": [
[
2025,
8,
6
]
],
"date-time": "2025-08-06T15:58:56Z",
"timestamp": 1754495936000
},
"deposited": {
"date-parts": [
[
2025,
9,
23
]
],
"date-time": "2025-09-23T08:42:49Z",
"timestamp": 1758616969000
},
"funder": [
{
"DOI": "10.13039/501100007088",
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100007088",
"id-type": "DOI"
}
],
"name": "Jagiellonian University"
},
{
"DOI": "10.13039/501100004382",
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100004382",
"id-type": "DOI"
}
],
"name": "Polish Academy of Sciences"
}
],
"indexed": {
"date-parts": [
[
2025,
9,
24
]
],
"date-time": "2025-09-24T00:10:42Z",
"timestamp": 1758672642106,
"version": "3.44.0"
},
"is-referenced-by-count": 0,
"issue": "9",
"issued": {
"date-parts": [
[
2025,
9
]
]
},
"journal-issue": {
"issue": "9",
"published-print": {
"date-parts": [
[
2025,
9
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://www.elsevier.com/tdm/userlicense/1.0/",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
9,
1
]
],
"date-time": "2025-09-01T00:00:00Z",
"timestamp": 1756684800000
}
},
{
"URL": "https://www.elsevier.com/legal/tdmrep-license",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
9,
1
]
],
"date-time": "2025-09-01T00:00:00Z",
"timestamp": 1756684800000
}
},
{
"URL": "http://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
8,
4
]
],
"date-time": "2025-08-04T00:00:00Z",
"timestamp": 1754265600000
}
}
],
"link": [
{
"URL": "https://api.elsevier.com/content/article/PII:S2589004225015792?httpAccept=text/xml",
"content-type": "text/xml",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://api.elsevier.com/content/article/PII:S2589004225015792?httpAccept=text/plain",
"content-type": "text/plain",
"content-version": "vor",
"intended-application": "text-mining"
}
],
"member": "78",
"original-title": [],
"page": "113318",
"prefix": "10.1016",
"published": {
"date-parts": [
[
2025,
9
]
]
},
"published-print": {
"date-parts": [
[
2025,
9
]
]
},
"publisher": "Elsevier BV",
"reference": [
{
"DOI": "10.1016/S0140-6736(20)30251-8",
"article-title": "Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding",
"author": "Lu",
"doi-asserted-by": "crossref",
"first-page": "565",
"journal-title": "Lancet",
"key": "10.1016/j.isci.2025.113318_bib1",
"volume": "395",
"year": "2020"
},
{
"article-title": "Distinctive Roles of Furin and TMPRSS2 in SARS-CoV-2 Infectivity",
"author": "Essalmani",
"journal-title": "J. Virol.",
"key": "10.1016/j.isci.2025.113318_bib2",
"volume": "96",
"year": "2022"
},
{
"DOI": "10.1126/science.abd2985",
"article-title": "Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity",
"author": "Cantuti-Castelvetri",
"doi-asserted-by": "crossref",
"first-page": "856",
"journal-title": "Science",
"key": "10.1016/j.isci.2025.113318_bib3",
"volume": "370",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2023.06.005",
"article-title": "TMEM106B is a receptor mediating ACE2-independent SARS-CoV-2 cell entry",
"author": "Baggen",
"doi-asserted-by": "crossref",
"first-page": "3427",
"journal-title": "Cell",
"key": "10.1016/j.isci.2025.113318_bib4",
"volume": "186",
"year": "2023"
},
{
"DOI": "10.1016/j.cell.2020.09.033",
"article-title": "SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2",
"author": "Clausen",
"doi-asserted-by": "crossref",
"first-page": "1043",
"journal-title": "Cell",
"key": "10.1016/j.isci.2025.113318_bib5",
"volume": "183",
"year": "2020"
},
{
"DOI": "10.1128/JVI.02232-10",
"article-title": "Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response",
"author": "Glowacka",
"doi-asserted-by": "crossref",
"first-page": "4122",
"journal-title": "J. Virol.",
"key": "10.1016/j.isci.2025.113318_bib6",
"volume": "85",
"year": "2011"
},
{
"DOI": "10.1128/JVI.01542-10",
"article-title": "Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2",
"author": "Matsuyama",
"doi-asserted-by": "crossref",
"first-page": "12658",
"journal-title": "J. Virol.",
"key": "10.1016/j.isci.2025.113318_bib7",
"volume": "84",
"year": "2010"
},
{
"DOI": "10.1128/JVI.00649-19",
"article-title": "TMPRSS2 Is the Major Activating Protease of Influenza A Virus in Primary Human Airway Cells and Influenza B Virus in Human Type II Pneumocytes",
"author": "Limburg",
"doi-asserted-by": "crossref",
"first-page": "e00649-19",
"journal-title": "J. Virol.",
"key": "10.1016/j.isci.2025.113318_bib8",
"volume": "93",
"year": "2019"
},
{
"DOI": "10.1016/j.biochi.2017.07.016",
"article-title": "TMPRSS2: A potential target for treatment of influenza virus and coronavirus infections",
"author": "Shen",
"doi-asserted-by": "crossref",
"first-page": "1",
"journal-title": "Biochimie",
"key": "10.1016/j.isci.2025.113318_bib9",
"volume": "142",
"year": "2017"
},
{
"DOI": "10.1016/j.biocel.2007.11.013",
"article-title": "Type II transmembrane serine proteases in development and disease",
"author": "Szabo",
"doi-asserted-by": "crossref",
"first-page": "1297",
"journal-title": "Int. J. Biochem. Cell Biol.",
"key": "10.1016/j.isci.2025.113318_bib10",
"volume": "40",
"year": "2008"
},
{
"DOI": "10.1158/1078-0432.CCR-18-0653",
"article-title": "Controls Luminal Epithelial Lineage and Antiandrogen Sensitivity",
"author": "Blee",
"doi-asserted-by": "crossref",
"first-page": "4551",
"journal-title": "Clin. Cancer Res.",
"key": "10.1016/j.isci.2025.113318_bib11",
"volume": "24",
"year": "2018"
},
{
"DOI": "10.1038/s41591-020-0868-6",
"article-title": "SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes",
"author": "Sungnak",
"doi-asserted-by": "crossref",
"first-page": "681",
"journal-title": "Nat. Med.",
"key": "10.1016/j.isci.2025.113318_bib12",
"volume": "26",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.05.042",
"article-title": "SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract",
"author": "Hou",
"doi-asserted-by": "crossref",
"first-page": "429",
"journal-title": "Cell",
"key": "10.1016/j.isci.2025.113318_bib13",
"volume": "182",
"year": "2020"
},
{
"DOI": "10.1164/rccm.202001-0179LE",
"article-title": "Single-Cell RNA Expression Profiling of ACE2, the Receptor of SARS-CoV-2",
"author": "Zhao",
"doi-asserted-by": "crossref",
"first-page": "756",
"journal-title": "Am. J. Respir. Crit. Care Med.",
"key": "10.1016/j.isci.2025.113318_bib14",
"volume": "202",
"year": "2020"
},
{
"DOI": "10.1016/j.coviro.2017.03.018",
"article-title": "Airway proteases: an emerging drug target for influenza and other respiratory virus infections",
"author": "Laporte",
"doi-asserted-by": "crossref",
"first-page": "16",
"journal-title": "Curr. Opin. Virol.",
"key": "10.1016/j.isci.2025.113318_bib15",
"volume": "24",
"year": "2017"
},
{
"DOI": "10.1016/j.jbc.2021.100306",
"article-title": "SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis",
"author": "Bayati",
"doi-asserted-by": "crossref",
"journal-title": "J. Biol. Chem.",
"key": "10.1016/j.isci.2025.113318_bib16",
"volume": "296",
"year": "2021"
},
{
"DOI": "10.1128/JVI.00253-07",
"article-title": "Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted",
"author": "Inoue",
"doi-asserted-by": "crossref",
"first-page": "8722",
"journal-title": "J. Virol.",
"key": "10.1016/j.isci.2025.113318_bib17",
"volume": "81",
"year": "2007"
},
{
"DOI": "10.1371/journal.ppat.1009212",
"article-title": "Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2",
"author": "Ou",
"doi-asserted-by": "crossref",
"journal-title": "PLoS Pathog.",
"key": "10.1016/j.isci.2025.113318_bib18",
"volume": "17",
"year": "2021"
},
{
"DOI": "10.1038/s41467-021-21213-4",
"article-title": "A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry",
"author": "Zhu",
"doi-asserted-by": "crossref",
"first-page": "961",
"journal-title": "Nat. Commun.",
"key": "10.1016/j.isci.2025.113318_bib19",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1128/JVI.00415-08",
"article-title": "Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide",
"author": "Bosch",
"doi-asserted-by": "crossref",
"first-page": "8887",
"journal-title": "J. Virol.",
"key": "10.1016/j.isci.2025.113318_bib20",
"volume": "82",
"year": "2008"
},
{
"DOI": "10.1074/jbc.M201107200",
"article-title": "Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells",
"author": "Ebert",
"doi-asserted-by": "crossref",
"first-page": "24609",
"journal-title": "J. Biol. Chem.",
"key": "10.1016/j.isci.2025.113318_bib21",
"volume": "277",
"year": "2002"
},
{
"DOI": "10.1126/science.1110656",
"article-title": "Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection",
"author": "Chandran",
"doi-asserted-by": "crossref",
"first-page": "1643",
"journal-title": "Science",
"key": "10.1016/j.isci.2025.113318_bib22",
"volume": "308",
"year": "2005"
},
{
"DOI": "10.1016/j.cell.2020.02.052",
"article-title": "SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor",
"author": "Hoffmann",
"doi-asserted-by": "crossref",
"first-page": "271",
"journal-title": "Cell",
"key": "10.1016/j.isci.2025.113318_bib23",
"volume": "181",
"year": "2020"
},
{
"DOI": "10.1073/pnas.0505577102",
"article-title": "Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry",
"author": "Simmons",
"doi-asserted-by": "crossref",
"first-page": "11876",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "10.1016/j.isci.2025.113318_bib24",
"volume": "102",
"year": "2005"
},
{
"DOI": "10.1038/s41586-020-2575-3",
"article-title": "Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2",
"author": "Hoffmann",
"doi-asserted-by": "crossref",
"first-page": "588",
"journal-title": "Nature",
"key": "10.1016/j.isci.2025.113318_bib25",
"volume": "585",
"year": "2020"
},
{
"DOI": "10.1038/s41586-022-04474-x",
"article-title": "Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity",
"author": "Meng",
"doi-asserted-by": "crossref",
"first-page": "706",
"journal-title": "Nature",
"key": "10.1016/j.isci.2025.113318_bib26",
"volume": "603",
"year": "2022"
},
{
"DOI": "10.1128/JVI.01815-18",
"article-title": "TMPRSS2 Contributes to Virus Spread and Immunopathology in the Airways of Murine Models after Coronavirus Infection",
"author": "Iwata-Yoshikawa",
"doi-asserted-by": "crossref",
"first-page": "e01815-18",
"journal-title": "J. Virol.",
"key": "10.1016/j.isci.2025.113318_bib27",
"volume": "93",
"year": "2019"
},
{
"DOI": "10.1128/JVI.00094-12",
"article-title": "Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry",
"author": "Kawase",
"doi-asserted-by": "crossref",
"first-page": "6537",
"journal-title": "J. Virol.",
"key": "10.1016/j.isci.2025.113318_bib28",
"volume": "86",
"year": "2012"
},
{
"DOI": "10.1128/jvi.00851-23",
"article-title": "SARS-CoV-2 Omicron entry is type II transmembrane serine protease-mediated in human airway and intestinal organoid models",
"author": "Mykytyn",
"doi-asserted-by": "crossref",
"journal-title": "J. Virol.",
"key": "10.1016/j.isci.2025.113318_bib29",
"volume": "97",
"year": "2023"
},
{
"DOI": "10.1126/sciadv.add3867",
"article-title": "Altered host protease determinants for SARS-CoV-2 Omicron",
"author": "Chan",
"doi-asserted-by": "crossref",
"journal-title": "Sci. Adv.",
"key": "10.1016/j.isci.2025.113318_bib30",
"volume": "9",
"year": "2023"
},
{
"DOI": "10.1177/002215540305100805",
"article-title": "Characterization of matriptase expression in normal human tissues",
"author": "Oberst",
"doi-asserted-by": "crossref",
"first-page": "1017",
"journal-title": "J. Histochem. Cytochem.",
"key": "10.1016/j.isci.2025.113318_bib31",
"volume": "51",
"year": "2003"
},
{
"DOI": "10.1152/ajplung.00437.2007",
"article-title": "Prostasin expression is regulated by airway surface liquid volume and is increased in cystic fibrosis",
"author": "Myerburg",
"doi-asserted-by": "crossref",
"first-page": "L932",
"journal-title": "Am. J. Physiol. Lung Cell. Mol. Physiol.",
"key": "10.1016/j.isci.2025.113318_bib32",
"volume": "294",
"year": "2008"
},
{
"DOI": "10.1074/jbc.REV120.015980",
"article-title": "Priming of SARS-CoV-2 S protein by several membrane-bound serine proteinases could explain enhanced viral infectivity and systemic COVID-19 infection",
"author": "Fuentes-Prior",
"doi-asserted-by": "crossref",
"journal-title": "J. Biol. Chem.",
"key": "10.1016/j.isci.2025.113318_bib33",
"volume": "296",
"year": "2021"
},
{
"DOI": "10.1128/JVI.03005-12",
"article-title": "Matriptase proteolytically activates influenza virus and promotes multicycle replication in the human airway epithelium",
"author": "Beaulieu",
"doi-asserted-by": "crossref",
"first-page": "4237",
"journal-title": "J. Virol.",
"key": "10.1016/j.isci.2025.113318_bib34",
"volume": "87",
"year": "2013"
},
{
"DOI": "10.1128/JVI.00421-17",
"article-title": "Kallikrein-Related Peptidase 5 Contributes to H3N2 Influenza Virus Infection in Human Lungs",
"author": "Magnen",
"doi-asserted-by": "crossref",
"first-page": "e00421-17",
"journal-title": "J. Virol.",
"key": "10.1016/j.isci.2025.113318_bib35",
"volume": "91",
"year": "2017"
},
{
"DOI": "10.1371/journal.pone.0102519",
"article-title": "Type II transmembrane serine protease gene variants associate with breast cancer",
"author": "Luostari",
"doi-asserted-by": "crossref",
"journal-title": "PLoS One",
"key": "10.1016/j.isci.2025.113318_bib36",
"volume": "9",
"year": "2014"
},
{
"DOI": "10.1111/febs.15663",
"article-title": "Hepsin: a multifunctional transmembrane serine protease in pathobiology",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "5252",
"journal-title": "FEBS J.",
"key": "10.1016/j.isci.2025.113318_bib37",
"volume": "288",
"year": "2021"
},
{
"article-title": "SARS-CoV-2 infects an",
"author": "Szlachcic",
"journal-title": "iScience",
"key": "10.1016/j.isci.2025.113318_bib38",
"volume": "25",
"year": "2022"
},
{
"DOI": "10.1038/s42255-022-00552-6",
"article-title": "Molecular consequences of SARS-CoV-2 liver tropism",
"author": "Wanner",
"doi-asserted-by": "crossref",
"first-page": "310",
"journal-title": "Nat. Metab.",
"key": "10.1016/j.isci.2025.113318_bib39",
"volume": "4",
"year": "2022"
},
{
"DOI": "10.1038/s42255-022-00554-4",
"article-title": "Infection of liver hepatocytes with SARS-CoV-2",
"author": "Barnes",
"doi-asserted-by": "crossref",
"first-page": "301",
"journal-title": "Nat. Metab.",
"key": "10.1016/j.isci.2025.113318_bib40",
"volume": "4",
"year": "2022"
},
{
"DOI": "10.1126/scisignal.aba9902",
"article-title": "Kallikrein 13 serves as a priming protease during infection by the human coronavirus HKU1",
"author": "Milewska",
"doi-asserted-by": "crossref",
"journal-title": "Sci. Signal.",
"key": "10.1016/j.isci.2025.113318_bib41",
"volume": "13",
"year": "2020"
},
{
"DOI": "10.1021/acsnanoscienceau.3c00060",
"article-title": "Single-Molecule Investigation of the Binding Interface Stability of SARS-CoV-2 Variants with ACE2",
"author": "Ray",
"doi-asserted-by": "crossref",
"first-page": "136",
"journal-title": "ACS Nanosci. Au",
"key": "10.1016/j.isci.2025.113318_bib42",
"volume": "4",
"year": "2024"
},
{
"DOI": "10.1038/s41564-022-01143-7",
"article-title": "SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway",
"author": "Willett",
"doi-asserted-by": "crossref",
"first-page": "1161",
"journal-title": "Nat. Microbiol.",
"key": "10.1016/j.isci.2025.113318_bib43",
"volume": "7",
"year": "2022"
},
{
"DOI": "10.1128/jvi.01853-24",
"article-title": "The respective roles of TMPRSS2 and cathepsins for SARS-CoV-2 infection in human respiratory organoids",
"author": "Kakizaki",
"doi-asserted-by": "crossref",
"journal-title": "J. Virol.",
"key": "10.1016/j.isci.2025.113318_bib44",
"volume": "99",
"year": "2025"
},
{
"DOI": "10.1080/22221751.2021.2023329",
"article-title": "SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells",
"author": "Zhao",
"doi-asserted-by": "crossref",
"first-page": "277",
"journal-title": "Emerg. Microbes Infect.",
"key": "10.1016/j.isci.2025.113318_bib45",
"volume": "11",
"year": "2022"
},
{
"DOI": "10.1016/j.isci.2022.105316",
"article-title": "Identification and differential usage of a host metalloproteinase entry pathway by SARS-CoV-2 Delta and Omicron",
"author": "Benlarbi",
"doi-asserted-by": "crossref",
"journal-title": "iScience",
"key": "10.1016/j.isci.2025.113318_bib46",
"volume": "25",
"year": "2022"
},
{
"DOI": "10.1016/B978-0-12-385504-6.00001-4",
"article-title": "Membrane-anchored serine proteases in health and disease",
"author": "Antalis",
"doi-asserted-by": "crossref",
"first-page": "1",
"journal-title": "Prog. Mol. Biol. Transl. Sci.",
"key": "10.1016/j.isci.2025.113318_bib47",
"volume": "99",
"year": "2011"
},
{
"DOI": "10.1126/scisignal.adn3785",
"article-title": "The host protease KLK5 primes and activates spike proteins to promote human betacoronavirus replication and lung inflammation",
"author": "Kim",
"doi-asserted-by": "crossref",
"journal-title": "Sci. Signal.",
"key": "10.1016/j.isci.2025.113318_bib48",
"volume": "17",
"year": "2024"
},
{
"DOI": "10.1038/s41467-024-45075-8",
"article-title": "Omicron Spike confers enhanced infectivity and interferon resistance to SARS-CoV-2 in human nasal tissue",
"author": "Shi",
"doi-asserted-by": "crossref",
"first-page": "889",
"journal-title": "Nat. Commun.",
"key": "10.1016/j.isci.2025.113318_bib49",
"volume": "15",
"year": "2024"
},
{
"DOI": "10.1128/jvi.02077-21",
"article-title": "Omicron: What Makes the Latest SARS-CoV-2 Variant of Concern So Concerning?",
"author": "Jung",
"doi-asserted-by": "crossref",
"journal-title": "J. Virol.",
"key": "10.1016/j.isci.2025.113318_bib50",
"volume": "96",
"year": "2022"
},
{
"DOI": "10.1016/S0140-6736(22)00017-4",
"article-title": "Early assessment of the clinical severity of the SARS-CoV-2 omicron variant in South Africa: a data linkage study",
"author": "Wolter",
"doi-asserted-by": "crossref",
"first-page": "437",
"journal-title": "Lancet",
"key": "10.1016/j.isci.2025.113318_bib51",
"volume": "399",
"year": "2022"
},
{
"DOI": "10.1016/j.amsu.2022.103737",
"article-title": "Understanding the omicron variant (B.1.1.529) of SARS-CoV-2: Mutational impacts, concerns, and the possible solutions",
"author": "Islam",
"doi-asserted-by": "crossref",
"journal-title": "Ann. Med. Surg.",
"key": "10.1016/j.isci.2025.113318_bib52",
"volume": "78",
"year": "2022"
},
{
"DOI": "10.1006/jmbi.1993.1626",
"article-title": "Comparative protein modelling by satisfaction of spatial restraints",
"author": "Sali",
"doi-asserted-by": "crossref",
"first-page": "779",
"journal-title": "J. Mol. Biol.",
"key": "10.1016/j.isci.2025.113318_bib54",
"volume": "234",
"year": "1993"
},
{
"DOI": "10.1021/acs.jcim.3c01153",
"article-title": "AmberTools",
"author": "Case",
"doi-asserted-by": "crossref",
"first-page": "6183",
"journal-title": "J. Chem. Inf. Model.",
"key": "10.1016/j.isci.2025.113318_bib55",
"volume": "63",
"year": "2023"
},
{
"DOI": "10.1002/pro.4792",
"article-title": "UCSF ChimeraX: Tools for structure building and analysis",
"author": "Meng",
"doi-asserted-by": "crossref",
"first-page": "e4792",
"journal-title": "Protein Sci.",
"key": "10.1016/j.isci.2025.113318_bib56",
"volume": "32",
"year": "2023"
},
{
"DOI": "10.1128/JVI.00957-20",
"article-title": "Replication of Severe Acute Respiratory Syndrome Coronavirus 2 in Human Respiratory Epithelium",
"author": "Milewska",
"doi-asserted-by": "crossref",
"first-page": "e00957-20",
"journal-title": "J. Virol.",
"key": "10.1016/j.isci.2025.113318_bib57",
"volume": "94",
"year": "2020"
},
{
"DOI": "10.1038/s41467-022-28528-w",
"article-title": "Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "871",
"journal-title": "Nat. Commun.",
"key": "10.1016/j.isci.2025.113318_bib58",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.1038/s41422-022-00672-4",
"article-title": "Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins",
"author": "Xu",
"doi-asserted-by": "crossref",
"first-page": "609",
"journal-title": "Cell Res.",
"key": "10.1016/j.isci.2025.113318_bib59",
"volume": "32",
"year": "2022"
},
{
"DOI": "10.1038/s41467-023-42527-5",
"article-title": "Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "7574",
"journal-title": "Nat. Commun.",
"key": "10.1016/j.isci.2025.113318_bib60",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.1042/BJ20041955",
"article-title": "Hepatocyte growth factor is a preferred in vitro substrate for human hepsin, a membrane-anchored serine protease implicated in prostate and ovarian cancers",
"author": "Herter",
"doi-asserted-by": "crossref",
"first-page": "125",
"journal-title": "Biochem. J.",
"key": "10.1016/j.isci.2025.113318_bib61",
"volume": "390",
"year": "2005"
},
{
"DOI": "10.1021/ja026939x",
"article-title": "HADDOCK: a protein-protein docking approach based on biochemical or biophysical information",
"author": "Dominguez",
"doi-asserted-by": "crossref",
"first-page": "1731",
"journal-title": "J. Am. Chem. Soc.",
"key": "10.1016/j.isci.2025.113318_bib62",
"volume": "125",
"year": "2003"
},
{
"DOI": "10.1016/j.jmb.2015.09.014",
"article-title": "The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes",
"author": "van Zundert",
"doi-asserted-by": "crossref",
"first-page": "720",
"journal-title": "J. Mol. Biol.",
"key": "10.1016/j.isci.2025.113318_bib63",
"volume": "428",
"year": "2016"
},
{
"DOI": "10.1021/jz501780a",
"article-title": "Building Water Models: A Different Approach",
"author": "Izadi",
"doi-asserted-by": "crossref",
"first-page": "3863",
"journal-title": "J. Phys. Chem. Lett.",
"key": "10.1016/j.isci.2025.113318_bib64",
"volume": "5",
"year": "2014"
},
{
"DOI": "10.1021/ct400314y",
"article-title": "Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald",
"author": "Salomon-Ferrer",
"doi-asserted-by": "crossref",
"first-page": "3878",
"journal-title": "J. Chem. Theory Comput.",
"key": "10.1016/j.isci.2025.113318_bib65",
"volume": "9",
"year": "2013"
},
{
"DOI": "10.1039/D0NR03969A",
"article-title": "Quantitative determination of mechanical stability in the novel coronavirus spike protein",
"author": "Moreira",
"doi-asserted-by": "crossref",
"first-page": "16409",
"journal-title": "Nanoscale",
"key": "10.1016/j.isci.2025.113318_bib53",
"volume": "12",
"year": "2020"
},
{
"DOI": "10.1063/5.0040966",
"article-title": "A compression strategy for particle mesh Ewald theory",
"author": "Simmonett",
"doi-asserted-by": "crossref",
"journal-title": "J. Chem. Phys.",
"key": "10.1016/j.isci.2025.113318_bib66",
"volume": "154",
"year": "2021"
},
{
"DOI": "10.1021/ct9001015",
"article-title": "Staggered Mesh Ewald: An extension of the Smooth Particle-Mesh Ewald method adding great versatility",
"author": "Cerutti",
"doi-asserted-by": "crossref",
"first-page": "2322",
"journal-title": "J. Chem. Theory Comput.",
"key": "10.1016/j.isci.2025.113318_bib67",
"volume": "5",
"year": "2009"
},
{
"DOI": "10.1021/ct800573m",
"article-title": "Bad Seeds Sprout Perilous Dynamics: Stochastic Thermostat Induced Trajectory Synchronization in Biomolecules",
"author": "Sindhikara",
"doi-asserted-by": "crossref",
"first-page": "1624",
"journal-title": "J. Chem. Theory Comput.",
"key": "10.1016/j.isci.2025.113318_bib68",
"volume": "5",
"year": "2009"
},
{
"DOI": "10.1016/j.cplett.2003.12.039",
"article-title": "Molecular dynamics simulations of water and biomolecules with a Monte Carlo constant pressure algorithm",
"author": "Åqvist",
"doi-asserted-by": "crossref",
"first-page": "288",
"journal-title": "Chem. Phys. Lett.",
"key": "10.1016/j.isci.2025.113318_bib69",
"volume": "384",
"year": "2004"
},
{
"DOI": "10.1016/0021-9991(77)90098-5",
"article-title": "Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes",
"author": "Jean-Paul",
"doi-asserted-by": "crossref",
"first-page": "327",
"journal-title": "J. Comput. Phys.",
"key": "10.1016/j.isci.2025.113318_bib70",
"volume": "23",
"year": "1977"
},
{
"DOI": "10.1371/journal.pcbi.1005659",
"article-title": "OpenMM 7: Rapid development of high performance algorithms for molecular dynamics",
"author": "Eastman",
"doi-asserted-by": "crossref",
"journal-title": "PLoS Comput. Biol.",
"key": "10.1016/j.isci.2025.113318_bib71",
"volume": "13",
"year": "2017"
},
{
"DOI": "10.1021/ct5010406",
"article-title": "Long-Time-Step Molecular Dynamics through Hydrogen Mass Repartitioning",
"author": "Hopkins",
"doi-asserted-by": "crossref",
"first-page": "1864",
"journal-title": "J. Chem. Theory Comput.",
"key": "10.1016/j.isci.2025.113318_bib72",
"volume": "11",
"year": "2015"
},
{
"author": "Moreira",
"key": "10.1016/j.isci.2025.113318_bib73",
"series-title": "All atom simulations snapshots and contact maps analysis scripts for SARS-CoV-2002 and SARS-CoV-2 spike proteins with and without ACE2 enzyme",
"year": "2020"
},
{
"DOI": "10.1021/acs.jctc.6b00986",
"article-title": "Combining the MARTINI and Structure-Based Coarse-Grained Approaches for the Molecular Dynamics Studies of Conformational Transitions in Proteins",
"author": "Poma",
"doi-asserted-by": "crossref",
"first-page": "1366",
"journal-title": "J. Chem. Theor. Comput.",
"key": "10.1016/j.isci.2025.113318_bib74",
"volume": "13",
"year": "2017"
}
],
"reference-count": 74,
"references-count": 74,
"relation": {},
"resource": {
"primary": {
"URL": "https://linkinghub.elsevier.com/retrieve/pii/S2589004225015792"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Evolution of the SARS-CoV-2 spike protein in utilizing host transmembrane serine proteases",
"type": "journal-article",
"update-policy": "https://doi.org/10.1016/elsevier_cm_policy",
"volume": "28"
}