The Enigmatic Conserved Q134-F135-N137 Triad in SARS-CoV-2 Spike Protein: A Conformational Transducer?
et al., Biomolecules, doi:10.3390/biom16010111, Jan 2026
Computational and structural study of the SARS-CoV-2 spike protein identifying a conserved amino acid triad (Q134-F135-N137) that remains unchanged across variants despite extensive mutations in surrounding regions. The study proposes this triad initiates conformational changes in the spike protein after binding to host cell gangliosides, leading to exposure of the receptor-binding domain necessary for viral entry. Using molecular modeling, authors identify a pathway of amino acids that could transmit conformational changes from the triad through the protein structure. They suggest this conserved region could serve as a therapeutic target since it appears essential for viral function yet resistant to mutation.
Lefebvre et al., 8 Jan 2026, France, peer-reviewed, 4 authors.
Contact: jacques.fantini@univ-amu.fr (corresponding author), marine.lefebvre@etu.univ-amu.fr, nouara.yahi@univ-amu.fr.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
The Enigmatic Conserved Q134-F135-N137 Triad in SARS-CoV-2 Spike Protein: A Conformational Transducer?
Biomolecules, doi:10.3390/biom16010111
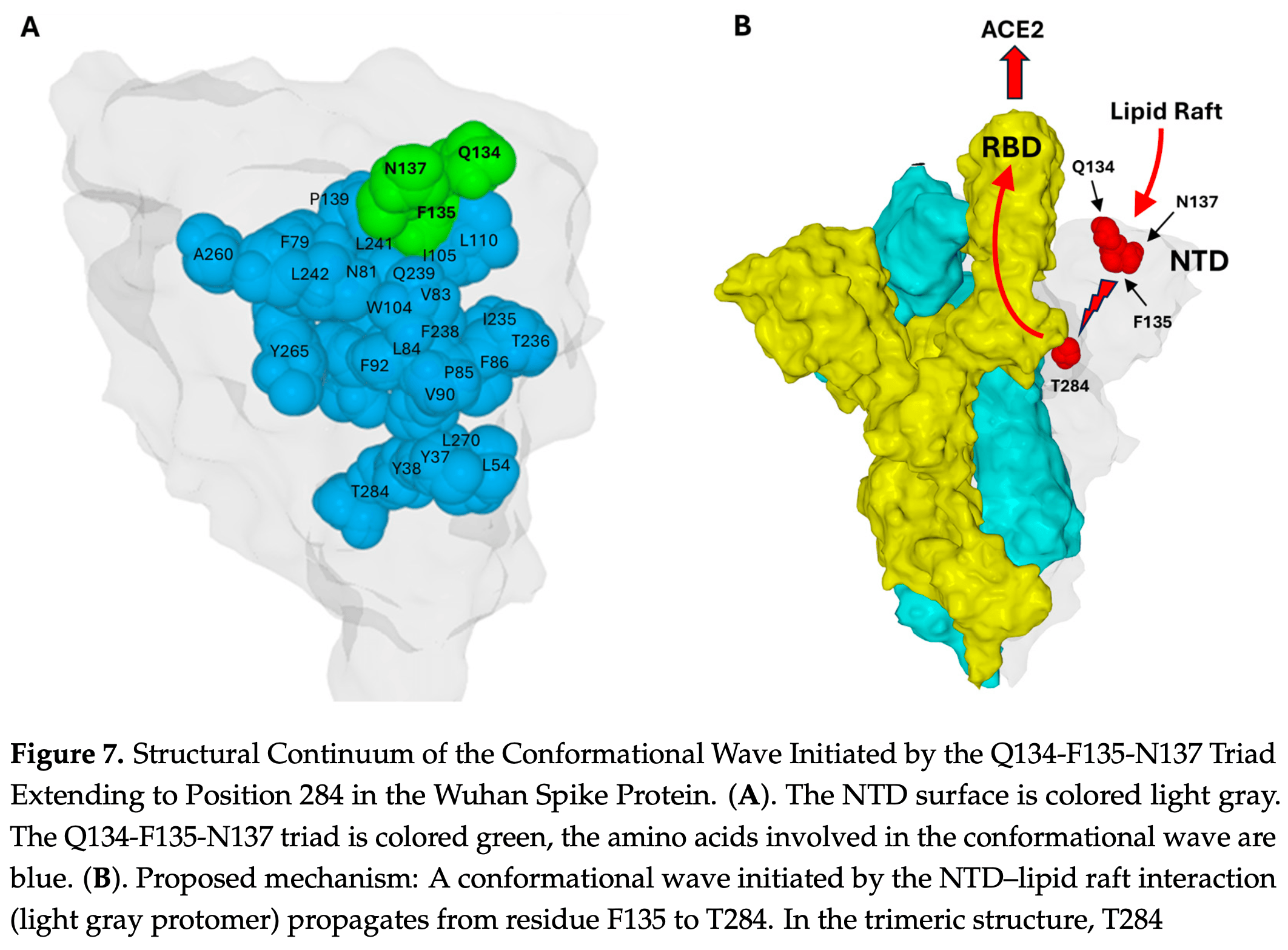
Lipid raft-associated gangliosides facilitate the early stages of SARS-CoV-2 entry by triggering the exposure of the receptor-binding domain (RBD) within the trimeric spike protein, which is initially sequestered. A broad range of in silico, cryoelectron microscopy and physicochemical approaches indicate that the RBD becomes accessible after a gangliosideinduced conformational rearrangement originating in the N-terminal domain (NTD) of one protomer and propagating to the neighboring RBD. We previously identified a triad of amino acids, Q134-F135-N137, as a strictly conserved element on the NTD. In the present review, we integrate a series of structural and experimental data revealing that this triad may act as a conformational transducer connected to a chain of residues that are capable of transmitting an internal conformational wave within the NTD. This wave is generated at the triad level after physical interactions with lipid raft gangliosides of the host cell membrane. It propagates inside the NTD and collides with the RBD of a neighboring protomer, triggering its unmasking. We also identify a chain of aromatic residues that are capable of controlling electron transfer through the NTD, leading us to hypothesize the existence of a dual conformational/quantum wave. In conclusion, the complete conservation of the Q134-F135-N137 triad despite six years of extensive NTD remodeling underscores its critical role in the viral life cycle. This triad represents a potential Achilles' heel within the hyper-variable NTD, offering a stable target for therapeutic or vaccinal interventions to disrupt the conformational wave and prevent infection. These possibilities are discussed.
Author Contributions: Conceptualization, J.F.; methodology, J.F., M.L., N.Y. and H.C.; validation, J.F., N.Y. and H.C.; draft preparation, J.F. and M.L.; writing-review and editing, J.F.; visualization, M.L.; supervision, J.F.; project administration, J.F. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest: The authors declare no conflict of interest.
Abbreviations The following abbreviations are used in this manuscript:
ACE-2 Angiotensin
References
Akke, Weininger, NMR Studies of Aromatic Ring Flips to Probe Conformational Fluctuations in Proteins, J. Phys. Chem. B, doi:10.1021/acs.jpcb.2c07258
Andrews, Harrison, Modeling conformational change in macromolecules as an elastic deformation, Proteins Struct. Funct. Bioinform, doi:10.1002/prot.340100210
Azzaz, Fantini, The epigenetic dimension of protein structure, Biomol. Concepts, doi:10.1515/bmc-2022-0006
Azzaz, Mazzarino, Chahinian, Yahi, Scala et al., Structure of the Myelin Sheath Proteolipid Plasmolipin (PLLP) in a Ganglioside-Containing Lipid Raft, Front. Biosci.-Landmark, doi:10.31083/j.fbl2808157
Baglivo, Baronio, Natalini, Beccari, Chiurazzi et al., Natural small molecules as inhibitors of coronavirus lipid-dependent attachment to host cells: A possible strategy for reducing SARS-CoV-2 infectivity?, Acta Bio Medica Atenei Parm
Bakillah, Hejji, Almasaud, Jami, Hawwari et al., Lipid raft integrity and cellular cholesterol homeostasis are critical for SARS-CoV-2 entry into cells, Nutrients, doi:10.3390/nu14163417
Banoun, Evolution of SARS-CoV-2: Review of mutations, role of the host immune system, Nephron
Barnes, West, Huey-Tubman, Hoffmann, Sharaf et al., Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies, Cell, doi:10.1016/j.cell.2020.06.025
Beitinger, Vogel, Möbius, Rahmann, Surface potentials and electric dipole moments of ganglioside and phospholipid monolayers: Contribution of the polar headgroup at the water/lipid interface, Biochim. Biophys. Acta (BBA)-Biomembr, doi:10.1016/0005-2736(89)90296-4
Benjamin, Gerhards, Solov'yov, Hore, Magnetosensitivity of Model Flavin-Tryptophan Radical Pairs in a Dynamic Protein Environment, J. Phys. Chem. B, doi:10.1021/acs.jpcb.5c01187
Božič, Podgornik, Changes in total charge on spike protein of SARS-CoV-2 in emerging lineages, Bioinform. Adv, doi:10.1093/bioadv/vbae053
Brown, London, Functions of lipid rafts in biological membranes, Annu. Rev. Cell Dev. Biol, doi:10.1146/annurev.cellbio.14.1.111
Cai, Zhang, Xiao, Peng, Sterling et al., Distinct conformational states of SARS-CoV-2 spike protein, Science, doi:10.1126/science.abd4251
Cerutti, Guo, Zhou, Gorman, Lee et al., Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite, Cell Host Microbe, doi:10.1016/j.chom.2021.03.005
Chahinian, Yahi, Fantini, Glutamate, Gangliosides, and the Synapse: Electrostatics at Work in the Brain, Int. J. Mol. Sci, doi:10.3390/ijms25168583
Chazal, Gerlier, Virus entry, assembly, budding, and membrane rafts, Microbiol. Mol. Biol. Rev, doi:10.1128/MMBR.67.2.226-237.2003
Chi, Yan, Zhang, Zhang, Zhang et al., A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2, Science, doi:10.1126/science.abc6952
Choi, Aizaki, Lai, Murine coronavirus requires lipid rafts for virus entry and cell-cell fusion but not for virus release, J. Virol, doi:10.1128/JVI.79.15.9862-9871.2005
Choi, Kim, Rhee, Park, Kim et al., Molecular Dynamics Studies on the structural characteristics for the stability prediction of SARS-CoV-2, Int. J. Mol. Sci, doi:10.3390/ijms22168714
Clarke, Sethi, Li, Kumar, Chang et al., Identifying allosteric hotspots with dynamics: Application to inter-and intra-species conservation, Structure, doi:10.1016/j.str.2016.03.008
Colson, Chaudet, Delerce, Pontarotti, Levasseur et al., Role of SARS-CoV-2 mutations in the evolution of the COVID-19 pandemic, J. Infect, doi:10.1016/j.jinf.2024.106150
Colson, Delerce, Beye, Levasseur, Boschi et al., First cases of infection with the 21L/BA.2 Omicron variant in Marseille, France, J. Med. Virol, doi:10.1002/jmv.27695
Colson, Fournier, Delerce, Million, Bedotto et al., Culture and identification of a "Deltamicron" SARS-CoV-2 in a three cases cluster in southern France, J. Med. Virol, doi:10.1002/jmv.27789
Colson, La Scola, Beye, Delerce, Raoult et al., Emergence of a second SARS-CoV-2 variant with a tremendous genetic leap from its ancestors, J. Med. Virol, doi:10.1002/jmv.29124
Cordonnier, Montagnier, Emerman, Single amino-acid changes in HIV envelope affect viral tropism and receptor binding, Nature, doi:10.1038/340571a0
Cosgriff, Brasier, Pi, Dogovski, Sarsero et al., A study of AroP-PheP chimeric proteins and identification of a residue involved in tryptophan transport, J. Bacteriol, doi:10.1128/JB.182.8.2207-2217.2000
Dalton, Lans, Giraldo, Quantifying conformational changes in GPCRs: Glimpse of a common functional mechanism, BMC Bioinform, doi:10.1186/s12859-015-0567-3
Das, Mukhopadhyay, Comparison and Possible Binding Orientations of SARS-CoV-2 Spike N-Terminal Domain for Gangliosides GM3 and GM1, J. Phys. Chem. B
Das, Mukhopadhyay, Identification of possible binding modes of SARS-CoV-2 spike N-terminal domain for ganglioside GM1, Chem. Phys. Lett, doi:10.1016/j.cplett.2022.140260
Deshpande, Harris, Martinez-Sobrido, Kobie, Walter, Epitope Classification and RBD Binding Properties of Neutralizing Antibodies Against SARS-CoV-2 Variants of Concern, Front. Immunol, doi:10.3389/fimmu.2021.691715
Dey, Sharma, Dhanawat, Gupta, Harshan et al., Synergistic binding of SARS-CoV-2 to ACE2 and gangliosides in native lipid membranes, ACS Infect. Dis, doi:10.1021/acsinfecdis.3c00519
Ding, Chen, Miao, Qi, Research advances on the role of lipids in the life cycle of human coronaviruses, Microorganisms, doi:10.3390/microorganisms12010063
Ding, Xu, Da Silva-Junior, Liu, Zhan, Medicinal chemistry insights into antiviral peptidomimetics, Drug Discov. Today, doi:10.1016/j.drudis.2022.103468
Díaz-Salinas, Jain, Durham, Munro, Single-molecule imaging reveals allosteric stimulation of SARS-CoV-2 spike receptor binding domain by host sialic acid, Sci. Adv, doi:10.1126/sciadv.adk4920
Espinosa-Gongora, Berg, Rehn, Varg, Dillner et al., Early detection of the emerging SARS-CoV-2 BA.2.86 lineage through integrated genomic surveillance of wastewater and COVID-19 cases in Sweden, weeks 31 to 38 2023, Euro Surveill. Bull. Eur. Sur Les. Mal. Transm. Eur. Commun. Dis. Bull, doi:10.2807/1560-7917.ES.2023.28.46.2300595
Fantini, Azzaz, Chahinian, Yahi, Electrostatic Surface Potential as a Key Parameter in Virus Transmission and Evolution: How to Manage Future Virus Pandemics in the Post-COVID-19 Era, Viruses, doi:10.3390/v15020284
Fantini, Chahinian, Yahi, Convergent Evolution Dynamics of SARS-CoV-2 and HIV Surface Envelope Glycoproteins Driven by Host Cell Surface Receptors and Lipid Rafts: Lessons for the Future, Int. J. Mol. Sci
Fantini, Chahinian, Yahi, Leveraging coronavirus binding to gangliosides for innovative vaccine and therapeutic strategies against COVID-19, Biochem. Biophys. Res. Commun, doi:10.1016/j.bbrc.2020.10.015
Fantini, Chahinian, Yahi, Synergistic antiviral effect of hydroxychloroquine and azithromycin in combination against SARS-CoV-2: What molecular dynamics studies of virus-host interactions reveal, Int. J. Antimicrob. Agents, doi:10.1016/j.ijantimicag.2020.106020
Fantini, Di Scala, Chahinian, Yahi, Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection, Int. J. Antimicrob. Agents, doi:10.1016/j.ijantimicag.2020.105960
Fantini, Lipid rafts and human diseases: Why we need to target gangliosides, FEBS Open Bio
Fantini, Matveeva, Lefebvre, Chahinian, What Is life? Rethinking Biology in Light of Fundamental Parameters, Life, doi:10.3390/life14030280
Fantini, Yahi, Azzaz, Chahinian, Structural dynamics of SARS-CoV-2 variants: A health monitoring strategy for anticipating Covid-19 outbreaks, J. Infect, doi:10.1016/j.jinf.2021.06.001
Fantini, Yahi, Colson, Chahinian, La Scola et al., The puzzling mutational landscape of the SARS-2-variant Omicron, J. Med. Virol
Fačkovec, Vondrášek, Optimal definition of inter-residual contact in globular proteins based on pairwise interaction energy calculations, its robustness, and applications, J. Phys. Chem. B, doi:10.1021/jp303088n
Goldswain, Dong, Penrice-Randal, Alruwaili, Shawli et al., The P323L substitution in the SARS-CoV-2 polymerase (NSP12) confers a selective advantage during infection, Genome Biol, doi:10.1186/s13059-023-02881-5
Gosline, Lillie, Carrington, Guerette, Ortlepp et al., Elastic proteins: Biological roles and mechanical properties, Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci, doi:10.1098/rstb.2001.1022
Gray, Winkler, Long-range electron transfer, Proc. Natl. Acad. Sci, doi:10.1073/pnas.0408029102
Guex, Peitsch, SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling, Electrophoresis, doi:10.1002/elps.1150181505
Guo, Huang, Yuan, Wei, Gao et al., The important role of lipid raft-mediated attachment in the infection of cultured cells by coronavirus infectious bronchitis virus beaudette strain, PLoS ONE, doi:10.1371/journal.pone.0170123
Guérin, Yahi, Azzaz, Chahinian, Sabatier et al., Structural dynamics of the SARS-CoV-2 spike protein: A 2-year retrospective analysis of SARS-CoV-2 variants (from Alpha to Omicron) reveals an early divergence between conserved and variable epitopes, Molecules, doi:10.3390/molecules27123851
Hamid, Chaudhary, Pandini, Khan, Allosteric Network Analysis Toolkit for Single-Domain Phosphoproteins
Haque, Chaurasia, Wessel, Iii; Iuvone, Dual regulation of cryptochrome 1 mRNA expression in chicken retina by light and circadian oscillators, Neuroreport, doi:10.1097/00001756-200212030-00016
Harvey, Carabelli, Jackson, Gupta, Thomson et al., SARS-CoV-2 variants, spike mutations and immune escape, Nat. Rev. Microbiol, doi:10.1038/s41579-021-00573-0
He, Guo, Guo, Chang, Wang et al., Femtosecond dynamics of short-range protein electron transfer in flavodoxin, Biochemistry, doi:10.1021/bi401137u
Howard Megan, Travanty Emily, Jeffers Scott, Smith, Wennier Sonia et al., Aromatic Amino Acids in the Juxtamembrane Domain of Severe Acute Respiratory Syndrome Coronavirus Spike Glycoprotein Are Important for Receptor-Dependent Virus Entry and Cell-Cell Fusion, J. Virol, doi:10.1128/JVI.01805-07
Ikonen, Roles of lipid rafts in membrane transport, Curr. Opin. Cell Biol, doi:10.1016/s0955-0674(00)00238-6
Jin, Hassan, Li, Liu, Marakhovskaia et al., Human coronavirus HKU1 spike structures reveal the basis for sialoglycan specificity and carbohydrate-promoted conformational changes, Nat. Commun, doi:10.1038/s41467-025-59137-y
Juhola, Postila, Rissanen, Lolicato, Vattulainen et al., Negatively Charged Gangliosides Promote Membrane Association of Amphipathic Neurotransmitters, Neuroscience, doi:10.1016/j.neuroscience.2018.05.035
Kaya, Lokits, Gilbert, Iverson, Meiler et al., A conserved phenylalanine as a relay between the α5 helix and the GDP binding region of heterotrimeric Gi protein α subunit, J. Biol. Chem
Ke, Oton, Qu, Cortese, Zila et al., Structures and distributions of SARS-CoV-2 spike proteins on intact virions, Nature, doi:10.1038/s41586-020-2665-2
Korber, Fischer, Gnanakaran, Yoon, Theiler et al., Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus, Cell, doi:10.1016/j.cell.2020.06.043
Krishnan, Aksimentiev, Lindsay, Matyushov, Long-Range Conductivity in Proteins Mediated by Aromatic Residues, ACS Phys. Chem. Au, doi:10.1021/acsphyschemau.3c00017
Kulkarni, Wiemer, Chang, Role of lipid rafts in pathogen-host interaction-a mini review, Front. Immunol, doi:10.3389/fimmu.2021.815020
Lam, Waman, Fraternali, Orengo, Lees, Structural and energetic analyses of SARS-CoV-2 N-terminal domain characterise sugar binding pockets and suggest putative impacts of variants on COVID-19 transmission, Comput. Struct. Biotechnol. J, doi:10.1016/j.csbj.2022.11.004
Lauster, Haag, Ballauff, Herrmann, Balancing stability and function: Impact of the surface charge of SARS-CoV-2 Omicron spike protein, npj Viruses, doi:10.1038/s44298-025-00104-1
Lauster, Osterrieder, Haag, Ballauff, Herrmann, Respiratory viruses interacting with cells: The importance of electrostatics, Front. Microbiol
Lefebvre, Chahinian, La Scola, Fantini, Characterization and Fluctuations of an Ivermectin Binding Site at the Lipid Raft Interface of the N-Terminal Domain (NTD) of the Spike Protein of SARS-CoV-2 Variants, Viruses, doi:10.3390/v16121836
Li, Zhu, Fan, Zhang, Peng et al., Dependence of SARS-CoV-2 infection on cholesterol-rich lipid raft and endosomal acidification, Comput. Struct. Biotechnol. J, doi:10.1016/j.csbj.2021.04.001
Lu, Liu, Tam, Lipid rafts are involved in SARS-CoV entry into Vero E6 cells, Biochem. Biophys. Res. Commun, doi:10.1016/j.bbrc.2008.02.023
Maggio, Cumar, Caputto, Surface behaviour of gangliosides and related glycosphingolipids, Biochem. J, doi:10.1042/bj1710559
Maggio, The surface behavior of glycosphingolipids in biomembranes: A new frontier of molecular ecology, Prog. Biophys. Mol. Biol, doi:10.1016/0079-6107(94)90006-X
Malhotra, Scott, Zavorotinskaya, Albritton, Analysis of the murine ecotropic leukemia virus receptor reveals a common biochemical determinant on diverse cell surface receptors that is essential to retrovirus entry, J. Virol, doi:10.1128/jvi.70.1.321-326.1996
Matveeva, Lefebvre, Chahinian, Yahi, Fantini, Host membranes as drivers of virus evolution, Viruses, doi:10.3390/v15091854
Mañes, Del Real, Martínez-A, Pathogens: Raft hijackers, Nat. Rev. Immunol, doi:10.1038/nri1129
Mccallum, De Marco, Lempp, Tortorici, Pinto et al., N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2, Cell, doi:10.1016/j.cell.2021.03.028
Mlinac-Jerkovic, Ilic, Zjalić, Mandić, Debeljak et al., Who's in, who's out? Re-evaluation of lipid raft residents, J. Neurochem
Nadezhdin, Neuberger, Trofimov, Krylov, Sinica et al., Structural mechanism of heat-induced opening of a temperature-sensitive TRP channel, Nat. Struct. Mol. Biol, doi:10.1038/s41594-021-00615-4
Nardacci, Colavita, Castilletti, Lapa, Matusali et al., Evidences for lipid involvement in SARS-CoV-2 cytopathogenesis, Cell Death Dis
Negi, Pandey, Potharaju, Jaiswal, Harshan et al., SARS-CoV-2 Evolved Variants Bind to Sialylated Gangliosides and Are Inhibited by a Tetravalent Sialo-Glycocluster, ACS Infect. Dis, doi:10.1021/acsinfecdis.5c00143
Negi, Sharma, Chaudhary, Gupta, Harshan et al., SARS-CoV-2 binding to terminal sialic acid of gangliosides embedded in lipid membranes, ACS Infect. Dis, doi:10.1021/acsinfecdis.3c00106
Nguyen, Mccord, Bui, Bouwman, Kitova et al., Sialic acid-containing glycolipids mediate binding and viral entry of SARS-CoV-2, Nat. Chem. Biol, doi:10.1038/s41589-021-00924-1
Nieto-Garai, Contreras, Arboleya, Lorizate, Role of protein-lipid interactions in viral entry, Adv. Biol, doi:10.1002/adbi.202101264
Noy-Porat, Mechaly, Levy, Makdasi, Alcalay et al., Therapeutic antibodies, targeting the SARS-CoV-2 spike N-terminal domain, protect lethally infected K18-hACE2 mice, Iscience
Nussinov, Tsai, Ma, The underappreciated role of allostery in the cellular network, Annu. Rev. Biophys, doi:10.1146/annurev-biophys-083012-130257
Ortega, Yang, Ames, Baudry, Parkinson et al., A phenylalanine rotameric switch for signal-state control in bacterial chemoreceptors, Nat. Commun, doi:10.1016/j.proche.2011.08.034
Ouyang, Tan, Lei, Song, Kieffer et al., Probing the biophysical constraints of SARS-CoV-2 spike N-terminal domain using deep mutational scanning, Sci. Adv, doi:10.1126/sciadv.add7221
Pachetti, Marini, Benedetti, Giudici, Mauro et al., Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant, J. Transl. Med
Palacios-Rapalo, De Jesus-Gonzalez, Cordero-Rivera, Farfan-Morales, Osuna-Ramos et al., Cholesterol-rich lipid rafts as platforms for SARS-CoV-2 entry, Front. Immunol, doi:10.3389/fimmu.2021.796855
Pandini, Kleinjung, Rasool, Khan, Coevolved mutations reveal distinct architectures for two core proteins in the bacterial flagellar motor, PLoS ONE, doi:10.1371/journal.pone.0142407
Paoli, Modesti, Magherini, Gamberi, Caselli et al., Site-directed mutagenesis of two aromatic residues lining the active site pocket of the yeast Ltp1, Biochim. Biophys. Acta, doi:10.1016/j.bbagen.2006.12.012
Pascarella, Ciccozzi, Bianchi, Benvenuto, Cauda et al., The value of electrostatic potentials of the spike receptor binding and N-terminal domains in addressing transmissibility and infectivity of SARS-CoV-2 variants of concern, J. Infect, doi:10.1016/j.jinf.2022.02.023
Pike, Rafts defined: A report on the Keystone Symposium on Lipid Rafts and Cell Function, J. Lipid Res, doi:10.1194/jlr.E600002-JLR200
Piovesan, Minervini, Tosatto, Hobza, Vondrášek, Identifying stabilizing key residues in proteins using interresidue interaction energy matrix, Proteins Struct. Funct. Bioinform, doi:10.1002/prot.21938
Pristerá, Okuse, Building excitable membranes: Lipid rafts and multiple controls on trafficking of electrogenic molecules, Neurosci, doi:10.1177/1073858410393977
Pérez-López, Nieto-Suárez, Mestres, Alsina, Haro et al., Behaviour of a peptide sequence from the GB virus C/hepatitis G virus E2 protein in Langmuir monolayers: Its interaction with phospholipid membrane models, Biophys. Chem, doi:10.1016/j.bpc.2009.01.007
Qing, Hantak, Perlman, Gallagher, Distinct Roles for Sialoside and Protein Receptors in Coronavirus Infection, mBio, doi:10.1128/mBio.02764-19
Reis, Holmberg, Watzke, Leser, Miller, Lipases at interfaces: A review, Adv. Colloid Interface Sci, doi:10.1016/j.cis.2008.06.001
Ripa, Andreu, López-Guerrero, Bello-Morales, Membrane rafts: Portals for viral entry, Front. Microbiol, doi:10.3389/fmicb.2021.631274
Sainz, Jr, Rausch, Gallaher, Garry et al., The aromatic domain of the coronavirus class I viral fusion protein induces membrane permeabilization: Putative role during viral entry, Biochemistry, doi:10.1021/bi048515g
Sato, Serizawa, Okahata, Binding of influenza A virus to monosialoganglioside (GM3) reconstituted in glucosylceramide and sphingomyelin membranes, Biochim. Biophys. Acta (BBA)-Biomembr, doi:10.1016/S0005-2736(96)00138-1
Secundo, Conformational changes of enzymes upon immobilisation, Chem. Soc. Rev, doi:10.1039/c3cs35495d
Seyran, Takayama, Uversky, Lundstrom, Palù et al., The structural basis of accelerated host cell entry by SARS-CoV-2, FEBS J, doi:10.1111/febs.15651
Sezgin, Levental, Mayor, Eggeling, The mystery of membrane organization: Composition, regulation and roles of lipid rafts, Nat. Rev. Mol. Cell Biol, doi:10.1038/nrm.2017.16
Sharma, Seven, Micic, Li, Leblanc, Surface chemistry and spectroscopic study of a cholera toxin B Langmuir monolayer, Langmuir ACS J. Surf. Colloids
Siddiqui, Musharaf, Gulumbe, The, 1 variant of COVID-19: Immune evasion, transmissibility, and implications for global health, Ther. Adv. Infect. Dis, doi:10.1177/20499361251314763
Simons, Ikonen, Functional rafts in cell membranes, Nature, doi:10.1038/42408
Simons, Toomre, Lipid rafts and signal transduction, Nat. Rev. Mol. Cell Biol, doi:10.1038/35036052
Sorice, Misasi, Riitano, Manganelli, Martellucci et al., Targeting lipid rafts as a strategy against coronavirus, Front. Cell Dev. Biol, doi:10.3389/fcell.2020.618296
Stahl, Sieber, An amino acid domino effect orchestrates ClpP's conformational states, Curr. Opin. Chem. Biol, doi:10.1016/j.cbpa.2017.08.007
Sun, The role of cell surface sialic acids for SARS-CoV-2 infection, Glycobiology, doi:10.1093/glycob/cwab032
Sviridov, Mukhamedova, Miller, Lipid rafts as a therapeutic target: Thematic review series: Biology of lipid rafts, J. Lipid Res, doi:10.1194/jlr.TR120000658
Takeda, Proteolytic activation of SARS-CoV-2 spike protein, Microbiol. Immunol
Tazhigulov, Gayvert, Wei, Bravaya, Emap, A web application for identifying and visualizing electron or hole hopping pathways in proteins, J. Phys. Chem. B, doi:10.1021/acs.jpcb.9b04816
Tee, Dong, Guarnera, Berezovsky, On the relationship between protein stability, thermostability, and allosteric signalling, J. Mol. Biol, doi:10.1016/j.jmb.2025.169537
Teixeira, Temerozo, Pereira-Dutra, Ferreira, Mattos et al., Simvastatin downregulates the SARS-CoV-2-induced inflammatory response and impairs viral infection through disruption of lipid rafts, Front. Immunol, doi:10.3389/fimmu.2022.820131
Tsai, Del Sol, Nussinov, Allostery, Absence of a Change in Shape Does Not Imply that Allostery Is Not at Play, J. Mol. Biol, doi:10.1016/j.jmb.2008.02.034
Van Stokkum, Kloz, Polli, Viola, Weißenborn et al., Vibronic dynamics resolved by global and target analysis of ultrafast transient absorption spectra, J. Chem. Phys, doi:10.1063/5.0060672
Vargas-Rosales, Caflisch, Domino Effect in Allosteric Signaling of Peptide Binding, J. Mol. Biol, doi:10.1016/j.jmb.2022.167661
Wang, Lu, Jiang, SARS-CoV-2 evolution from the BA.2.86 to JN.1 variants: Unexpected consequences, Trends Immunol, doi:10.1016/j.it.2024.01.003
Wang, Xu, Yu, A phenylalanine dynamic switch controls the interfacial activation of Rhizopus chinensis lipase, Int. J. Biol. Macromol, doi:10.1016/j.ijbiomac.2021.01.086
Wang, Yang, Liu, Guo, Zhang et al., SARS coronavirus entry into host cells through a novel clathrin-and caveolae-independent endocytic pathway, Cell Res, doi:10.1038/cr.2008.15
Winger, Caspari, The Spike of Concern-The Novel Variants of SARS-CoV-2, Viruses, doi:10.3390/v13061002
Wrapp, Wang, Corbett, Goldsmith, Hsieh et al., Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation, Science, doi:10.1126/science.abb2507
Xu, Jarocha, Zollitsch, Konowalczyk, Henbest et al., Magnetic sensitivity of cryptochrome 4 from a migratory songbird, Nature, doi:10.1038/s41586-021-03618-9
Xu, Wang, Liu, Zhang, Han et al., Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM, Sci. Adv, doi:10.1126/sciadv.abe5575
Xu, Zhang, How significant is a protein structure similarity with TM-score = 0.5?, Bioinformatics, doi:10.1093/bioinformatics/btq066
Yang, Jiao, Fan, Ye, Lv et al., Macrocyclic Compounds: Unveiling Their Distinctive Antiviral Advantages in Medicinal Research, J. Med. Chem
Yang, Li, Shen, Liu, Influenza A virus entry inhibitors targeting the hemagglutinin, Viruses, doi:10.3390/v5010352
Yu, Zheng, Zhou, Li, Chen et al., SARS-CoV-2 spike engagement of ACE2 primes S2 ′ site cleavage and fusion initiation, Proc. Natl. Acad. Sci, doi:10.1073/pnas.2111199119
Zandi, Hosseini, Soltani, Rasooli, Moghadami et al., The role of lipids in the pathophysiology of coronavirus infections, Osong Public Health Res. Perspect, doi:10.24171/j.phrp.2021.0153
Zhang, Cai, Xiao, Lu, Peng et al., Structural impact on SARS-CoV-2 spike protein by D614G substitution, Science, doi:10.1126/science.abf2303
Zhang, Skolnick, Scoring function for automated assessment of protein structure template quality, Proteins, doi:10.1002/prot.20264
Zhang, Xiao, Cai, Chen, Structure of SARS-CoV-2 spike protein, Curr. Opin. Virol, doi:10.1016/j.coviro.2021.08.010
DOI record:
{
"DOI": "10.3390/biom16010111",
"ISSN": [
"2218-273X"
],
"URL": "http://dx.doi.org/10.3390/biom16010111",
"abstract": "<jats:p>Lipid raft-associated gangliosides facilitate the early stages of SARS-CoV-2 entry by triggering the exposure of the receptor-binding domain (RBD) within the trimeric spike protein, which is initially sequestered. A broad range of in silico, cryoelectron microscopy and physicochemical approaches indicate that the RBD becomes accessible after a ganglioside-induced conformational rearrangement originating in the N-terminal domain (NTD) of one protomer and propagating to the neighboring RBD. We previously identified a triad of amino acids, Q134-F135-N137, as a strictly conserved element on the NTD. In the present review, we integrate a series of structural and experimental data revealing that this triad may act as a conformational transducer connected to a chain of residues that are capable of transmitting an internal conformational wave within the NTD. This wave is generated at the triad level after physical interactions with lipid raft gangliosides of the host cell membrane. It propagates inside the NTD and collides with the RBD of a neighboring protomer, triggering its unmasking. We also identify a chain of aromatic residues that are capable of controlling electron transfer through the NTD, leading us to hypothesize the existence of a dual conformational/quantum wave. In conclusion, the complete conservation of the Q134-F135-N137 triad despite six years of extensive NTD remodeling underscores its critical role in the viral life cycle. This triad represents a potential Achilles’ heel within the hyper-variable NTD, offering a stable target for therapeutic or vaccinal interventions to disrupt the conformational wave and prevent infection. These possibilities are discussed.</jats:p>",
"alternative-id": [
"biom16010111"
],
"author": [
{
"affiliation": [
{
"name": "IHU Méditerranée Infection, 19-21 Boulevard Jean Moulin, 13005 Marseille, France"
},
{
"name": "Microbes Evolution Phylogeny and Infections (MEPHI), Aix-Marseille University, 27 Boulevard Jean Moulin, 13005 Marseille, France"
}
],
"family": "Lefebvre",
"given": "Marine",
"sequence": "first"
},
{
"ORCID": "https://orcid.org/0000-0002-9516-4168",
"affiliation": [
{
"name": "Department of Biology, Faculty of Medicine, INSERM UA16, Aix-Marseille University, 13015 Marseille, France"
}
],
"authenticated-orcid": false,
"family": "Chahinian",
"given": "Henri",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-2800-5458",
"affiliation": [
{
"name": "Department of Biology, Faculty of Medicine, INSERM UA16, Aix-Marseille University, 13015 Marseille, France"
}
],
"authenticated-orcid": false,
"family": "Yahi",
"given": "Nouara",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-8653-5521",
"affiliation": [
{
"name": "Department of Biology, Faculty of Medicine, INSERM UA16, Aix-Marseille University, 13015 Marseille, France"
}
],
"authenticated-orcid": false,
"family": "Fantini",
"given": "Jacques",
"sequence": "additional"
}
],
"container-title": "Biomolecules",
"container-title-short": "Biomolecules",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2026,
1,
8
]
],
"date-time": "2026-01-08T10:10:50Z",
"timestamp": 1767867050000
},
"deposited": {
"date-parts": [
[
2026,
1,
8
]
],
"date-time": "2026-01-08T10:27:56Z",
"timestamp": 1767868076000
},
"indexed": {
"date-parts": [
[
2026,
1,
8
]
],
"date-time": "2026-01-08T12:37:11Z",
"timestamp": 1767875831198,
"version": "3.49.0"
},
"is-referenced-by-count": 0,
"issue": "1",
"issued": {
"date-parts": [
[
2026,
1,
8
]
]
},
"journal-issue": {
"issue": "1",
"published-online": {
"date-parts": [
[
2026,
1
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2026,
1,
8
]
],
"date-time": "2026-01-08T00:00:00Z",
"timestamp": 1767830400000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/2218-273X/16/1/111/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "111",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2026,
1,
8
]
]
},
"published-online": {
"date-parts": [
[
2026,
1,
8
]
]
},
"publisher": "MDPI AG",
"reference": [
{
"DOI": "10.1038/42408",
"article-title": "Functional rafts in cell membranes",
"author": "Simons",
"doi-asserted-by": "crossref",
"first-page": "569",
"journal-title": "Nature",
"key": "ref_1",
"volume": "387",
"year": "1997"
},
{
"DOI": "10.1038/35036052",
"article-title": "Lipid rafts and signal transduction",
"author": "Simons",
"doi-asserted-by": "crossref",
"first-page": "31",
"journal-title": "Nat. Rev. Mol. Cell Biol.",
"key": "ref_2",
"volume": "1",
"year": "2000"
},
{
"article-title": "Building excitable membranes: Lipid rafts and multiple controls on trafficking of electrogenic molecules",
"author": "Okuse",
"first-page": "70",
"journal-title": "Neurosci",
"key": "ref_3",
"volume": "18",
"year": "2012"
},
{
"DOI": "10.1016/S0955-0674(00)00238-6",
"article-title": "Roles of lipid rafts in membrane transport",
"author": "Ikonen",
"doi-asserted-by": "crossref",
"first-page": "470",
"journal-title": "Curr. Opin. Cell Biol.",
"key": "ref_4",
"volume": "13",
"year": "2001"
},
{
"DOI": "10.1146/annurev.cellbio.14.1.111",
"article-title": "Functions of lipid rafts in biological membranes",
"author": "Brown",
"doi-asserted-by": "crossref",
"first-page": "111",
"journal-title": "Annu. Rev. Cell Dev. Biol.",
"key": "ref_5",
"volume": "14",
"year": "1998"
},
{
"DOI": "10.1194/jlr.E600002-JLR200",
"article-title": "Rafts defined: A report on the Keystone Symposium on Lipid Rafts and Cell Function",
"author": "Pike",
"doi-asserted-by": "crossref",
"first-page": "1597",
"journal-title": "J. Lipid Res.",
"key": "ref_6",
"volume": "47",
"year": "2006"
},
{
"DOI": "10.1111/jnc.15446",
"article-title": "Who’s in, who’s out? Re-evaluation of lipid raft residents",
"author": "Ilic",
"doi-asserted-by": "crossref",
"first-page": "657",
"journal-title": "J. Neurochem.",
"key": "ref_7",
"volume": "158",
"year": "2021"
},
{
"DOI": "10.1038/nrm.2017.16",
"article-title": "The mystery of membrane organization: Composition, regulation and roles of lipid rafts",
"author": "Sezgin",
"doi-asserted-by": "crossref",
"first-page": "361",
"journal-title": "Nat. Rev. Mol. Cell Biol.",
"key": "ref_8",
"volume": "18",
"year": "2017"
},
{
"DOI": "10.1016/0005-2736(89)90296-4",
"article-title": "Surface potentials and electric dipole moments of ganglioside and phospholipid monolayers: Contribution of the polar headgroup at the water/lipid interface",
"author": "Beitinger",
"doi-asserted-by": "crossref",
"first-page": "293",
"journal-title": "Biochim. Biophys. Acta (BBA)—Biomembr.",
"key": "ref_9",
"volume": "984",
"year": "1989"
},
{
"DOI": "10.1042/bj1710559",
"article-title": "Surface behaviour of gangliosides and related glycosphingolipids",
"author": "Maggio",
"doi-asserted-by": "crossref",
"first-page": "559",
"journal-title": "Biochem. J.",
"key": "ref_10",
"volume": "171",
"year": "1978"
},
{
"DOI": "10.1016/0079-6107(94)90006-X",
"article-title": "The surface behavior of glycosphingolipids in biomembranes: A new frontier of molecular ecology",
"author": "Maggio",
"doi-asserted-by": "crossref",
"first-page": "55",
"journal-title": "Prog. Biophys. Mol. Biol.",
"key": "ref_11",
"volume": "62",
"year": "1994"
},
{
"DOI": "10.3390/ijms25168583",
"doi-asserted-by": "crossref",
"key": "ref_12",
"unstructured": "Chahinian, H., Yahi, N., and Fantini, J. (2024). Glutamate, Gangliosides, and the Synapse: Electrostatics at Work in the Brain. Int. J. Mol. Sci., 25."
},
{
"DOI": "10.1016/j.neuroscience.2018.05.035",
"article-title": "Negatively Charged Gangliosides Promote Membrane Association of Amphipathic Neurotransmitters",
"author": "Juhola",
"doi-asserted-by": "crossref",
"first-page": "214",
"journal-title": "Neuroscience",
"key": "ref_13",
"volume": "384",
"year": "2018"
},
{
"DOI": "10.3390/v15091854",
"doi-asserted-by": "crossref",
"key": "ref_14",
"unstructured": "Matveeva, M., Lefebvre, M., Chahinian, H., Yahi, N., and Fantini, J. (2023). Host membranes as drivers of virus evolution. Viruses, 15."
},
{
"DOI": "10.3390/v15020284",
"doi-asserted-by": "crossref",
"key": "ref_15",
"unstructured": "Fantini, J., Azzaz, F., Chahinian, H., and Yahi, N. (2023). Electrostatic Surface Potential as a Key Parameter in Virus Transmission and Evolution: How to Manage Future Virus Pandemics in the Post-COVID-19 Era. Viruses, 15."
},
{
"DOI": "10.3389/fmicb.2023.1169547",
"doi-asserted-by": "crossref",
"key": "ref_16",
"unstructured": "Lauster, D., Osterrieder, K., Haag, R., Ballauff, M., and Herrmann, A. (2023). Respiratory viruses interacting with cells: The importance of electrostatics. Front. Microbiol., 14."
},
{
"DOI": "10.1002/adbi.202101264",
"article-title": "Role of protein–lipid interactions in viral entry",
"author": "Contreras",
"doi-asserted-by": "crossref",
"first-page": "2101264",
"journal-title": "Adv. Biol.",
"key": "ref_17",
"volume": "6",
"year": "2022"
},
{
"DOI": "10.1038/s44298-025-00104-1",
"article-title": "Balancing stability and function: Impact of the surface charge of SARS-CoV-2 Omicron spike protein",
"author": "Lauster",
"doi-asserted-by": "crossref",
"first-page": "23",
"journal-title": "npj Viruses",
"key": "ref_18",
"volume": "3",
"year": "2025"
},
{
"DOI": "10.1093/bioadv/vbae053",
"article-title": "Changes in total charge on spike protein of SARS-CoV-2 in emerging lineages",
"author": "Podgornik",
"doi-asserted-by": "crossref",
"first-page": "vbae053",
"journal-title": "Bioinform. Adv.",
"key": "ref_19",
"volume": "4",
"year": "2024"
},
{
"DOI": "10.3390/ijms24031923",
"article-title": "Convergent Evolution Dynamics of SARS-CoV-2 and HIV Surface Envelope Glycoproteins Driven by Host Cell Surface Receptors and Lipid Rafts: Lessons for the Future",
"author": "Fantini",
"doi-asserted-by": "crossref",
"first-page": "1923",
"journal-title": "Int. J. Mol. Sci.",
"key": "ref_20",
"volume": "24",
"year": "2023"
},
{
"DOI": "10.3389/fcell.2020.618296",
"doi-asserted-by": "crossref",
"key": "ref_21",
"unstructured": "Sorice, M., Misasi, R., Riitano, G., Manganelli, V., Martellucci, S., Longo, A., Garofalo, T., and Mattei, V. (2021). Targeting lipid rafts as a strategy against coronavirus. Front. Cell Dev. Biol., 8."
},
{
"DOI": "10.1128/JVI.79.15.9862-9871.2005",
"article-title": "Murine coronavirus requires lipid rafts for virus entry and cell-cell fusion but not for virus release",
"author": "Choi",
"doi-asserted-by": "crossref",
"first-page": "9862",
"journal-title": "J. Virol.",
"key": "ref_22",
"volume": "79",
"year": "2005"
},
{
"DOI": "10.1016/j.bbrc.2008.02.023",
"article-title": "Lipid rafts are involved in SARS-CoV entry into Vero E6 cells",
"author": "Lu",
"doi-asserted-by": "crossref",
"first-page": "344",
"journal-title": "Biochem. Biophys. Res. Commun.",
"key": "ref_23",
"volume": "369",
"year": "2008"
},
{
"DOI": "10.1194/jlr.TR120000658",
"article-title": "Lipid rafts as a therapeutic target: Thematic review series: Biology of lipid rafts",
"author": "Sviridov",
"doi-asserted-by": "crossref",
"first-page": "687",
"journal-title": "J. Lipid Res.",
"key": "ref_24",
"volume": "61",
"year": "2020"
},
{
"DOI": "10.3389/fmicb.2021.631274",
"doi-asserted-by": "crossref",
"key": "ref_25",
"unstructured": "Ripa, I., Andreu, S., López-Guerrero, J.A., and Bello-Morales, R. (2021). Membrane rafts: Portals for viral entry. Front. Microbiol., 12."
},
{
"DOI": "10.1128/MMBR.67.2.226-237.2003",
"article-title": "Virus entry, assembly, budding, and membrane rafts",
"author": "Chazal",
"doi-asserted-by": "crossref",
"first-page": "226",
"journal-title": "Microbiol. Mol. Biol. Rev.",
"key": "ref_26",
"volume": "67",
"year": "2003"
},
{
"DOI": "10.1038/nri1129",
"article-title": "Pathogens: Raft hijackers",
"doi-asserted-by": "crossref",
"first-page": "557",
"journal-title": "Nat. Rev. Immunol.",
"key": "ref_27",
"volume": "3",
"year": "2003"
},
{
"DOI": "10.1021/acsinfecdis.3c00106",
"article-title": "SARS-CoV-2 binding to terminal sialic acid of gangliosides embedded in lipid membranes",
"author": "Negi",
"doi-asserted-by": "crossref",
"first-page": "1346",
"journal-title": "ACS Infect. Dis.",
"key": "ref_28",
"volume": "9",
"year": "2023"
},
{
"DOI": "10.1038/s41589-021-00924-1",
"article-title": "Sialic acid-containing glycolipids mediate binding and viral entry of SARS-CoV-2",
"author": "Nguyen",
"doi-asserted-by": "crossref",
"first-page": "81",
"journal-title": "Nat. Chem. Biol.",
"key": "ref_29",
"volume": "18",
"year": "2022"
},
{
"DOI": "10.1128/mBio.02764-19",
"doi-asserted-by": "crossref",
"key": "ref_30",
"unstructured": "Qing, E., Hantak, M., Perlman, S., and Gallagher, T. (2020). Distinct Roles for Sialoside and Protein Receptors in Coronavirus Infection. mBio, 11."
},
{
"DOI": "10.1021/acsinfecdis.3c00519",
"article-title": "Synergistic binding of SARS-CoV-2 to ACE2 and gangliosides in native lipid membranes",
"author": "Dey",
"doi-asserted-by": "crossref",
"first-page": "907",
"journal-title": "ACS Infect. Dis.",
"key": "ref_31",
"volume": "10",
"year": "2024"
},
{
"DOI": "10.1093/glycob/cwab032",
"article-title": "The role of cell surface sialic acids for SARS-CoV-2 infection",
"author": "Sun",
"doi-asserted-by": "crossref",
"first-page": "1245",
"journal-title": "Glycobiology",
"key": "ref_32",
"volume": "31",
"year": "2021"
},
{
"DOI": "10.1021/acs.jpcb.3c02286",
"article-title": "Comparison and Possible Binding Orientations of SARS-CoV-2 Spike N-Terminal Domain for Gangliosides GM3 and GM1",
"author": "Das",
"doi-asserted-by": "crossref",
"first-page": "6940",
"journal-title": "J. Phys. Chem. B",
"key": "ref_33",
"volume": "127",
"year": "2023"
},
{
"DOI": "10.1038/s41467-025-59137-y",
"article-title": "Human coronavirus HKU1 spike structures reveal the basis for sialoglycan specificity and carbohydrate-promoted conformational changes",
"author": "Jin",
"doi-asserted-by": "crossref",
"first-page": "4158",
"journal-title": "Nat. Commun.",
"key": "ref_34",
"volume": "16",
"year": "2025"
},
{
"DOI": "10.1016/j.coviro.2021.08.010",
"article-title": "Structure of SARS-CoV-2 spike protein",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "173",
"journal-title": "Curr. Opin. Virol.",
"key": "ref_35",
"volume": "50",
"year": "2021"
},
{
"DOI": "10.1038/s41586-020-2665-2",
"article-title": "Structures and distributions of SARS-CoV-2 spike proteins on intact virions",
"author": "Ke",
"doi-asserted-by": "crossref",
"first-page": "498",
"journal-title": "Nature",
"key": "ref_36",
"volume": "588",
"year": "2020"
},
{
"DOI": "10.1126/sciadv.abe5575",
"article-title": "Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM",
"author": "Xu",
"doi-asserted-by": "crossref",
"first-page": "eabe5575",
"journal-title": "Sci. Adv.",
"key": "ref_37",
"volume": "7",
"year": "2021"
},
{
"DOI": "10.1126/science.abc6952",
"article-title": "A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2",
"author": "Chi",
"doi-asserted-by": "crossref",
"first-page": "650",
"journal-title": "Science",
"key": "ref_38",
"volume": "369",
"year": "2020"
},
{
"DOI": "10.1016/j.chom.2021.03.005",
"article-title": "Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite",
"author": "Cerutti",
"doi-asserted-by": "crossref",
"first-page": "819",
"journal-title": "Cell Host Microbe",
"key": "ref_39",
"volume": "29",
"year": "2021"
},
{
"DOI": "10.1016/j.isci.2021.102479",
"article-title": "Therapeutic antibodies, targeting the SARS-CoV-2 spike N-terminal domain, protect lethally infected K18-hACE2 mice",
"author": "Mechaly",
"doi-asserted-by": "crossref",
"first-page": "102479",
"journal-title": "Iscience",
"key": "ref_40",
"volume": "24",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2021.03.028",
"article-title": "N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2",
"author": "McCallum",
"doi-asserted-by": "crossref",
"first-page": "2332",
"journal-title": "Cell",
"key": "ref_41",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.3389/fimmu.2021.691715",
"doi-asserted-by": "crossref",
"key": "ref_42",
"unstructured": "Deshpande, A., Harris, B.D., Martinez-Sobrido, L., Kobie, J.J., and Walter, M.R. (2021). Epitope Classification and RBD Binding Properties of Neutralizing Antibodies Against SARS-CoV-2 Variants of Concern. Front. Immunol., 12."
},
{
"DOI": "10.1016/j.cell.2020.06.025",
"article-title": "Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies",
"author": "Barnes",
"doi-asserted-by": "crossref",
"first-page": "828",
"journal-title": "Cell",
"key": "ref_43",
"volume": "182",
"year": "2020"
},
{
"DOI": "10.1002/2211-5463.13612",
"article-title": "Lipid rafts and human diseases: Why we need to target gangliosides",
"author": "Fantini",
"doi-asserted-by": "crossref",
"first-page": "1636",
"journal-title": "FEBS Open Bio",
"key": "ref_44",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.1111/febs.15651",
"article-title": "The structural basis of accelerated host cell entry by SARS-CoV-2",
"author": "Seyran",
"doi-asserted-by": "crossref",
"first-page": "5010",
"journal-title": "FEBS J.",
"key": "ref_45",
"volume": "288",
"year": "2021"
},
{
"DOI": "10.1021/acsinfecdis.5c00143",
"article-title": "SARS-CoV-2 Evolved Variants Bind to Sialylated Gangliosides and Are Inhibited by a Tetravalent Sialo-Glycocluster",
"author": "Negi",
"doi-asserted-by": "crossref",
"first-page": "3036",
"journal-title": "ACS Infect. Dis.",
"key": "ref_46",
"volume": "11",
"year": "2025"
},
{
"DOI": "10.1126/sciadv.adk4920",
"article-title": "Single-molecule imaging reveals allosteric stimulation of SARS-CoV-2 spike receptor binding domain by host sialic acid",
"author": "Jain",
"doi-asserted-by": "crossref",
"first-page": "eadk4920",
"journal-title": "Sci. Adv.",
"key": "ref_47",
"volume": "10",
"year": "2024"
},
{
"DOI": "10.1016/j.csbj.2021.04.001",
"article-title": "Dependence of SARS-CoV-2 infection on cholesterol-rich lipid raft and endosomal acidification",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "1933",
"journal-title": "Comput. Struct. Biotechnol. J.",
"key": "ref_48",
"volume": "19",
"year": "2021"
},
{
"DOI": "10.3389/fimmu.2022.820131",
"doi-asserted-by": "crossref",
"key": "ref_49",
"unstructured": "Teixeira, L., Temerozo, J.R., Pereira-Dutra, F.S., Ferreira, A.C., Mattos, M., Gonçalves, B.S., Sacramento, C.Q., Palhinha, L., Cunha-Fernandes, T., and Dias, S.S. (2022). Simvastatin downregulates the SARS-CoV-2-induced inflammatory response and impairs viral infection through disruption of lipid rafts. Front. Immunol., 13."
},
{
"DOI": "10.3390/nu14163417",
"doi-asserted-by": "crossref",
"key": "ref_50",
"unstructured": "Bakillah, A., Hejji, F.A., Almasaud, A., Jami, H.A., Hawwari, A., Qarni, A.A., Iqbal, J., and Alharbi, N.K. (2022). Lipid raft integrity and cellular cholesterol homeostasis are critical for SARS-CoV-2 entry into cells. Nutrients, 14."
},
{
"DOI": "10.1038/s41419-021-03527-9",
"article-title": "Evidences for lipid involvement in SARS-CoV-2 cytopathogenesis",
"author": "Nardacci",
"doi-asserted-by": "crossref",
"first-page": "263",
"journal-title": "Cell Death Dis.",
"key": "ref_51",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.24171/j.phrp.2021.0153",
"article-title": "The role of lipids in the pathophysiology of coronavirus infections",
"author": "Zandi",
"doi-asserted-by": "crossref",
"first-page": "278",
"journal-title": "Osong Public Health Res. Perspect.",
"key": "ref_52",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1016/j.csbj.2022.11.004",
"article-title": "Structural and energetic analyses of SARS-CoV-2 N-terminal domain characterise sugar binding pockets and suggest putative impacts of variants on COVID-19 transmission",
"author": "Lam",
"doi-asserted-by": "crossref",
"first-page": "6302",
"journal-title": "Comput. Struct. Biotechnol. J.",
"key": "ref_53",
"volume": "20",
"year": "2022"
},
{
"DOI": "10.1016/j.cplett.2022.140260",
"article-title": "Identification of possible binding modes of SARS-CoV-2 spike N-terminal domain for ganglioside GM1",
"author": "Das",
"doi-asserted-by": "crossref",
"first-page": "140260",
"journal-title": "Chem. Phys. Lett.",
"key": "ref_54",
"volume": "812",
"year": "2023"
},
{
"DOI": "10.1016/j.ijantimicag.2020.105960",
"article-title": "Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection",
"author": "Fantini",
"doi-asserted-by": "crossref",
"first-page": "105960",
"journal-title": "Int. J. Antimicrob. Agents",
"key": "ref_55",
"volume": "55",
"year": "2020"
},
{
"DOI": "10.1016/j.bpc.2009.01.007",
"article-title": "Behaviour of a peptide sequence from the GB virus C/hepatitis G virus E2 protein in Langmuir monolayers: Its interaction with phospholipid membrane models",
"author": "Mestres",
"doi-asserted-by": "crossref",
"first-page": "153",
"journal-title": "Biophys. Chem.",
"key": "ref_56",
"volume": "141",
"year": "2009"
},
{
"DOI": "10.1021/acs.langmuir.7b04252",
"article-title": "Surface chemistry and spectroscopic study of a cholera toxin B Langmuir monolayer",
"author": "Sharma",
"doi-asserted-by": "crossref",
"first-page": "2557",
"journal-title": "Langmuir ACS J. Surf. Colloids",
"key": "ref_57",
"volume": "34",
"year": "2018"
},
{
"DOI": "10.1016/S0005-2736(96)00138-1",
"article-title": "Binding of influenza A virus to monosialoganglioside (GM3) reconstituted in glucosylceramide and sphingomyelin membranes",
"author": "Sato",
"doi-asserted-by": "crossref",
"first-page": "14",
"journal-title": "Biochim. Biophys. Acta (BBA)-Biomembr.",
"key": "ref_58",
"volume": "1285",
"year": "1996"
},
{
"DOI": "10.3390/v16121836",
"doi-asserted-by": "crossref",
"key": "ref_59",
"unstructured": "Lefebvre, M., Chahinian, H., La Scola, B., and Fantini, J. (2024). Characterization and Fluctuations of an Ivermectin Binding Site at the Lipid Raft Interface of the N-Terminal Domain (NTD) of the Spike Protein of SARS-CoV-2 Variants. Viruses, 16."
},
{
"DOI": "10.1016/j.cis.2008.06.001",
"article-title": "Lipases at interfaces: A review",
"author": "Reis",
"doi-asserted-by": "crossref",
"first-page": "237",
"journal-title": "Adv. Colloid Interface Sci.",
"key": "ref_60",
"volume": "147–148",
"year": "2009"
},
{
"DOI": "10.1002/elps.1150181505",
"article-title": "SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling",
"author": "Guex",
"doi-asserted-by": "crossref",
"first-page": "2714",
"journal-title": "Electrophoresis",
"key": "ref_61",
"volume": "18",
"year": "1997"
},
{
"DOI": "10.1126/science.abb2507",
"article-title": "Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation",
"author": "Wrapp",
"doi-asserted-by": "crossref",
"first-page": "1260",
"journal-title": "Science",
"key": "ref_62",
"volume": "367",
"year": "2020"
},
{
"DOI": "10.1126/science.abd4251",
"article-title": "Distinct conformational states of SARS-CoV-2 spike protein",
"author": "Cai",
"doi-asserted-by": "crossref",
"first-page": "1586",
"journal-title": "Science",
"key": "ref_63",
"volume": "369",
"year": "2020"
},
{
"DOI": "10.3390/molecules27123851",
"doi-asserted-by": "crossref",
"key": "ref_64",
"unstructured": "Guérin, P., Yahi, N., Azzaz, F., Chahinian, H., Sabatier, J.-M., and Fantini, J. (2022). Structural dynamics of the SARS-CoV-2 spike protein: A 2-year retrospective analysis of SARS-CoV-2 variants (from Alpha to Omicron) reveals an early divergence between conserved and variable epitopes. Molecules, 27."
},
{
"DOI": "10.1016/j.cell.2020.06.043",
"article-title": "Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus",
"author": "Korber",
"doi-asserted-by": "crossref",
"first-page": "812",
"journal-title": "Cell",
"key": "ref_65",
"volume": "182",
"year": "2020"
},
{
"DOI": "10.1016/j.jinf.2024.106150",
"article-title": "Role of SARS-CoV-2 mutations in the evolution of the COVID-19 pandemic",
"author": "Colson",
"doi-asserted-by": "crossref",
"first-page": "106150",
"journal-title": "J. Infect.",
"key": "ref_66",
"volume": "88",
"year": "2024"
},
{
"DOI": "10.1126/science.abf2303",
"article-title": "Structural impact on SARS-CoV-2 spike protein by D614G substitution",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "525",
"journal-title": "Science",
"key": "ref_67",
"volume": "372",
"year": "2021"
},
{
"DOI": "10.3390/ijms22168714",
"doi-asserted-by": "crossref",
"key": "ref_68",
"unstructured": "Choi, K.-E., Kim, J.-M., Rhee, J., Park, A.K., Kim, E.-J., and Kang, N.S. (2021). Molecular Dynamics Studies on the structural characteristics for the stability prediction of SARS-CoV-2. Int. J. Mol. Sci., 22."
},
{
"DOI": "10.1111/1348-0421.12945",
"article-title": "Proteolytic activation of SARS-CoV-2 spike protein",
"author": "Takeda",
"doi-asserted-by": "crossref",
"first-page": "15",
"journal-title": "Microbiol. Immunol.",
"key": "ref_69",
"volume": "66",
"year": "2022"
},
{
"DOI": "10.1073/pnas.2111199119",
"article-title": "SARS-CoV-2 spike engagement of ACE2 primes S2′ site cleavage and fusion initiation",
"author": "Yu",
"doi-asserted-by": "crossref",
"first-page": "e2111199119",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_70",
"volume": "119",
"year": "2022"
},
{
"DOI": "10.3389/fimmu.2021.796855",
"doi-asserted-by": "crossref",
"key": "ref_71",
"unstructured": "Palacios-Rapalo, S.N., De Jesus-Gonzalez, L.A., Cordero-Rivera, C.D., Farfan-Morales, C.N., Osuna-Ramos, J.F., Martinez-Mier, G., Quistian-Galvan, J., Munoz-Perez, A., Bernal-Dolores, V., and Del Ángel, R.M. (2021). Cholesterol-rich lipid rafts as platforms for SARS-CoV-2 entry. Front. Immunol., 12."
},
{
"DOI": "10.1371/journal.pone.0170123",
"doi-asserted-by": "crossref",
"key": "ref_72",
"unstructured": "Guo, H., Huang, M., Yuan, Q., Wei, Y., Gao, Y., Mao, L., Gu, L., Tan, Y.W., Zhong, Y., and Liu, D. (2017). The important role of lipid raft-mediated attachment in the infection of cultured cells by coronavirus infectious bronchitis virus beaudette strain. PLoS ONE, 12."
},
{
"article-title": "Natural small molecules as inhibitors of coronavirus lipid-dependent attachment to host cells: A possible strategy for reducing SARS-CoV-2 infectivity?",
"author": "Baglivo",
"first-page": "161",
"journal-title": "Acta Bio Medica Atenei Parm.",
"key": "ref_73",
"volume": "91",
"year": "2020"
},
{
"DOI": "10.3389/fimmu.2021.815020",
"doi-asserted-by": "crossref",
"key": "ref_74",
"unstructured": "Kulkarni, R., Wiemer, E.A., and Chang, W. (2022). Role of lipid rafts in pathogen-host interaction-a mini review. Front. Immunol., 12."
},
{
"DOI": "10.1038/cr.2008.15",
"article-title": "SARS coronavirus entry into host cells through a novel clathrin-and caveolae-independent endocytic pathway",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "290",
"journal-title": "Cell Res.",
"key": "ref_75",
"volume": "18",
"year": "2008"
},
{
"DOI": "10.3390/microorganisms12010063",
"doi-asserted-by": "crossref",
"key": "ref_76",
"unstructured": "Ding, C., Chen, Y., Miao, G., and Qi, Z. (2023). Research advances on the role of lipids in the life cycle of human coronaviruses. Microorganisms, 12."
},
{
"DOI": "10.1016/j.jmb.2022.167661",
"article-title": "Domino Effect in Allosteric Signaling of Peptide Binding",
"author": "Caflisch",
"doi-asserted-by": "crossref",
"first-page": "167661",
"journal-title": "J. Mol. Biol.",
"key": "ref_77",
"volume": "434",
"year": "2022"
},
{
"DOI": "10.1016/j.cbpa.2017.08.007",
"article-title": "An amino acid domino effect orchestrates ClpP’s conformational states",
"author": "Stahl",
"doi-asserted-by": "crossref",
"first-page": "102",
"journal-title": "Curr. Opin. Chem. Biol.",
"key": "ref_78",
"volume": "40",
"year": "2017"
},
{
"DOI": "10.31083/j.fbl2808157",
"doi-asserted-by": "crossref",
"key": "ref_79",
"unstructured": "Azzaz, F., Mazzarino, M., Chahinian, H., Yahi, N., Scala, C.D., and Fantini, J. (2023). Structure of the Myelin Sheath Proteolipid Plasmolipin (PLLP) in a Ganglioside-Containing Lipid Raft. Front. Biosci.-Landmark, 28."
},
{
"DOI": "10.1038/s41594-021-00615-4",
"article-title": "Structural mechanism of heat-induced opening of a temperature-sensitive TRP channel",
"author": "Nadezhdin",
"doi-asserted-by": "crossref",
"first-page": "564",
"journal-title": "Nat. Struct. Mol. Biol.",
"key": "ref_80",
"volume": "28",
"year": "2021"
},
{
"DOI": "10.1016/j.ijbiomac.2021.01.086",
"article-title": "A phenylalanine dynamic switch controls the interfacial activation of Rhizopus chinensis lipase",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "1",
"journal-title": "Int. J. Biol. Macromol.",
"key": "ref_81",
"volume": "173",
"year": "2021"
},
{
"DOI": "10.1016/j.jinf.2021.06.001",
"article-title": "Structural dynamics of SARS-CoV-2 variants: A health monitoring strategy for anticipating Covid-19 outbreaks",
"author": "Fantini",
"doi-asserted-by": "crossref",
"first-page": "197",
"journal-title": "J. Infect.",
"key": "ref_82",
"volume": "83",
"year": "2021"
},
{
"DOI": "10.1186/s12967-020-02344-6",
"article-title": "Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant",
"author": "Pachetti",
"doi-asserted-by": "crossref",
"first-page": "179",
"journal-title": "J. Transl. Med.",
"key": "ref_83",
"volume": "18",
"year": "2020"
},
{
"DOI": "10.1186/s13059-023-02881-5",
"article-title": "The P323L substitution in the SARS-CoV-2 polymerase (NSP12) confers a selective advantage during infection",
"author": "Goldswain",
"doi-asserted-by": "crossref",
"first-page": "47",
"journal-title": "Genome Biol.",
"key": "ref_84",
"volume": "24",
"year": "2023"
},
{
"DOI": "10.1159/000515417",
"article-title": "Evolution of SARS-CoV-2: Review of mutations, role of the host immune system",
"author": "Banoun",
"doi-asserted-by": "crossref",
"first-page": "392",
"journal-title": "Nephron",
"key": "ref_85",
"volume": "145",
"year": "2021"
},
{
"DOI": "10.3390/v13061002",
"doi-asserted-by": "crossref",
"key": "ref_86",
"unstructured": "Winger, A., and Caspari, T. (2021). The Spike of Concern-The Novel Variants of SARS-CoV-2. Viruses, 13."
},
{
"DOI": "10.1038/s41579-021-00573-0",
"article-title": "SARS-CoV-2 variants, spike mutations and immune escape",
"author": "Harvey",
"doi-asserted-by": "crossref",
"first-page": "409",
"journal-title": "Nat. Rev. Microbiol.",
"key": "ref_87",
"volume": "19",
"year": "2021"
},
{
"DOI": "10.1126/sciadv.add7221",
"article-title": "Probing the biophysical constraints of SARS-CoV-2 spike N-terminal domain using deep mutational scanning",
"author": "Ouyang",
"doi-asserted-by": "crossref",
"first-page": "eadd7221",
"journal-title": "Sci. Adv.",
"key": "ref_88",
"volume": "8",
"year": "2022"
},
{
"DOI": "10.1002/jmv.27577",
"article-title": "The puzzling mutational landscape of the SARS-2-variant Omicron",
"author": "Fantini",
"doi-asserted-by": "crossref",
"first-page": "2019",
"journal-title": "J. Med. Virol.",
"key": "ref_89",
"volume": "94",
"year": "2022"
},
{
"DOI": "10.1002/jmv.29124",
"article-title": "Emergence of a second SARS-CoV-2 variant with a tremendous genetic leap from its ancestors",
"author": "Colson",
"doi-asserted-by": "crossref",
"first-page": "e29124",
"journal-title": "J. Med. Virol.",
"key": "ref_90",
"volume": "95",
"year": "2023"
},
{
"DOI": "10.1016/j.jinf.2022.02.023",
"article-title": "The value of electrostatic potentials of the spike receptor binding and N-terminal domains in addressing transmissibility and infectivity of SARS-CoV-2 variants of concern",
"author": "Pascarella",
"doi-asserted-by": "crossref",
"first-page": "e62",
"journal-title": "J. Infect.",
"key": "ref_91",
"volume": "84",
"year": "2022"
},
{
"DOI": "10.1002/jmv.27695",
"article-title": "First cases of infection with the 21L/BA.2 Omicron variant in Marseille, France",
"author": "Colson",
"doi-asserted-by": "crossref",
"first-page": "3421",
"journal-title": "J. Med. Virol.",
"key": "ref_92",
"volume": "94",
"year": "2022"
},
{
"DOI": "10.1002/jmv.27789",
"article-title": "Culture and identification of a “Deltamicron” SARS-CoV-2 in a three cases cluster in southern France",
"author": "Colson",
"doi-asserted-by": "crossref",
"first-page": "3739",
"journal-title": "J. Med. Virol.",
"key": "ref_93",
"volume": "94",
"year": "2022"
},
{
"article-title": "Early detection of the emerging SARS-CoV-2 BA.2.86 lineage through integrated genomic surveillance of wastewater and COVID-19 cases in Sweden, weeks 31 to 38 2023",
"author": "Berg",
"first-page": "2300595",
"journal-title": "Euro Surveill. Bull. Eur. Sur Les. Mal. Transm. Eur. Commun. Dis. Bull.",
"key": "ref_94",
"volume": "28",
"year": "2023"
},
{
"DOI": "10.1016/j.it.2024.01.003",
"article-title": "SARS-CoV-2 evolution from the BA.2.86 to JN.1 variants: Unexpected consequences",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "81",
"journal-title": "Trends Immunol.",
"key": "ref_95",
"volume": "45",
"year": "2024"
},
{
"DOI": "10.1016/j.ijantimicag.2020.106020",
"article-title": "Synergistic antiviral effect of hydroxychloroquine and azithromycin in combination against SARS-CoV-2: What molecular dynamics studies of virus-host interactions reveal",
"author": "Fantini",
"doi-asserted-by": "crossref",
"first-page": "106020",
"journal-title": "Int. J. Antimicrob. Agents",
"key": "ref_96",
"volume": "56",
"year": "2020"
},
{
"DOI": "10.1016/j.bbrc.2020.10.015",
"article-title": "Leveraging coronavirus binding to gangliosides for innovative vaccine and therapeutic strategies against COVID-19",
"author": "Fantini",
"doi-asserted-by": "crossref",
"first-page": "132",
"journal-title": "Biochem. Biophys. Res. Commun.",
"key": "ref_97",
"volume": "538",
"year": "2021"
},
{
"article-title": "The JN. 1 variant of COVID-19: Immune evasion, transmissibility, and implications for global health",
"author": "Musharaf",
"first-page": "20499361251314763",
"journal-title": "Ther. Adv. Infect. Dis.",
"key": "ref_98",
"volume": "12",
"year": "2025"
},
{
"DOI": "10.1093/nar/gkw315",
"article-title": "The RING 2.0 web server for high quality residue interaction networks",
"author": "Piovesan",
"doi-asserted-by": "crossref",
"first-page": "W367",
"journal-title": "Nucleic Acids Res.",
"key": "ref_99",
"volume": "44",
"year": "2016"
},
{
"DOI": "10.1002/prot.21938",
"article-title": "Identifying stabilizing key residues in proteins using interresidue interaction energy matrix",
"author": "Hobza",
"doi-asserted-by": "crossref",
"first-page": "402",
"journal-title": "Proteins Struct. Funct. Bioinform.",
"key": "ref_100",
"volume": "72",
"year": "2008"
},
{
"DOI": "10.1021/jp303088n",
"article-title": "Optimal definition of inter-residual contact in globular proteins based on pairwise interaction energy calculations, its robustness, and applications",
"doi-asserted-by": "crossref",
"first-page": "12651",
"journal-title": "J. Phys. Chem. B",
"key": "ref_101",
"volume": "116",
"year": "2012"
},
{
"key": "ref_102",
"unstructured": "Khan, S.M., and Pazos, F. (2026). Allosteric Network Analysis Toolkit for Single-Domain Phosphoproteins. Protein Evolution: Methods and Protocols, Springer."
},
{
"DOI": "10.1371/journal.pone.0142407",
"doi-asserted-by": "crossref",
"key": "ref_103",
"unstructured": "Pandini, A., Kleinjung, J., Rasool, S., and Khan, S. (2015). Coevolved mutations reveal distinct architectures for two core proteins in the bacterial flagellar motor. PLoS ONE, 10."
},
{
"DOI": "10.1146/annurev-biophys-083012-130257",
"article-title": "The underappreciated role of allostery in the cellular network",
"author": "Nussinov",
"doi-asserted-by": "crossref",
"first-page": "169",
"journal-title": "Annu. Rev. Biophys.",
"key": "ref_104",
"volume": "42",
"year": "2013"
},
{
"DOI": "10.1016/j.str.2016.03.008",
"article-title": "Identifying allosteric hotspots with dynamics: Application to inter-and intra-species conservation",
"author": "Clarke",
"doi-asserted-by": "crossref",
"first-page": "826",
"journal-title": "Structure",
"key": "ref_105",
"volume": "24",
"year": "2016"
},
{
"DOI": "10.1093/bioinformatics/btq066",
"article-title": "How significant is a protein structure similarity with TM-score = 0.5?",
"author": "Xu",
"doi-asserted-by": "crossref",
"first-page": "889",
"journal-title": "Bioinformatics",
"key": "ref_106",
"volume": "26",
"year": "2010"
},
{
"DOI": "10.1186/s12859-015-0567-3",
"doi-asserted-by": "crossref",
"key": "ref_107",
"unstructured": "Dalton, J.A., Lans, I., and Giraldo, J. (2015). Quantifying conformational changes in GPCRs: Glimpse of a common functional mechanism. BMC Bioinform., 16."
},
{
"DOI": "10.1039/c3cs35495d",
"article-title": "Conformational changes of enzymes upon immobilisation",
"author": "Secundo",
"doi-asserted-by": "crossref",
"first-page": "6250",
"journal-title": "Chem. Soc. Rev.",
"key": "ref_108",
"volume": "42",
"year": "2013"
},
{
"DOI": "10.1002/prot.340100210",
"article-title": "Modeling conformational change in macromolecules as an elastic deformation",
"author": "Andrews",
"doi-asserted-by": "crossref",
"first-page": "162",
"journal-title": "Proteins Struct. Funct. Bioinform.",
"key": "ref_109",
"volume": "10",
"year": "1991"
},
{
"article-title": "Elastic proteins: Biological roles and mechanical properties",
"author": "Gosline",
"first-page": "121",
"journal-title": "Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci.",
"key": "ref_110",
"volume": "357",
"year": "2002"
},
{
"key": "ref_111",
"unstructured": "Tee, W.-V., Dong, B., Guarnera, E., and Berezovsky, I.N. (J. Mol. Biol., 2025). On the relationship between protein stability, thermostability, and allosteric signalling, J. Mol. Biol., in press."
},
{
"DOI": "10.1016/j.jmb.2008.02.034",
"article-title": "Allostery: Absence of a Change in Shape Does Not Imply that Allostery Is Not at Play",
"author": "Tsai",
"doi-asserted-by": "crossref",
"first-page": "1",
"journal-title": "J. Mol. Biol.",
"key": "ref_112",
"volume": "378",
"year": "2008"
},
{
"DOI": "10.1002/prot.20264",
"article-title": "Scoring function for automated assessment of protein structure template quality",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "702",
"journal-title": "Proteins",
"key": "ref_113",
"volume": "57",
"year": "2004"
},
{
"DOI": "10.1128/JVI.01805-07",
"article-title": "Aromatic Amino Acids in the Juxtamembrane Domain of Severe Acute Respiratory Syndrome Coronavirus Spike Glycoprotein Are Important for Receptor-Dependent Virus Entry and Cell-Cell Fusion",
"author": "Smith",
"doi-asserted-by": "crossref",
"first-page": "2883",
"journal-title": "J. Virol.",
"key": "ref_114",
"volume": "82",
"year": "2008"
},
{
"DOI": "10.1021/bi048515g",
"article-title": "The aromatic domain of the coronavirus class I viral fusion protein induces membrane permeabilization: Putative role during viral entry",
"author": "Sainz",
"doi-asserted-by": "crossref",
"first-page": "947",
"journal-title": "Biochemistry",
"key": "ref_115",
"volume": "44",
"year": "2005"
},
{
"DOI": "10.1038/340571a0",
"article-title": "Single amino-acid changes in HIV envelope affect viral tropism and receptor binding",
"author": "Cordonnier",
"doi-asserted-by": "crossref",
"first-page": "571",
"journal-title": "Nature",
"key": "ref_116",
"volume": "340",
"year": "1989"
},
{
"DOI": "10.1128/jvi.70.1.321-326.1996",
"article-title": "Analysis of the murine ecotropic leukemia virus receptor reveals a common biochemical determinant on diverse cell surface receptors that is essential to retrovirus entry",
"author": "Malhotra",
"doi-asserted-by": "crossref",
"first-page": "321",
"journal-title": "J. Virol.",
"key": "ref_117",
"volume": "70",
"year": "1996"
},
{
"DOI": "10.3390/v5010352",
"article-title": "Influenza A virus entry inhibitors targeting the hemagglutinin",
"author": "Yang",
"doi-asserted-by": "crossref",
"first-page": "352",
"journal-title": "Viruses",
"key": "ref_118",
"volume": "5",
"year": "2013"
},
{
"DOI": "10.1021/acs.jpcb.2c07258",
"article-title": "NMR Studies of Aromatic Ring Flips to Probe Conformational Fluctuations in Proteins",
"author": "Akke",
"doi-asserted-by": "crossref",
"first-page": "591",
"journal-title": "J. Phys. Chem. B",
"key": "ref_119",
"volume": "127",
"year": "2023"
},
{
"DOI": "10.1074/jbc.M114.572875",
"article-title": "A conserved phenylalanine as a relay between the α5 helix and the GDP binding region of heterotrimeric Gi protein α subunit",
"author": "Kaya",
"doi-asserted-by": "crossref",
"first-page": "24475",
"journal-title": "J. Biol. Chem.",
"key": "ref_120",
"volume": "289",
"year": "2014"
},
{
"DOI": "10.1038/ncomms3881",
"article-title": "A phenylalanine rotameric switch for signal-state control in bacterial chemoreceptors",
"author": "Ortega",
"doi-asserted-by": "crossref",
"first-page": "2881",
"journal-title": "Nat. Commun.",
"key": "ref_121",
"volume": "4",
"year": "2013"
},
{
"DOI": "10.1016/j.proche.2011.08.034",
"article-title": "Quantum effects in biology: Bird navigation",
"author": "Ritz",
"doi-asserted-by": "crossref",
"first-page": "262",
"journal-title": "Procedia Chem.",
"key": "ref_122",
"volume": "3",
"year": "2011"
},
{
"DOI": "10.1038/s41586-021-03618-9",
"article-title": "Magnetic sensitivity of cryptochrome 4 from a migratory songbird",
"author": "Xu",
"doi-asserted-by": "crossref",
"first-page": "535",
"journal-title": "Nature",
"key": "ref_123",
"volume": "594",
"year": "2021"
},
{
"DOI": "10.1097/00001756-200212030-00016",
"article-title": "Dual regulation of cryptochrome 1 mRNA expression in chicken retina by light and circadian oscillators",
"author": "Haque",
"doi-asserted-by": "crossref",
"first-page": "2247",
"journal-title": "Neuroreport",
"key": "ref_124",
"volume": "13",
"year": "2002"
},
{
"DOI": "10.1021/acs.jpcb.5c01187",
"article-title": "Magnetosensitivity of Model Flavin–Tryptophan Radical Pairs in a Dynamic Protein Environment",
"author": "Benjamin",
"doi-asserted-by": "crossref",
"first-page": "5937",
"journal-title": "J. Phys. Chem. B",
"key": "ref_125",
"volume": "129",
"year": "2025"
},
{
"DOI": "10.1021/acsphyschemau.3c00017",
"article-title": "Long-Range Conductivity in Proteins Mediated by Aromatic Residues",
"author": "Krishnan",
"doi-asserted-by": "crossref",
"first-page": "444",
"journal-title": "ACS Phys. Chem. Au",
"key": "ref_126",
"volume": "3",
"year": "2023"
},
{
"DOI": "10.1073/pnas.0408029102",
"article-title": "Long-range electron transfer",
"author": "Gray",
"doi-asserted-by": "crossref",
"first-page": "3534",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_127",
"volume": "102",
"year": "2005"
},
{
"DOI": "10.1021/acs.jpcb.9b04816",
"article-title": "eMap: A web application for identifying and visualizing electron or hole hopping pathways in proteins",
"author": "Tazhigulov",
"doi-asserted-by": "crossref",
"first-page": "6946",
"journal-title": "J. Phys. Chem. B",
"key": "ref_128",
"volume": "123",
"year": "2019"
},
{
"DOI": "10.1016/j.bbagen.2006.12.012",
"article-title": "Site-directed mutagenesis of two aromatic residues lining the active site pocket of the yeast Ltp1",
"author": "Paoli",
"doi-asserted-by": "crossref",
"first-page": "753",
"journal-title": "Biochim. Biophys. Acta",
"key": "ref_129",
"volume": "1770",
"year": "2007"
},
{
"DOI": "10.1128/JB.182.8.2207-2217.2000",
"article-title": "A study of AroP-PheP chimeric proteins and identification of a residue involved in tryptophan transport",
"author": "Cosgriff",
"doi-asserted-by": "crossref",
"first-page": "2207",
"journal-title": "J. Bacteriol.",
"key": "ref_130",
"volume": "182",
"year": "2000"
},
{
"DOI": "10.1021/bi401137u",
"article-title": "Femtosecond dynamics of short-range protein electron transfer in flavodoxin",
"author": "He",
"doi-asserted-by": "crossref",
"first-page": "9120",
"journal-title": "Biochemistry",
"key": "ref_131",
"volume": "52",
"year": "2013"
},
{
"DOI": "10.1063/5.0060672",
"article-title": "Vibronic dynamics resolved by global and target analysis of ultrafast transient absorption spectra",
"author": "Kloz",
"doi-asserted-by": "crossref",
"first-page": "114113",
"journal-title": "J. Chem. Phys.",
"key": "ref_132",
"volume": "155",
"year": "2021"
},
{
"DOI": "10.1515/bmc-2022-0006",
"article-title": "The epigenetic dimension of protein structure",
"author": "Azzaz",
"doi-asserted-by": "crossref",
"first-page": "55",
"journal-title": "Biomol. Concepts",
"key": "ref_133",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.3390/life14030280",
"doi-asserted-by": "crossref",
"key": "ref_134",
"unstructured": "Fantini, J., Matveeva, M., Lefebvre, M., and Chahinian, H. (2024). What Is life? Rethinking Biology in Light of Fundamental Parameters. Life, 14."
},
{
"DOI": "10.1016/j.drudis.2022.103468",
"article-title": "Medicinal chemistry insights into antiviral peptidomimetics",
"author": "Ding",
"doi-asserted-by": "crossref",
"first-page": "103468",
"journal-title": "Drug Discov. Today",
"key": "ref_135",
"volume": "28",
"year": "2023"
},
{
"DOI": "10.1021/acs.jmedchem.5c02430",
"article-title": "Macrocyclic Compounds: Unveiling Their Distinctive Antiviral Advantages in Medicinal Research",
"author": "Yang",
"doi-asserted-by": "crossref",
"first-page": "21035",
"journal-title": "J. Med. Chem.",
"key": "ref_136",
"volume": "68",
"year": "2025"
}
],
"reference-count": 136,
"references-count": 136,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.mdpi.com/2218-273X/16/1/111"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "The Enigmatic Conserved Q134-F135-N137 Triad in SARS-CoV-2 Spike Protein: A Conformational Transducer?",
"type": "journal-article",
"update-policy": "https://doi.org/10.3390/mdpi_crossmark_policy",
"volume": "16"
}