SARS-CoV-2 journey: from alpha variant to omicron and its sub-variants
et al., Infection, doi:10.1007/s15010-024-02223-y, Mar 2024
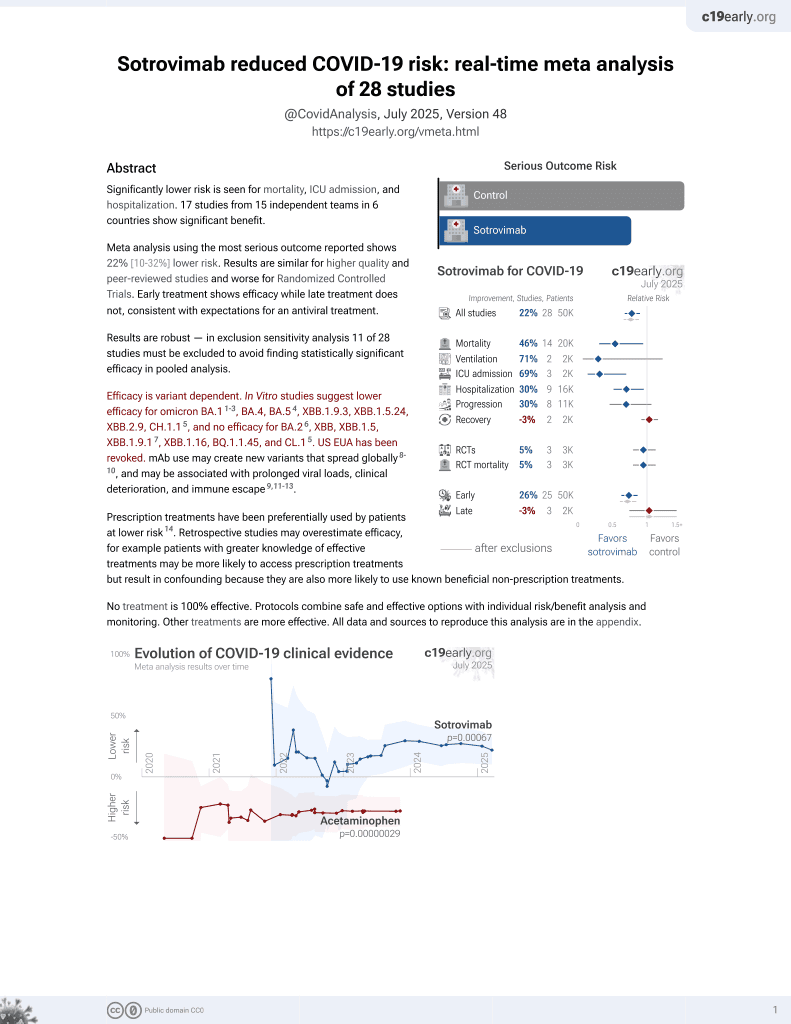
Sotrovimab for COVID-19
45th treatment shown to reduce risk in
August 2022, now with p = 0.00048 from 29 studies, recognized in 42 countries.
Efficacy is variant dependent.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
Review of SARS-CoV-2 variants showing increased transmissibility, disease severity, and immune escape with mutations in the spike protein receptor binding domain. Authors cover variants from the initial D614G mutation through omicron sub-variants and recombinants. Extensive mutations enable the variants to evade neutralizing antibodies from prior infection, vaccination, and monoclonal antibody treatments.
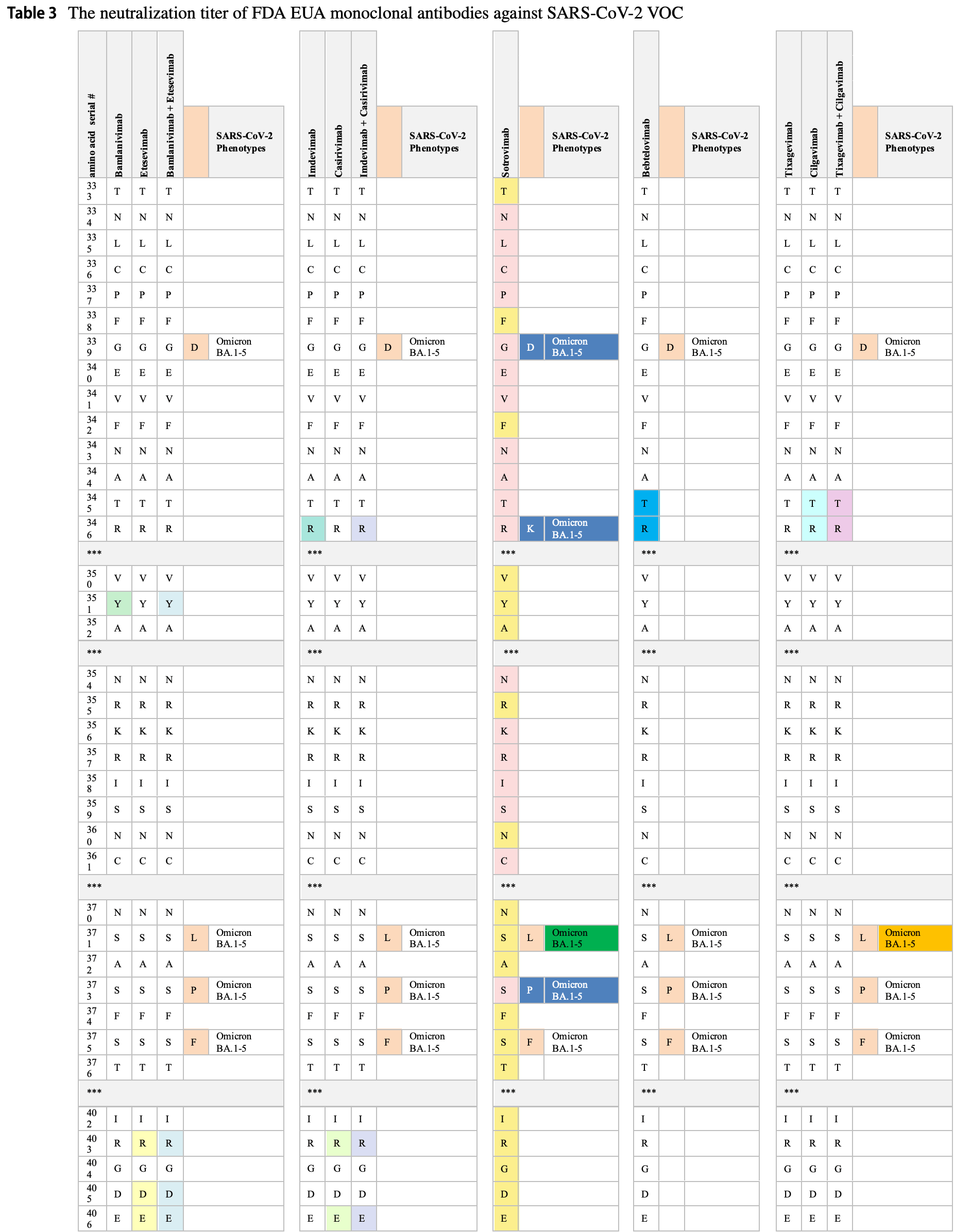
Efficacy is variant dependent. In Vitro studies predict lower efficacy for BA.11-3, BA.4, BA.54, XBB.1.9.3, XBB.1.5.24, XBB.2.9, CH.1.15, and no efficacy for BA.26, XBB, XBB.1.5, ХВВ.1.9.17, XBB.1.16, BQ.1.1.45, and CL.15. US EUA has been revoked.
Review covers casirivimab/imdevimab, bamlanivimab/etesevimab, sotrovimab, tixagevimab/cilgavimab, and bebtelovimab.
1.
Liu et al., Striking Antibody Evasion Manifested by the Omicron Variant of SARS-CoV-2, bioRxiv, doi:10.1101/2021.12.14.472719.
2.
Sheward et al., Variable loss of antibody potency against SARS-CoV-2 B.1.1.529 (Omicron), bioRxiv, doi:10.1101/2021.12.19.473354.
3.
VanBlargan et al., An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by several therapeutic monoclonal antibodies, bioRxiv, doi:10.1101/2021.12.15.472828.
4.
Haars et al., Prevalence of SARS-CoV-2 Omicron Sublineages and Spike Protein Mutations Conferring Resistance against Monoclonal Antibodies in a Swedish Cohort during 2022–2023, Microorganisms, doi:10.3390/microorganisms11102417.
5.
Pochtovyi et al., In Vitro Efficacy of Antivirals and Monoclonal Antibodies against SARS-CoV-2 Omicron Lineages XBB.1.9.1, XBB.1.9.3, XBB.1.5, XBB.1.16, XBB.2.4, BQ.1.1.45, CH.1.1, and CL.1, Vaccines, doi:10.3390/vaccines11101533.
Hattab et al., 30 Mar 2024, peer-reviewed, 4 authors.
Contact: athirah.bakhtiar@monash.edu.
SARS-CoV-2 journey: from alpha variant to omicron and its sub-variants
Infection, doi:10.1007/s15010-024-02223-y
The COVID-19 pandemic has affected hundreds of millions of individuals and caused more than six million deaths. The prolonged pandemic duration and the continual inter-individual transmissibility have contributed to the emergence of a wide variety of SARS-CoV-2 variants. Genomic surveillance and phylogenetic studies have shown that substantial mutations in crucial supersites of spike glycoprotein modulate the binding affinity of the evolved SARS-COV-2 lineages to ACE2 receptors and modify the binding of spike protein with neutralizing antibodies. The immunological spike mutations have been associated with differential transmissibility, infectivity, and therapeutic efficacy of the vaccines and the immunological therapies among the new variants. This review highlights the diverse genetic mutations assimilated in various SARS-CoV-2 variants. The implications of the acquired mutations related to viral transmission, infectivity, and COVID-19 severity are discussed. This review also addresses the effectiveness of human neutralizing antibodies induced by SARS-CoV-2 infection or immunization and the therapeutic antibodies against the ascended variants.
Author contribution DH had the idea of the review and performed the literature review and data analysis. DH wrote the original draft of the manuscript. MA, ZA-A, and AB reviewed and edited the manuscript. All authors have read and approved the final article.
Declarations Conflict of interest The authors declare no conflict of interest. Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http:// creat iveco mmons. org/ licen ses/ by/4. 0/.
References
Annavajhala, Mohri, Wang, Nair, Zucker et al., Emergence and expansion of SARS-CoV-2 B.1.526 after identification in New York, Nature, doi:10.1038/s41586-021-03908-2
Arora, Kempf, Nehlmeier, Graichen, Sidarovich et al., Delta variant (B.1.617.2) sublineages do not show increased neutralization resistance, Cell Mol Immunol
Baral, Bhattarai, Hossen, Stebliankin, Gerstman et al., Mutation-induced changes in the receptorbinding interface of the SARS-CoV-2 Delta variant B.1.617.2 and implications for immune evasion, Biochem Biophys Res Commun, doi:10.1016/j.bbrc.2021.08.036
Barnes, Jette, Abernathy, Dam, Esswein et al., SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies, Nature, doi:10.1038/s41586-020-2852-1
Bernal, Andrews, Gower, Gallagher, Simmons et al., Effectiveness of Covid-19 Vaccines against the B16172 (Delta) Variant, N Engl J Med
Candido, Claro, De Jesus, Souza, Moreira et al., Evolution and epidemic spread of SARS-CoV-2 in Brazil, Science
Cao, Wang, Jian, Song, Yisimayi, Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies, Nature
Cathcart, Havenar-Daughton, Lempp, Ma, Schmid, The dual function monoclonal antibodies VIR-7831 and VIR-7832 demonstrate potent in vitro and in vivo activity against SARS-CoV-2, bioRxiv, doi:10.1101/2021.03.09.434607
Cerutti, Guo, Zhou, Gorman, Lee et al., Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite, Cell Host Microbe, doi:10.1016/j.chom.2021.03.005
Chadha, Khullar, Mittal, Facing the wrath of enigmatic mutations: a review on the emergence of severe acute respiratory syndrome coronavirus 2 variants amid coronavirus disease-19 pandemic, Environ Microbiol, doi:10.1111/1462-2920.15687
Chan, Kok, Zhu, Chu, To et al., Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan, Emerg Microbes Infect
Chen, Chen, Azman, Sun, Lu, Neutralizing antibodies against severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) variants induced by natural infection or vaccination: a systematic review and pooled analysis, Clin Infect Dis, doi:10.1093/cid/ciab646
Chen, Wang, Wang, Wei, Mutations Strengthened SARS-CoV-2 Infectivity, J Mol Biol, doi:10.1016/j.jmb.2020.07.009
Cherian, Potdar, Jadhav, Yadav, Gupta et al., in the Second Wave of COVID-19 in Maharashtra India, Microorganism
Chowdhury, Bappy, Chowdhury, Chowdhury, Chowdhury, COVID-19 Induced Cardiovascular Complications and Recent Therapeutic Advances, Eur J Med Heal Sci
Chowdhury, Bappy, On the Delta Plus Variant of SARS-CoV-2, Eur J Med Heal Sci
Collier, Marco, Ferreira, Meng, Datir et al., mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants, bioRxiv, doi:10.1101/2021.01.25.427948
Dan, Mateus, Kato, Hastie, Yu et al., Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection, Science
Davies, Abbott, Barnard, Jarvis, Kucharski et al., Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England, Science
Deng, Garcia-Knight, Khalid, Servellita, Wang, Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation, medRxiv, doi:10.1101/2021.03.07.21252647
Edara, Floyd, Lai, Gardner, Hudson et al., Infection and mRNA-1273 vaccine antibodies neutralize SARS-CoV-2 UK variant, medRxiv Prepr Serv Heal Sci
Edara, Pinsky, Suthar, Lai, Gardner et al., Infection and vaccine-induced neutralizing-antibody responses to the SARS-CoV-2 B. 1.617 variants, N Engl J Med
Faria, Claro, Candido, Franco, Andrade et al., Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings
Ferreira, Datir, Papa, Kemp, Meng et al., SARS-CoV-2 B.1.617 emergence and sensitivity to vaccine-elicited antibodies, bioRxiv
Focosi, Mcconnell, Casadevall, Cappello, Valdiserra et al., Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant, Lancet Infect Dis, doi:10.1016/j.cell.2020.09.032
Focosi, Quiroga, Mcconnell, Johnson, Casadevall, Convergent evolution in SARS-CoV-2 spike creates a variant soup from which new COVID-19 waves emerge, Int J Mol Sci
Focosi, Quiroga, Mcconnell, Johnson, Casadevall, Convergent evolution in SARS-CoV-2 spike creates a variant soup from which new COVID-19 waves emerge, Int J Mol Sci, doi:10.3390/ijms24032264
Garcia-Beltran, Lam, Astudillo, Yang, Miller et al., COVID-19-neutralizing antibodies predict disease severity and survival, Cell, doi:10.1016/j.cell.2020.12.015
Garcia-Beltran, Lam, St, Denis, Nitido et al., Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity, Cell, doi:10.1016/j.cell.2021.03.013
Garcia-Beltran, St, Denis, Hoelzemer, Lam et al., mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant, Cell, doi:10.1016/j.cell.2021.12.033
Grannis, Rowley, Ong, Stenehjem, Klein et al., Interim estimates of COVID-19 vaccine effectiveness against COVID-19-associated emergency department or urgent care clinic encounters and hospitalizations among adults during SARS-CoV-2 B.1617. 2 (Delta) variant predominance-Nine States, Morb Mortal Wkly Rep
Greaney, Starr, Barnes, Weisblum, Schmidt et al., Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies, Nat Commun, doi:10.1038/s41467-021-24435-8
Greaney, Starr, Gilchuk, Zost, Binshtein et al., Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition, Cell Host Microbe, doi:10.1016/j.chom.2020.11.007
Gruell, Vanshylla, Tober-Lau, Hillus, Sander et al., Neutralisation sensitivity of the SARS-CoV-2 omicron BA.2.75 sublineage, Lancet Infect Dis
Gräf, Bello, Venas, Pereira, Paixão, Identification of a novel SARS-CoV-2 P.1 sub-lineage in Brazil provides new insights about the mechanisms of emergence of variants of concern, Virus Evol, doi:10.1093/ve/veab091
Gupta, Kaur, Yadav, Mukhopadhyay, Sahay, Clinical characterization and genomic analysis of samples from COVID-19 breakthrough infections during the second wave among the various states of India, Viruses, doi:10.3390/v13091782
Hasan, Kalikiri, Mirza, Sundararaju, Sharma et al., Real-Time SARS-CoV-2 Genotyping by High-Throughput Multiplex PCR Reveals the Epidemiology of the Variants of Concern in Qatar, Int J Infect Dis
Hoffmann, Arora, Groß, Seidel, Hörnich et al., SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies, Cell
Hoffmann, Hofmann-Winkler, Krüger, Kempf, Nehlmeier et al., SARS-CoV-2 variant B1617 is resistant to bamlanivimab and evades antibodies induced by infection and vaccination, Cell Rep
Hu, Peng, Wang, Fang, Yang, Emerging SARS-CoV-2 variants reduce neutralization sensitivity to convalescent sera and monoclonal antibodies, Cell Mol Immunol, doi:10.1038/s41423-021-00648-1
Iketani, Liu, Guo, Liu, Chan et al., Antibody evasion properties of SARS-CoV-2 Omicron sublineages, Nature
Ito, Suzuki, Uriu, Itakura, Zahradnik et al., Convergent evolution of SARS-CoV-2 Omicron subvariants leading to the emergence of BQ11 variant, Nat Commun
Kahn, Mcintosh, Discussion, Pediatr Infect Dis J
Kannan, Spratt, Cohen, Naqvi, Chand et al., Evolutionary analysis of the Delta and Delta Plus variants of the SARS-CoV-2 viruses, J Autoimmun, doi:10.1016/j.jaut.2021.102715
Kim, Lee, Yang, Kim, Kim et al., The Architecture of SARS-CoV-2 Transcriptome, Cell, doi:10.1016/j.cell.2020.04.011
Korber, Fischer, Gnanakaran, Yoon, Theiler et al., Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus, Cell
Kurhade, Zou, Xia, Cai, Yang et al., Neutralization of Omicron BA. 1, BA. 2, and BA. 3 SARS-CoV-2 by 3 doses of BNT162b2 vaccine, Nat Commun
Lai, Bergna, Caucci, Clementi, Vicenti et al., Molecular tracing of SARS-CoV-2 in Italy in the first three months of the epidemic, Viruses
Laiton-Donato, Franco-Muñoz, Da, Ruiz-Moreno, Ciro, Characterization of the emerging B.1.621 variant of interest of SARS-CoV-2, Infect Genet Evol, doi:10.1016/j.meegid.2021.105038
Li, Wu, Nie, Zhang, Hao et al., The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity, Cell, doi:10.1016/j.cell.2020.07.012
Li, Zhang, Liang, Zhang, Wu et al., Antigenicity comparison of SARS-CoV-2 Omicron sublineages with other variants contained multiple mutations in RBD, MedComm
Liu, Ginn, Dejnirattisai, Supasa, Wang et al., Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum, Cell
Liu, Liu, Xia, Zhang, Fontes-Garfias et al., Neutralizing activity of BNT162b2-elicited serum, N Engl J Med
Mariano, Farthing, Lale-Farjat, Bergeron, Structural characterization of SARS-CoV-2: where we are, and where we need to be, Front Mol Biosci, doi:10.3389/fmolb.2020.605236
Mccallum, Marco, Lempp, Tortorici, Pinto et al., N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2, Cell, doi:10.1016/j.cell.2021.03.028
Me, Lai, Wali, Samaha, Solis et al., Neutralization against BA. 2.75. 2, BQ.1.1, and XBB from mRNA Bivalent Booster, N Engl J Med
Meng, Abdullahi, Ferreira, Goonawardane, Saito et al., Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity, Nature
Meng, Kemp, Papa, Datir, Ferreira, Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7, Cell Rep, doi:10.1016/j.celrep.2021.109292
Miller, Clark, Raman, Sasisekharan, Insights on the mutational landscape of the SARS-CoV-2 Omicron variant receptor-binding domain, Cell Reports Med
Mohapatra, Kandi, Sarangi, Verma, Tuli et al., The recently emerged BA. 4 and BA. 5 lineages of Omicron and their global health concerns amid the ongoing wave of COVID-19 pandemic-Correspondence, Int J Surg
Mohapatra, Mahal, Kutikuppala, Pal, Kandi et al., Renewed global threat by the novel SARS-CoV-2 variants, Front Virol
Naveca, Cd, Nascimento, Souza, Corado, SARS-CoV-2 reinfection by the new Variant of Concern (VOC) P.1 in Amazonas, Brazil, Virological
Nemet, Kliker, Lustig, Zuckerman, Erster, Third BNT162b2 vaccination neutralization of SARS-CoV-2 Omicron infection, doi:10.1056/NEJMc2119358
O'horo, Challener, Speicher, Bosch, Seville et al., Effectiveness of Monoclonal Antibodies in Preventing Severe COVID-19 With Emergence of the Delta Variant, Mayo Clin Proc, doi:10.1016/j.mayocp.2021.12.002
Ou, Liu, Lei, Li, Mi et al., Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV, Nat Commun, doi:10.1038/s41467-020-15562-9
Paiva, Guedes, Docena, Bezerra, Dezordi et al., Multiple introductions followed by ongoing community spread of sars-cov-2 at one of the largest metropolitan areas of northeast brazil, Viruses
Parums, The XBB. 1.5 ('Kraken') Subvariant of Omicron SARS-CoV-2 and its Rapid Global Spread, Med Sci Monit Int Med J Exp Clin Res
Pearson, Russell, Davies, Kucharski, Covid-19 Working Group et al., Estimates of severity and transmissibility of novel South Africa SARS-CoV-2 variant 501Y.V2, Preprint
Peiris, Lai, Poon, Guan, Yam et al., Coronavirus as a possible cause of severe acute respiratory syndrome, Lancet
Pereira, Tosta, Lima, De Oliveira, Da Silva et al., Genomic surveillance activities unveil the introduction of the SARS-CoV-2 B.1.525 variant of interest in Brazil: Case report, J Med Virol
Planas, Veyer, Baidaliuk, Staropoli, Guivel-Benhassine et al., Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization, Nature, doi:10.1038/s41586-021-03777-9
Pulliam, Schalkwyk C Van, Govender, Gottberg A Von, Cohen, Groome, Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa
Resende, Bezerra, Vasconcelos, Arantes, Appolinario, Spike E484K mutation in the first SARS-CoV-2 reinfection case confirmed in Brazil, Virological
Saito, Tamura, Zahradnik, Deguchi, Tabata, Virological characteristics of the SARS-CoV-2 Omicron BA275 variant, Cell Host Microbe, doi:10.1016/j.chom.2022.10.003
Sapkal, Yadav, Sahay, Deshpande, Gupta et al., Neutralization of Delta variant with sera of Covishield™ vaccinees and COVID-19-recovered vaccinated individuals, J Travel Med
Scott, Hsiao, Moyo, Singh, Tegally et al., Track Omicron's spread with molecular data, Science
Sheikh, Mcmenamin, Taylor, Robertson, SARS-CoV-2 Delta VOC in Scotland: demographics, risk of hospital admission, and vaccine effectiveness, Lancet, doi:10.1016/S0140-6736(21)01358-1
Shuai, Chan, Hu, Chai, Yuen et al., Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1. 529 Omicron, Nature
Starr, Greaney, Addetia, Hannon, Choudhary et al., Prospective mapping of viral mutations that escape antibodies used to treat COVID-19, Science
Starr, Greaney, Hilton, Ellis, Crawford et al., Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding, Cell, doi:10.1016/j.cell.2020.08.012
Suthar, Zimmerman, Kauffman, Mantus, Linderman et al., Rapid Generation of Neutralizing Antibody Responses in COVID-19 Patients, Cell Reports Med, doi:10.1016/j.xcrm.2020.100040
Tegally, Moir, Everatt, Giovanetti, Scheepers, Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa, Nat Med, doi:10.1038/s41591-022-01911-2
Tegally, Wilkinson, Giovanetti, Iranzadeh, Fonseca, Detection of a SARS-CoV-2 variant of concern in South Africa, Nature, doi:10.1038/s41586-021-03402-9
Tenforde, Effectiveness of a Third Dose of Pfizer-BioNTech and Moderna Vaccines in Preventing COVID-19 Hospitalization Among Immunocompetent and Immunocompromised Adults-United States, August-December 2021, MMWR Morb Mortal Wkly Rep
Tuccori, Ferraro, Convertino, Cappello, Valdiserra et al., Anti-SARS-CoV-2 neutralizing monoclonal antibodies: clinical pipeline, MAbs, doi:10.1080/19420862.2020.1854149
Uraki, Ito, Furusawa, Yamayoshi, Iwatsuki-Horimoto et al., Humoral immune evasion of the omicron subvariants BQ. 1.1 and XBB, Lancet Infect Dis
Vanblargan, Errico, Halfmann, Zost, Crowe et al., An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies, Nat Med, doi:10.1038/s41591-021-01678-y
Volz, Hill, Mccrone, Price, Jorgensen et al., Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity, Cell
Volz, Mishra, Chand, Barrett, Johnson et al., Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England, Nature
Walensky, Walke, Fauci, SARS-CoV-2 Variants of Concern in the United States-Challenges and Opportunities, JAMA J Am Med Assoc
Wang, Casner, Nair, Wang, Yu et al., Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization, Cell Host Microbe, doi:10.1016/j.chom.2021.04.007
Wang, Liu, Chen, Huang, Xu et al., The establishment of reference sequence for SARS-CoV-2 and variation analysis, J Med Virol
Wang, Nair, Liu, Iketani, Luo et al., Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1, Nature, doi:10.1038/s41586-021-03398-2
Weisblum, Schmidt, Zhang, Dasilva, Poston et al., Escape from neutralizing antibodies 1 by SARS-CoV-2 spike protein variants, Elife
West, Wertheim, Wang, Vasylyeva, Havens et al., Detection and characterization of the SARS-CoV-2 lineage B.1.526 in New York, Nat Commun, doi:10.1038/s41467-021-25168-4
Westendorf, Žentelis, Wang, Foster, Vaillancourt et al., LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants, Cell Rep
Wibmer, Ayres, Hermanus, Madzivhandila, Kgagudi et al., SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma, Nat Med, doi:10.1038/s41591-021-01285-x
Widge, Rouphael, Jackson, Anderson, Roberts et al., Durability of responses after SARS-CoV-2 mRNA-1273 vaccination, N Engl J Med
Xia, Zhu, Liu, Lan, Xu et al., Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein, Cell Mol Immunol
Yadav, Sapkal, Abraham, Ella, Deshpande et al., Neutralization of Variant Under Investigation B.1.617.1 With Sera of BBV152 Vaccinees, Clin Infect Dis, doi:10.1093/cid/ciab411
Yamasoba, Kimura, Nasser, Morioka, Nao, Virological characteristics of the SARS-CoV-2 Omicron BA.2 spike, Cell, doi:10.1016/j.cell.2022.04.035
Yamasoba, Kosugi, Kimura, Fujita, Uriu et al., Neutralisation sensitivity of SARS-CoV-2 omicron subvariants to therapeutic monoclonal antibodies, Lancet Infect Dis, doi:10.2807/1560-7917.ES.2021.26.28.2100573
Zahradník, Marciano, Shemesh, Zoler, Harari et al., SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution, Nat Microbiol, doi:10.1038/s41564-021-00954-4
Zhang, Liang, Yu, Du, Cheng et al., A systematic review of Vaccine Breakthrough Infections by SARS-CoV-2 Delta Variant, Int J Biol Sci
Zhou, Dcosta, Landau, Tada, Resistance of SARS-CoV-2 Omicron BA 1 and BA 2 Variants to Vaccine-Elicited Sera and Therapeutic Monoclonal Antibodies, Viruses
Zhou, Dcosta, Samanovic, Mulligan, Landau et al., B. 1.526 SARS-CoV-2 variants identified in New York City are neutralized by vaccine-elicited and therapeutic monoclonal antibodies, MBio
Zhou, Lou, Wang, Hu, Zhang et al., A pneumonia outbreak associated with a new coronavirus of probable bat origin, Nature, doi:10.1038/s41586-020-2012-7
DOI record:
{
"DOI": "10.1007/s15010-024-02223-y",
"ISSN": [
"0300-8126",
"1439-0973"
],
"URL": "http://dx.doi.org/10.1007/s15010-024-02223-y",
"abstract": "<jats:title>Abstract</jats:title><jats:p>The COVID-19 pandemic has affected hundreds of millions of individuals and caused more than six million deaths. The prolonged pandemic duration and the continual inter-individual transmissibility have contributed to the emergence of a wide variety of SARS-CoV-2 variants. Genomic surveillance and phylogenetic studies have shown that substantial mutations in crucial supersites of spike glycoprotein modulate the binding affinity of the evolved SARS-COV-2 lineages to ACE2 receptors and modify the binding of spike protein with neutralizing antibodies. The immunological spike mutations have been associated with differential transmissibility, infectivity, and therapeutic efficacy of the vaccines and the immunological therapies among the new variants. This review highlights the diverse genetic mutations assimilated in various SARS-CoV-2 variants. The implications of the acquired mutations related to viral transmission, infectivity, and COVID-19 severity are discussed. This review also addresses the effectiveness of human neutralizing antibodies induced by SARS-CoV-2 infection or immunization and the therapeutic antibodies against the ascended variants.</jats:p>",
"alternative-id": [
"2223"
],
"assertion": [
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Received",
"name": "received",
"order": 1,
"value": "27 December 2023"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "22 February 2024"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "First Online",
"name": "first_online",
"order": 3,
"value": "30 March 2024"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Change Date",
"name": "change_date",
"order": 4,
"value": "29 April 2024"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Change Type",
"name": "change_type",
"order": 5,
"value": "Correction"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Change Details",
"name": "change_details",
"order": 6,
"value": "A Correction to this paper has been published:"
},
{
"URL": "https://doi.org/10.1007/s15010-024-02283-0",
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Change Details",
"name": "change_details",
"order": 7,
"value": "https://doi.org/10.1007/s15010-024-02283-0"
},
{
"group": {
"label": "Declarations",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 1
},
{
"group": {
"label": "Conflict of interest",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 2,
"value": "The authors declare no conflict of interest."
}
],
"author": [
{
"affiliation": [],
"family": "Hattab",
"given": "Dima",
"sequence": "first"
},
{
"affiliation": [],
"family": "Amer",
"given": "Mumen F. A.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Al-Alami",
"given": "Zina M.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Bakhtiar",
"given": "Athirah",
"sequence": "additional"
}
],
"container-title": "Infection",
"container-title-short": "Infection",
"content-domain": {
"crossmark-restriction": false,
"domain": [
"link.springer.com"
]
},
"created": {
"date-parts": [
[
2024,
3,
30
]
],
"date-time": "2024-03-30T06:01:52Z",
"timestamp": 1711778512000
},
"deposited": {
"date-parts": [
[
2024,
5,
31
]
],
"date-time": "2024-05-31T18:35:37Z",
"timestamp": 1717180537000
},
"funder": [
{
"DOI": "10.13039/501100001779",
"doi-asserted-by": "crossref",
"name": "Monash University"
}
],
"indexed": {
"date-parts": [
[
2024,
6,
1
]
],
"date-time": "2024-06-01T00:21:02Z",
"timestamp": 1717201262107
},
"is-referenced-by-count": 1,
"issue": "3",
"issued": {
"date-parts": [
[
2024,
3,
30
]
]
},
"journal-issue": {
"issue": "3",
"published-print": {
"date-parts": [
[
2024,
6
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
3,
30
]
],
"date-time": "2024-03-30T00:00:00Z",
"timestamp": 1711756800000
}
},
{
"URL": "https://creativecommons.org/licenses/by/4.0",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
3,
30
]
],
"date-time": "2024-03-30T00:00:00Z",
"timestamp": 1711756800000
}
}
],
"link": [
{
"URL": "https://link.springer.com/content/pdf/10.1007/s15010-024-02223-y.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://link.springer.com/article/10.1007/s15010-024-02223-y/fulltext.html",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://link.springer.com/content/pdf/10.1007/s15010-024-02223-y.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "297",
"original-title": [],
"page": "767-786",
"prefix": "10.1007",
"published": {
"date-parts": [
[
2024,
3,
30
]
]
},
"published-online": {
"date-parts": [
[
2024,
3,
30
]
]
},
"published-print": {
"date-parts": [
[
2024,
6
]
]
},
"publisher": "Springer Science and Business Media LLC",
"reference": [
{
"DOI": "10.1016/S0140-6736(03)13077-2",
"author": "JSM Peiris",
"doi-asserted-by": "publisher",
"first-page": "1319",
"journal-title": "Lancet",
"key": "2223_CR1",
"unstructured": "Peiris JSM, Lai ST, Poon LLM, Guan Y, Yam LYC, Lim W, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–25.",
"volume": "361",
"year": "2003"
},
{
"DOI": "10.1097/01.inf.0000188166.17324.60",
"author": "JS Kahn",
"doi-asserted-by": "publisher",
"first-page": "223",
"journal-title": "Pediatr Infect Dis J",
"key": "2223_CR2",
"unstructured": "Kahn JS, McIntosh K. Discussion. Pediatr Infect Dis J. 2005;24:223–7.",
"volume": "24",
"year": "2005"
},
{
"DOI": "10.1016/j.cell.2020.04.011",
"author": "D Kim",
"doi-asserted-by": "publisher",
"journal-title": "Cell",
"key": "2223_CR3",
"unstructured": "Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. The Architecture of SARS-CoV-2 Transcriptome. Cell. 2020. https://doi.org/10.1016/j.cell.2020.04.011.",
"year": "2020"
},
{
"DOI": "10.3389/fmolb.2020.605236",
"author": "G Mariano",
"doi-asserted-by": "publisher",
"journal-title": "Front Mol Biosci.",
"key": "2223_CR4",
"unstructured": "Mariano G, Farthing RJ, Lale-Farjat SLM, Bergeron JRC. Structural characterization of SARS-CoV-2: where we are, and where we need to be. Front Mol Biosci. 2020;7:605236. https://doi.org/10.3389/fmolb.2020.605236.",
"volume": "7",
"year": "2020"
},
{
"DOI": "10.1080/22221751.2020.1719902",
"author": "JFW Chan",
"doi-asserted-by": "publisher",
"first-page": "221",
"journal-title": "Emerg Microbes Infect",
"key": "2223_CR5",
"unstructured": "Chan JFW, Kok KH, Zhu Z, Chu H, To KKW, Yuan S, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–36.",
"volume": "9",
"year": "2020"
},
{
"DOI": "10.1038/s41423-020-0374-2",
"author": "S Xia",
"doi-asserted-by": "publisher",
"first-page": "765",
"journal-title": "Cell Mol Immunol",
"key": "2223_CR6",
"unstructured": "Xia S, Zhu Y, Liu M, Lan Q, Xu W, Wu Y, et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol. 2020;17:765–7.",
"volume": "17",
"year": "2020"
},
{
"DOI": "10.1038/s41586-020-2012-7",
"author": "P Zhou",
"doi-asserted-by": "publisher",
"journal-title": "Nature",
"key": "2223_CR7",
"unstructured": "Zhou P, Lou YX, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020. https://doi.org/10.1038/s41586-020-2012-7.",
"year": "2020"
},
{
"DOI": "10.1038/s41467-020-15562-9",
"author": "X Ou",
"doi-asserted-by": "publisher",
"journal-title": "Nat Commun",
"key": "2223_CR8",
"unstructured": "Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020. https://doi.org/10.1038/s41467-020-15562-9.",
"year": "2020"
},
{
"key": "2223_CR9",
"unstructured": "World Health Organization. Tracking SARS-CoV-2 variants [Internet]. 2019. Available from: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/"
},
{
"key": "2223_CR10",
"unstructured": "Centers for Disease Control and Prevention. SARS variants classifications and Definitions [Internet]. 2019. Available from: www.cdc.gov/coronavirus/2019-ncov/more/scienceand-research/scientific-brief-emerging-variants.html"
},
{
"DOI": "10.3390/v12080798",
"author": "A Lai",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Viruses",
"key": "2223_CR11",
"unstructured": "Lai A, Bergna A, Caucci S, Clementi N, Vicenti I, Dragoni F, et al. Molecular tracing of SARS-CoV-2 in Italy in the first three months of the epidemic. Viruses. 2020;12:1–13.",
"volume": "12",
"year": "2020"
},
{
"DOI": "10.1016/j.celrep.2021.109292",
"author": "B Meng",
"doi-asserted-by": "publisher",
"issue": "13",
"journal-title": "Cell Rep.",
"key": "2223_CR12",
"unstructured": "Meng B, Kemp SA, Papa G, Datir R, Ferreira ATM, et al. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Rep. 2021;35(13):109292. https://doi.org/10.1016/j.celrep.2021.109292.",
"volume": "35",
"year": "2021"
},
{
"DOI": "10.1001/jama.2021.2294",
"author": "RP Walensky",
"doi-asserted-by": "publisher",
"first-page": "1037",
"journal-title": "JAMA J Am Med Assoc",
"key": "2223_CR13",
"unstructured": "Walensky RP, Walke HT, Fauci AS. SARS-CoV-2 Variants of Concern in the United States-Challenges and Opportunities. JAMA J Am Med Assoc. 2021;325:1037–8.",
"volume": "325",
"year": "2021"
},
{
"key": "2223_CR14",
"unstructured": "Resende PC, Bezerra JF, Vasconcelos RHT, Arantes I, Appolinario L, et al. Spike E484K mutation in the first SARS-CoV-2 reinfection case confirmed in Brazil. Virological.org; 2020. https://virological.org/t/spike-e484k-mutation-in-the-first-sars-cov-2-reinfection-case-confirmed-in-brazil-2020/584."
},
{
"DOI": "10.1038/s41423-021-00648-1",
"author": "J Hu",
"doi-asserted-by": "publisher",
"journal-title": "Cell Mol Immunol",
"key": "2223_CR15",
"unstructured": "Hu J, Peng P, Wang K, Fang L, Yang LF, Jin A, shun, et al. Emerging SARS-CoV-2 variants reduce neutralization sensitivity to convalescent sera and monoclonal antibodies. Cell Mol Immunol. 2021. https://doi.org/10.1038/s41423-021-00648-1.",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2021.03.013",
"author": "WF Garcia-Beltran",
"doi-asserted-by": "publisher",
"journal-title": "Cell",
"key": "2223_CR16",
"unstructured": "Garcia-Beltran WF, Lam EC, St. Denis K, Nitido AD, Garcia ZH, Hauser BM, et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021. https://doi.org/10.1016/j.cell.2021.03.013.",
"year": "2021"
},
{
"DOI": "10.1016/j.xcrm.2022.100527",
"author": "NL Miller",
"doi-asserted-by": "publisher",
"first-page": "2",
"journal-title": "Cell Reports Med",
"key": "2223_CR17",
"unstructured": "Miller NL, Clark T, Raman R, Sasisekharan R. Insights on the mutational landscape of the SARS-CoV-2 Omicron variant receptor-binding domain. Cell Reports Med. 2022;3:2.",
"volume": "3",
"year": "2022"
},
{
"DOI": "10.1101/2022.07.14.500041",
"doi-asserted-by": "crossref",
"key": "2223_CR18",
"unstructured": "Yamasoba D, Kosugi Y, Kimura I, Fujita S, Uriu K, Ito J, et al. (2022) Neutralisation sensitivity of SARS-CoV-2 omicron subvariants to therapeutic monoclonal antibodies. Lancet Infect Dis"
},
{
"DOI": "10.2807/1560-7917.ES.2021.26.28.2100573",
"author": "S Alizon",
"doi-asserted-by": "publisher",
"journal-title": "Euro Surveill",
"key": "2223_CR19",
"unstructured": "Alizon S, Haim-Boukobza S, Foulongne V, Verdurme L, Trombert-Paolantoni S, Lecorche E, et al. Rapid spread of the SARS-CoV-2 Delta variant in some French regions, June 2021. Euro Surveill. 2021. https://doi.org/10.2807/1560-7917.ES.2021.26.28.2100573.",
"year": "2021"
},
{
"DOI": "10.1002/jmv.25762",
"author": "C Wang",
"doi-asserted-by": "publisher",
"first-page": "667",
"journal-title": "J Med Virol",
"key": "2223_CR20",
"unstructured": "Wang C, Liu Z, Chen Z, Huang X, Xu M, He T, et al. The establishment of reference sequence for SARS-CoV-2 and variation analysis. J Med Virol. 2020;92:667–74.",
"volume": "92",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.11.020",
"author": "E Volz",
"doi-asserted-by": "publisher",
"first-page": "64",
"journal-title": "Cell",
"key": "2223_CR21",
"unstructured": "Volz E, Hill V, McCrone JT, Price A, Jorgensen D, O’Toole Á, et al. Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell. 2021;184:64-75.e11.",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2020.06.043",
"author": "B Korber",
"doi-asserted-by": "publisher",
"first-page": "812",
"journal-title": "Cell",
"key": "2223_CR22",
"unstructured": "Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812-827.e19.",
"volume": "182",
"year": "2020"
},
{
"DOI": "10.1126/science.abg3055",
"author": "NG Davies",
"doi-asserted-by": "publisher",
"first-page": "01",
"journal-title": "Science",
"key": "2223_CR23",
"unstructured": "Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372:01–10.",
"volume": "372",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2020.08.012",
"author": "TN Starr",
"doi-asserted-by": "publisher",
"journal-title": "Cell",
"key": "2223_CR24",
"unstructured": "Starr TN, Greaney AJ, Hilton SK, Ellis D, Crawford KHD, Dingens AS, et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell. 2020. https://doi.org/10.1016/j.cell.2020.08.012.",
"year": "2020"
},
{
"DOI": "10.1038/s41586-021-03402-9",
"author": "H Tegally",
"doi-asserted-by": "publisher",
"first-page": "438",
"journal-title": "Nature",
"key": "2223_CR25",
"unstructured": "Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–43. https://doi.org/10.1038/s41586-021-03402-9.",
"volume": "592",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2021.03.028",
"author": "M McCallum",
"doi-asserted-by": "publisher",
"journal-title": "Cell",
"key": "2223_CR26",
"unstructured": "McCallum M, De Marco A, Lempp FA, Tortorici MA, Pinto D, Walls AC, et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021. https://doi.org/10.1016/j.cell.2021.03.028.",
"year": "2021"
},
{
"DOI": "10.1038/s41591-021-01285-x",
"author": "CK Wibmer",
"doi-asserted-by": "publisher",
"journal-title": "Nat Med",
"key": "2223_CR27",
"unstructured": "Wibmer CK, Ayres F, Hermanus T, Madzivhandila M, Kgagudi P, Oosthuysen B, et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med. 2021. https://doi.org/10.1038/s41591-021-01285-x.",
"year": "2021"
},
{
"DOI": "10.1016/j.chom.2021.03.005",
"author": "G Cerutti",
"doi-asserted-by": "publisher",
"journal-title": "Cell Host Microbe",
"key": "2223_CR28",
"unstructured": "Cerutti G, Guo Y, Zhou T, Gorman J, Lee M, Rapp M, et al. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe. 2021. https://doi.org/10.1016/j.chom.2021.03.005.",
"year": "2021"
},
{
"DOI": "10.1126/science.abd2161",
"author": "DS Candido",
"doi-asserted-by": "publisher",
"first-page": "1255",
"journal-title": "Science (-80)",
"key": "2223_CR29",
"unstructured": "Candido DS, Claro IM, de Jesus JG, Souza WM, Moreira FRR, Dellicour S, et al. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science (-80). 2020;369:1255–60.",
"volume": "369",
"year": "2020"
},
{
"key": "2223_CR30",
"unstructured": "Faria NR, Claro IM, Candido D, Franco LAM, Andrade PS, Thais M, et al. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. VirologicalOrg [Internet]. 2021;1–9. Available from: https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586"
},
{
"DOI": "10.21203/rs.3.rs-318392/v1",
"doi-asserted-by": "crossref",
"key": "2223_CR31",
"unstructured": "Naveca F, Costa Cd, Nascimento V, Souza V, Corado A, et al. SARS-CoV-2 reinfection by the new Variant of Concern (VOC) P.1 in Amazonas, Brazil. Virological.org; 2021. https://virological.org/t/sars-cov-2-reinfection-by-the-new-variant-of-concern-voc-p-1-in-amazonas-brazil/596."
},
{
"DOI": "10.3390/v12121414",
"author": "MHS Paiva",
"doi-asserted-by": "publisher",
"first-page": "12",
"journal-title": "Viruses",
"key": "2223_CR32",
"unstructured": "Paiva MHS, Guedes DRD, Docena C, Bezerra MF, Dezordi FZ, Machado LC, et al. Multiple introductions followed by ongoing community spread of sars-cov-2 at one of the largest metropolitan areas of northeast brazil. Viruses. 2020;12:12.",
"volume": "12",
"year": "2020"
},
{
"DOI": "10.1101/2021.03.07.21252647",
"doi-asserted-by": "publisher",
"key": "2223_CR33",
"unstructured": "Deng X, Garcia-Knight MA, Khalid MM, Servellita V, Wang C, et al. Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation. medRxiv. 2021;2021.03.07.21252647. https://doi.org/10.1101/2021.03.07.21252647."
},
{
"DOI": "10.1002/jmv.27086",
"author": "F Pereira",
"doi-asserted-by": "publisher",
"first-page": "5523",
"journal-title": "J Med Virol",
"key": "2223_CR34",
"unstructured": "Pereira F, Tosta S, Lima MM, de Oliveira R, da Silva L, Nardy VB, Gómez MKA, et al. Genomic surveillance activities unveil the introduction of the SARS-CoV-2 B.1.525 variant of interest in Brazil: Case report. J Med Virol. 2021;93:5523–6.",
"volume": "93",
"year": "2021"
},
{
"DOI": "10.1038/s41586-021-03908-2",
"author": "MK Annavajhala",
"doi-asserted-by": "publisher",
"journal-title": "Nature",
"key": "2223_CR35",
"unstructured": "Annavajhala MK, Mohri H, Wang P, Nair M, Zucker JE, Sheng Z, et al. Emergence and expansion of SARS-CoV-2 B.1.526 after identification in New York. Nature. 2021. https://doi.org/10.1038/s41586-021-03908-2.",
"year": "2021"
},
{
"DOI": "10.1038/s41564-021-00954-4",
"author": "J Zahradník",
"doi-asserted-by": "publisher",
"journal-title": "Nat Microbiol",
"key": "2223_CR36",
"unstructured": "Zahradník J, Marciano S, Shemesh M, Zoler E, Harari D, Chiaravalli J, et al. SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat Microbiol. 2021. https://doi.org/10.1038/s41564-021-00954-4.",
"year": "2021"
},
{
"DOI": "10.1016/j.meegid.2021.105038",
"author": "K Laiton-Donato",
"doi-asserted-by": "publisher",
"journal-title": "Infect Genet Evol.",
"key": "2223_CR37",
"unstructured": "Laiton-Donato K, Franco-Muñoz C, Álvarez-Díaz DA, Ruiz-Moreno HA, Usme-Ciro JA, et al. Characterization of the emerging B.1.621 variant of interest of SARS-CoV-2. Infect Genet Evol. 2021;95:105038. https://doi.org/10.1016/j.meegid.2021.105038.",
"volume": "95",
"year": "2021"
},
{
"DOI": "10.1016/j.jmb.2020.07.009",
"author": "J Chen",
"doi-asserted-by": "publisher",
"journal-title": "J Mol Biol",
"key": "2223_CR38",
"unstructured": "Chen J, Wang R, Wang M, Wei GW. Mutations Strengthened SARS-CoV-2 Infectivity. J Mol Biol. 2020. https://doi.org/10.1016/j.jmb.2020.07.009.",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.07.012",
"author": "Q Li",
"doi-asserted-by": "publisher",
"journal-title": "Cell",
"key": "2223_CR39",
"unstructured": "Li Q, Wu J, Nie J, Zhang L, Hao H, Liu S, et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell. 2020. https://doi.org/10.1016/j.cell.2020.07.012.",
"year": "2020"
},
{
"author": "S Cherian",
"first-page": "1",
"journal-title": "Microorganism",
"key": "2223_CR40",
"unstructured": "Cherian S, Potdar V, Jadhav S, Yadav P, Gupta N, Das M, et al. in the Second Wave of COVID-19 in Maharashtra India. Microorganism. 2021;2:1–11.",
"volume": "2",
"year": "2021"
},
{
"DOI": "10.1093/cid/ciab646",
"author": "X Chen",
"doi-asserted-by": "publisher",
"first-page": "734",
"issue": "4",
"journal-title": "Clin Infect Dis.",
"key": "2223_CR41",
"unstructured": "Chen X, Chen Z, Azman AS, Sun R, Lu W, et al. Neutralizing antibodies against severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) variants induced by natural infection or vaccination: a systematic review and pooled analysis. Clin Infect Dis. 2022;74(4):734–42. https://doi.org/10.1093/cid/ciab646.",
"volume": "74",
"year": "2022"
},
{
"DOI": "10.1093/cid/ciab411",
"author": "PD Yadav",
"doi-asserted-by": "publisher",
"journal-title": "Clin Infect Dis",
"key": "2223_CR42",
"unstructured": "Yadav PD, Sapkal GN, Abraham P, Ella R, Deshpande G, Patil DY, et al. Neutralization of Variant Under Investigation B.1.617.1 With Sera of BBV152 Vaccinees. Clin Infect Dis. 2022. https://doi.org/10.1093/cid/ciab411.",
"year": "2022"
},
{
"DOI": "10.1056/NEJMc2107799",
"author": "V-V Edara",
"doi-asserted-by": "publisher",
"first-page": "664",
"journal-title": "N Engl J Med",
"key": "2223_CR43",
"unstructured": "Edara V-V, Pinsky BA, Suthar MS, Lai L, Davis-Gardner ME, Floyd K, et al. Infection and vaccine-induced neutralizing-antibody responses to the SARS-CoV-2 B. 1.617 variants. N Engl J Med. 2021;385:664–6.",
"volume": "385",
"year": "2021"
},
{
"DOI": "10.1016/j.bbrc.2021.08.036",
"author": "P Baral",
"doi-asserted-by": "publisher",
"journal-title": "Biochem Biophys Res Commun",
"key": "2223_CR44",
"unstructured": "Baral P, Bhattarai N, Hossen ML, Stebliankin V, Gerstman BS, Narasimhan G, et al. Mutation-induced changes in the receptor-binding interface of the SARS-CoV-2 Delta variant B.1.617.2 and implications for immune evasion. Biochem Biophys Res Commun. 2021. https://doi.org/10.1016/j.bbrc.2021.08.036.",
"year": "2021"
},
{
"author": "S Chowdhury",
"first-page": "52",
"journal-title": "Eur J Med Heal Sci",
"key": "2223_CR45",
"unstructured": "Chowdhury S, Bappy MH. On the Delta Plus Variant of SARS-CoV-2. Eur J Med Heal Sci. 2021;3:52–5.",
"volume": "3",
"year": "2021"
},
{
"DOI": "10.1016/j.jaut.2021.102715",
"author": "SR Kannan",
"doi-asserted-by": "publisher",
"journal-title": "J Autoimmun",
"key": "2223_CR46",
"unstructured": "Kannan SR, Spratt AN, Cohen AR, Naqvi SH, Chand HS, Quinn TP, et al. Evolutionary analysis of the Delta and Delta Plus variants of the SARS-CoV-2 viruses. J Autoimmun. 2021. https://doi.org/10.1016/j.jaut.2021.102715.",
"year": "2021"
},
{
"DOI": "10.1101/2021.11.11.21266068",
"doi-asserted-by": "crossref",
"key": "2223_CR47",
"unstructured": "Pulliam JRC, Schalkwyk C van, Govender N, Gottberg A von, Cohen C, Groome MJ, et al. (2021) Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa. medRxiv [Internet]"
},
{
"DOI": "10.1038/s41591-022-01911-2",
"author": "H Tegally",
"doi-asserted-by": "publisher",
"first-page": "1785",
"journal-title": "Nat Med.",
"key": "2223_CR48",
"unstructured": "Tegally H, Moir M, Everatt J, Giovanetti M, Scheepers C, et al. Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nat Med. 2022;28:1785–90. https://doi.org/10.1038/s41591-022-01911-2.",
"volume": "28",
"year": "2022"
},
{
"DOI": "10.1016/j.chom.2022.10.003",
"author": "A Saito",
"doi-asserted-by": "publisher",
"first-page": "1540",
"issue": "11",
"journal-title": "Cell Host Microbe.",
"key": "2223_CR49",
"unstructured": "Saito A, Tamura T, Zahradnik J, Deguchi S, Tabata K, et al. Virological characteristics of the SARS-CoV-2 Omicron BA275 variant. Cell Host Microbe. 2022;30(11):1540–55.e15. https://doi.org/10.1016/j.chom.2022.10.003.",
"volume": "30",
"year": "2022"
},
{
"DOI": "10.1126/science.abn4543",
"author": "L Scott",
"doi-asserted-by": "publisher",
"first-page": "1454",
"journal-title": "Science (-80)",
"key": "2223_CR50",
"unstructured": "Scott L, Hsiao N, Moyo S, Singh L, Tegally H, Dor G, et al. Track Omicron’s spread with molecular data. Science (-80). 2021;374:1454–5.",
"volume": "374",
"year": "2021"
},
{
"DOI": "10.3389/fviro.2022.1077155",
"doi-asserted-by": "crossref",
"key": "2223_CR51",
"unstructured": "Mohapatra RK, Mahal A, Kutikuppala L V, Pal M, Kandi V, Sarangi AK, et al. (2022) Renewed global threat by the novel SARS-CoV-2 variants ‘XBB, BF 7, BQ 1, BA. 275, BA 46’: A discussion. Front Virol 2:1077155"
},
{
"author": "DV Parums",
"first-page": "e939580",
"journal-title": "Med Sci Monit Int Med J Exp Clin Res",
"key": "2223_CR52",
"unstructured": "Parums DV. The XBB. 1.5 (‘Kraken’) Subvariant of Omicron SARS-CoV-2 and its Rapid Global Spread. Med Sci Monit Int Med J Exp Clin Res. 2023;29:e939580–1.",
"volume": "29",
"year": "2023"
},
{
"DOI": "10.3390/ijms24032264",
"author": "D Focosi",
"doi-asserted-by": "publisher",
"first-page": "2264",
"issue": "3",
"journal-title": "Int J Mol Sci.",
"key": "2223_CR53",
"unstructured": "Focosi D, Quiroga R, McConnell S, Johnson MC, Casadevall A. Convergent evolution in SARS-CoV-2 spike creates a variant soup from which new COVID-19 waves emerge. Int J Mol Sci. 2023;24(3):2264. https://doi.org/10.3390/ijms24032264.",
"volume": "24",
"year": "2023"
},
{
"DOI": "10.1016/S1473-3099(22)00311-5",
"author": "D Focosi",
"doi-asserted-by": "publisher",
"first-page": "e311",
"issue": "11",
"journal-title": "Lancet Infect Dis.",
"key": "2223_CR54",
"unstructured": "Focosi D, McConnell S, Casadevall A, Cappello E, Valdiserra G, Tuccori M. Monoclonal antibody therapies against SARS-CoV-2. Lancet Infect Dis. 2022;22(11):e311–26. https://doi.org/10.1016/S1473-3099(22)00311-5.",
"volume": "22",
"year": "2022"
},
{
"DOI": "10.1016/j.cell.2020.09.032",
"author": "L Yurkovetskiy",
"doi-asserted-by": "publisher",
"journal-title": "Cell",
"key": "2223_CR55",
"unstructured": "Yurkovetskiy L, Wang X, Pascal KE, Tomkins-Tinch C, Nyalile TP, Wang Y, et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell. 2020. https://doi.org/10.1016/j.cell.2020.09.032.",
"year": "2020"
},
{
"DOI": "10.1038/s41586-021-03470-x",
"author": "E Volz",
"doi-asserted-by": "publisher",
"first-page": "266",
"journal-title": "Nature",
"key": "2223_CR56",
"unstructured": "Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593:266–9.",
"volume": "593",
"year": "2021"
},
{
"key": "2223_CR57",
"unstructured": "Pearson CA, Russell TW, Davies NG, Kucharski AJ, CMMID COVID-19 working group, Edmunds WJ, et al. (2021) Estimates of severity and transmissibility of novel South Africa SARS-CoV-2 variant 501Y.V2. Preprint 50:1–4"
},
{
"DOI": "10.1093/ve/veab091",
"author": "T Gräf",
"doi-asserted-by": "publisher",
"first-page": "veab091",
"issue": "2",
"journal-title": "Virus Evol.",
"key": "2223_CR58",
"unstructured": "Gräf T, Bello G, Venas TMM, Pereira EC, Paixão ACD, et al. Identification of a novel SARS-CoV-2 P.1 sub-lineage in Brazil provides new insights about the mechanisms of emergence of variants of concern. Virus Evol. 2021;7(2):veab091. https://doi.org/10.1093/ve/veab091.",
"volume": "7",
"year": "2021"
},
{
"DOI": "10.1111/1462-2920.15687",
"author": "J Chadha",
"doi-asserted-by": "publisher",
"first-page": "2615",
"issue": "6",
"journal-title": "Environ Microbiol.",
"key": "2223_CR59",
"unstructured": "Chadha J, Khullar L, Mittal N. Facing the wrath of enigmatic mutations: a review on the emergence of severe acute respiratory syndrome coronavirus 2 variants amid coronavirus disease-19 pandemic. Environ Microbiol. 2022;24(6):2615–29. https://doi.org/10.1111/1462-2920.15687.",
"volume": "24",
"year": "2022"
},
{
"DOI": "10.1093/jtm/taab119",
"author": "GN Sapkal",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "J Travel Med",
"key": "2223_CR60",
"unstructured": "Sapkal GN, Yadav PD, Sahay RR, Deshpande G, Gupta N, Nyayanit DA, et al. Neutralization of Delta variant with sera of Covishield™ vaccinees and COVID-19-recovered vaccinated individuals. J Travel Med. 2021;28:1–3.",
"volume": "28",
"year": "2021"
},
{
"DOI": "10.1016/S0140-6736(21)01358-1",
"author": "A Sheikh",
"doi-asserted-by": "publisher",
"journal-title": "Lancet",
"key": "2223_CR61",
"unstructured": "Sheikh A, McMenamin J, Taylor B, Robertson C. SARS-CoV-2 Delta VOC in Scotland: demographics, risk of hospital admission, and vaccine effectiveness. Lancet. 2021. https://doi.org/10.1016/S0140-6736(21)01358-1.",
"year": "2021"
},
{
"DOI": "10.1016/j.mayocp.2021.12.002",
"author": "JC O’Horo",
"doi-asserted-by": "publisher",
"journal-title": "Mayo Clin Proc",
"key": "2223_CR62",
"unstructured": "O’Horo JC, Challener DW, Speicher L, Bosch W, Seville MT, Bierle DM, et al. Effectiveness of Monoclonal Antibodies in Preventing Severe COVID-19 With Emergence of the Delta Variant. Mayo Clin Proc. 2022. https://doi.org/10.1016/j.mayocp.2021.12.002.",
"year": "2022"
},
{
"author": "S Chowdhury",
"first-page": "17",
"journal-title": "Eur J Med Heal Sci",
"key": "2223_CR63",
"unstructured": "Chowdhury S, Bappy MH, Chowdhury S, Chowdhury MS, Chowdhury NS. COVID-19 Induced Cardiovascular Complications and Recent Therapeutic Advances. Eur J Med Heal Sci. 2021;3:17–22.",
"volume": "3",
"year": "2021"
},
{
"DOI": "10.1038/s41586-022-04474-x",
"author": "B Meng",
"doi-asserted-by": "publisher",
"first-page": "706",
"journal-title": "Nature",
"key": "2223_CR64",
"unstructured": "Meng B, Abdullahi A, Ferreira IATM, Goonawardane N, Saito A, Kimura I, et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature. 2022;603:706–14.",
"volume": "603",
"year": "2022"
},
{
"DOI": "10.1038/s41586-022-04442-5",
"author": "H Shuai",
"doi-asserted-by": "publisher",
"first-page": "693",
"journal-title": "Nature",
"key": "2223_CR65",
"unstructured": "Shuai H, Chan JF-W, Hu B, Chai Y, Yuen TT-T, Yin F, et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1. 529 Omicron. Nature. 2022;603:693–9.",
"volume": "603",
"year": "2022"
},
{
"DOI": "10.1016/j.cell.2022.04.035",
"author": "D Yamasoba",
"doi-asserted-by": "publisher",
"first-page": "2103",
"issue": "12",
"journal-title": "Cell.",
"key": "2223_CR66",
"unstructured": "Yamasoba D, Kimura I, Nasser H, Morioka Y, Nao N, et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2 spike. Cell. 2022;185(12):2103–115.e19. https://doi.org/10.1016/j.cell.2022.04.035.",
"volume": "185",
"year": "2022"
},
{
"DOI": "10.1016/j.ijsu.2022.106698",
"author": "RK Mohapatra",
"doi-asserted-by": "publisher",
"first-page": "106698",
"journal-title": "Int J Surg",
"key": "2223_CR67",
"unstructured": "Mohapatra RK, Kandi V, Sarangi AK, Verma S, Tuli HS, Chakraborty S, et al. The recently emerged BA. 4 and BA. 5 lineages of Omicron and their global health concerns amid the ongoing wave of COVID-19 pandemic–Correspondence. Int J Surg. 2022;103:106698.",
"volume": "103",
"year": "2022"
},
{
"DOI": "10.3390/ijms24032264",
"author": "D Focosi",
"doi-asserted-by": "publisher",
"first-page": "2264",
"journal-title": "Int J Mol Sci",
"key": "2223_CR68",
"unstructured": "Focosi D, Quiroga R, McConnell S, Johnson MC, Casadevall A. Convergent evolution in SARS-CoV-2 spike creates a variant soup from which new COVID-19 waves emerge. Int J Mol Sci. 2023;24:2264.",
"volume": "24",
"year": "2023"
},
{
"DOI": "10.1038/s41467-023-38188-z",
"author": "J Ito",
"doi-asserted-by": "publisher",
"first-page": "2671",
"journal-title": "Nat Commun",
"key": "2223_CR69",
"unstructured": "Ito J, Suzuki R, Uriu K, Itakura Y, Zahradnik J, Kimura KT, et al. Convergent evolution of SARS-CoV-2 Omicron subvariants leading to the emergence of BQ11 variant. Nat Commun. 2023;14:2671.",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.1016/j.xcrm.2020.100040",
"author": "MS Suthar",
"doi-asserted-by": "publisher",
"journal-title": "Cell Reports Med",
"key": "2223_CR70",
"unstructured": "Suthar MS, Zimmerman MG, Kauffman RC, Mantus G, Linderman SL, Hudson WH, et al. Rapid Generation of Neutralizing Antibody Responses in COVID-19 Patients. Cell Reports Med. 2020. https://doi.org/10.1016/j.xcrm.2020.100040.",
"year": "2020"
},
{
"author": "JM Dan",
"first-page": "6529",
"journal-title": "Science (-80)",
"key": "2223_CR71",
"unstructured": "Dan JM, Mateus J, Kato Y, Hastie KM, Yu ED, Faliti CE, et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science (-80). 2021;371:6529.",
"volume": "371",
"year": "2021"
},
{
"DOI": "10.1056/NEJMc2032195",
"author": "AT Widge",
"doi-asserted-by": "publisher",
"first-page": "80",
"journal-title": "N Engl J Med",
"key": "2223_CR72",
"unstructured": "Widge AT, Rouphael NG, Jackson LA, Anderson EJ, Roberts PC, Makhene M, et al. Durability of responses after SARS-CoV-2 mRNA-1273 vaccination. N Engl J Med. 2021;384:80–2.",
"volume": "384",
"year": "2021"
},
{
"DOI": "10.1080/19420862.2020.1854149",
"author": "M Tuccori",
"doi-asserted-by": "publisher",
"journal-title": "MAbs",
"key": "2223_CR73",
"unstructured": "Tuccori M, Ferraro S, Convertino I, Cappello E, Valdiserra G, Blandizzi C, et al. (2020) Anti-SARS-CoV-2 neutralizing monoclonal antibodies: clinical pipeline. MAbs. 2020. https://doi.org/10.1080/19420862.2020.1854149.",
"year": "2020"
},
{
"DOI": "10.1038/s41586-021-04385-3",
"author": "Y Cao",
"doi-asserted-by": "publisher",
"first-page": "657",
"journal-title": "Nature",
"key": "2223_CR74",
"unstructured": "Cao Y, Wang J, Jian F, Xiao T, Song W, Yisimayi A, et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–63.",
"volume": "602",
"year": "2022"
},
{
"DOI": "10.1038/s41586-020-2852-1",
"author": "CO Barnes",
"doi-asserted-by": "publisher",
"journal-title": "Nature",
"key": "2223_CR75",
"unstructured": "Barnes CO, Jette CA, Abernathy ME, Dam KMA, Esswein SR, Gristick HB, et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020. https://doi.org/10.1038/s41586-020-2852-1.",
"year": "2020"
},
{
"DOI": "10.1038/s41467-021-24435-8",
"author": "AJ Greaney",
"doi-asserted-by": "publisher",
"journal-title": "Nat Commun",
"key": "2223_CR76",
"unstructured": "Greaney AJ, Starr TN, Barnes CO, Weisblum Y, Schmidt F, Caskey M, et al. Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nat Commun. 2021. https://doi.org/10.1038/s41467-021-24435-8.",
"year": "2021"
},
{
"DOI": "10.1101/2021.03.09.434607",
"author": "AL Cathcart",
"doi-asserted-by": "publisher",
"journal-title": "bioRxiv",
"key": "2223_CR77",
"unstructured": "Cathcart AL, Havenar-Daughton C, Lempp FA, Ma D, Schmid MA, et al. The dual function monoclonal antibodies VIR-7831 and VIR-7832 demonstrate potent in vitro and in vivo activity against SARS-CoV-2. bioRxiv. 2021. https://doi.org/10.1101/2021.03.09.434607.",
"year": "2021"
},
{
"DOI": "10.1016/j.celrep.2022.110812",
"author": "K Westendorf",
"doi-asserted-by": "publisher",
"journal-title": "Cell Rep",
"key": "2223_CR78",
"unstructured": "Westendorf K, Žentelis S, Wang L, Foster D, Vaillancourt P, Wiggin M, et al. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. Cell Rep. 2022;39: 110812.",
"volume": "39",
"year": "2022"
},
{
"DOI": "10.1016/j.cell.2020.12.015",
"author": "WF Garcia-Beltran",
"doi-asserted-by": "publisher",
"journal-title": "Cell",
"key": "2223_CR79",
"unstructured": "Garcia-Beltran WF, Lam EC, Astudillo MG, Yang D, Miller TE, Feldman J, et al. COVID-19-neutralizing antibodies predict disease severity and survival. Cell. 2021. https://doi.org/10.1016/j.cell.2020.12.015.",
"year": "2021"
},
{
"DOI": "10.1101/2021.02.02.21250799",
"doi-asserted-by": "crossref",
"key": "2223_CR80",
"unstructured": "Edara VV, Floyd K, Lai L, Gardner M, Hudson W, Piantadosi A, et al. Infection and mRNA-1273 vaccine antibodies neutralize SARS-CoV-2 UK variant. medRxiv Prepr Serv Heal Sci [Internet]. 2021 Feb 5;2021.02.02.21250799. Available from: https://pubmed.ncbi.nlm.nih.gov/33564782"
},
{
"DOI": "10.1038/s41586-021-03398-2",
"author": "P Wang",
"doi-asserted-by": "publisher",
"journal-title": "Nature",
"key": "2223_CR81",
"unstructured": "Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y, et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021. https://doi.org/10.1038/s41586-021-03398-2.",
"year": "2021"
},
{
"DOI": "10.1101/2021.01.19.21249840",
"doi-asserted-by": "crossref",
"key": "2223_CR82",
"unstructured": "Collier D, De Marco A, Ferreira I, Meng B, Datir R, Walls AC, et al. (2021) SARS-CoV-2 B. 1.1. 7 sensitivity to mRNA vaccine-elicited, convalescent and monoclonal antibodies. MedRxiv"
},
{
"DOI": "10.1101/2021.01.25.427948",
"doi-asserted-by": "publisher",
"key": "2223_CR83",
"unstructured": "Wu K, Werner AP, Moliva JI, Koch M, Choi A, et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv. 2021;2021.01.25.427948. https://doi.org/10.1101/2021.01.25.427948."
},
{
"DOI": "10.1016/j.cell.2021.03.036",
"author": "M Hoffmann",
"doi-asserted-by": "publisher",
"first-page": "2384",
"journal-title": "Cell",
"key": "2223_CR84",
"unstructured": "Hoffmann M, Arora P, Groß R, Seidel A, Hörnich BF, Hahn AS, et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384-2393.e12.",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.7554/eLife.61312",
"author": "Y Weisblum",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Elife",
"key": "2223_CR85",
"unstructured": "Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, Lorenzi JCC, et al. Escape from neutralizing antibodies 1 by SARS-CoV-2 spike protein variants. Elife. 2020;9:1.",
"volume": "9",
"year": "2020"
},
{
"DOI": "10.1056/NEJMc2102017",
"author": "Y Liu",
"doi-asserted-by": "publisher",
"first-page": "1466",
"journal-title": "N Engl J Med",
"key": "2223_CR86",
"unstructured": "Liu Y, Liu J, Xia H, Zhang X, Fontes-Garfias CR, Swanson KA, et al. Neutralizing activity of BNT162b2-elicited serum. N Engl J Med. 2021;384:1466–8.",
"volume": "384",
"year": "2021"
},
{
"DOI": "10.1016/j.chom.2021.04.007",
"author": "P Wang",
"doi-asserted-by": "publisher",
"journal-title": "Cell Host Microbe",
"key": "2223_CR87",
"unstructured": "Wang P, Casner RG, Nair MS, Wang M, Yu J, Cerutti G, et al. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe. 2021. https://doi.org/10.1016/j.chom.2021.04.007.",
"year": "2021"
},
{
"DOI": "10.1016/j.chom.2020.11.007",
"author": "AJ Greaney",
"doi-asserted-by": "publisher",
"journal-title": "Cell Host Microbe",
"key": "2223_CR88",
"unstructured": "Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, et al. Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition. Cell Host Microbe. 2021. https://doi.org/10.1016/j.chom.2020.11.007.",
"year": "2021"
},
{
"DOI": "10.1128/mBio.01386-21",
"author": "H Zhou",
"doi-asserted-by": "publisher",
"first-page": "e01386",
"journal-title": "MBio",
"key": "2223_CR89",
"unstructured": "Zhou H, Dcosta BM, Samanovic MI, Mulligan MJ, Landau NR, Tada T. B. 1.526 SARS-CoV-2 variants identified in New York City are neutralized by vaccine-elicited and therapeutic monoclonal antibodies. MBio. 2021;12:e01386-e1421.",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1038/s41467-021-25168-4",
"author": "AP West",
"doi-asserted-by": "publisher",
"journal-title": "Nat Commun",
"key": "2223_CR90",
"unstructured": "West AP, Wertheim JO, Wang JC, Vasylyeva TI, Havens JL, Chowdhury MA, et al. Detection and characterization of the SARS-CoV-2 lineage B.1.526 in New York. Nat Commun. 2021. https://doi.org/10.1038/s41467-021-25168-4.",
"year": "2021"
},
{
"DOI": "10.1016/j.celrep.2021.109415",
"author": "M Hoffmann",
"doi-asserted-by": "publisher",
"first-page": "3",
"journal-title": "Cell Rep",
"key": "2223_CR91",
"unstructured": "Hoffmann M, Hofmann-Winkler H, Krüger N, Kempf A, Nehlmeier I, Graichen L, et al. SARS-CoV-2 variant B1617 is resistant to bamlanivimab and evades antibodies induced by infection and vaccination. Cell Rep. 2021;36:3.",
"volume": "36",
"year": "2021"
},
{
"key": "2223_CR92",
"unstructured": "Ferreira I, Datir R, Papa G, Kemp S, Meng B, Rakshit P, et al. SARS-CoV-2 B.1.617 emergence and sensitivity to vaccine-elicited antibodies. bioRxiv [Internet]. 2021 Jan 1;2021.05.08.443253. Available from: http://biorxiv.org/content/early/2021/05/09/2021.05.08.443253.abstract"
},
{
"DOI": "10.3390/v13091782",
"author": "N Gupta",
"doi-asserted-by": "publisher",
"first-page": "1782",
"issue": "9",
"journal-title": "Viruses.",
"key": "2223_CR93",
"unstructured": "Gupta N, Kaur H, Yadav PD, Mukhopadhyay L, Sahay RR, et al. Clinical characterization and genomic analysis of samples from COVID-19 breakthrough infections during the second wave among the various states of India. Viruses. 2021;13(9):1782. https://doi.org/10.3390/v13091782.",
"volume": "13",
"year": "2021"
},
{
"DOI": "10.7150/ijbs.68973",
"author": "M Zhang",
"doi-asserted-by": "publisher",
"first-page": "889",
"journal-title": "Int J Biol Sci",
"key": "2223_CR94",
"unstructured": "Zhang M, Liang Y, Yu D, Du B, Cheng W, Li L, et al. A systematic review of Vaccine Breakthrough Infections by SARS-CoV-2 Delta Variant. Int J Biol Sci. 2022;18:889–900.",
"volume": "18",
"year": "2022"
},
{
"DOI": "10.1016/j.ijid.2021.09.006",
"author": "MR Hasan",
"doi-asserted-by": "publisher",
"first-page": "52",
"journal-title": "Int J Infect Dis",
"key": "2223_CR95",
"unstructured": "Hasan MR, Kalikiri MKR, Mirza F, Sundararaju S, Sharma A, Xaba T, et al. Real-Time SARS-CoV-2 Genotyping by High-Throughput Multiplex PCR Reveals the Epidemiology of the Variants of Concern in Qatar. Int J Infect Dis. 2021;112:52–4.",
"volume": "112",
"year": "2021"
},
{
"DOI": "10.1056/NEJMoa2108891",
"author": "J Lopez Bernal",
"doi-asserted-by": "publisher",
"first-page": "585",
"journal-title": "N Engl J Med",
"key": "2223_CR96",
"unstructured": "Lopez Bernal J, Andrews N, Gower C, Gallagher E, Simmons R, Thelwall S, et al. Effectiveness of Covid-19 Vaccines against the B16172 (Delta) Variant. N Engl J Med. 2021;385:585–94.",
"volume": "385",
"year": "2021"
},
{
"DOI": "10.1038/s41586-021-03777-9",
"author": "D Planas",
"doi-asserted-by": "publisher",
"journal-title": "Nature",
"key": "2223_CR97",
"unstructured": "Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021. https://doi.org/10.1038/s41586-021-03777-9.",
"year": "2021"
},
{
"DOI": "10.15585/mmwr.mm7104a2",
"doi-asserted-by": "crossref",
"key": "2223_CR98",
"unstructured": "Tenforde MW (2022) Effectiveness of a Third Dose of Pfizer-BioNTech and Moderna Vaccines in Preventing COVID-19 Hospitalization Among Immunocompetent and Immunocompromised Adults—United States, August–December 2021. MMWR Morb Mortal Wkly Rep 71"
},
{
"DOI": "10.1016/j.cell.2021.12.033",
"author": "WF Garcia-Beltran",
"doi-asserted-by": "publisher",
"journal-title": "Cell",
"key": "2223_CR99",
"unstructured": "Garcia-Beltran WF, St. Denis KJ, Hoelzemer A, Lam EC, Nitido AD, Sheehan ML, et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022. https://doi.org/10.1016/j.cell.2021.12.033.",
"year": "2022"
},
{
"DOI": "10.15585/mmwr.mm7037e2",
"author": "SJ Grannis",
"doi-asserted-by": "publisher",
"first-page": "1291",
"journal-title": "Morb Mortal Wkly Rep",
"key": "2223_CR100",
"unstructured": "Grannis SJ, Rowley EA, Ong TC, Stenehjem E, Klein NP, DeSilva MB, et al. Interim estimates of COVID-19 vaccine effectiveness against COVID-19–associated emergency department or urgent care clinic encounters and hospitalizations among adults during SARS-CoV-2 B.1617. 2 (Delta) variant predominance—Nine States, June–August 202. Morb Mortal Wkly Rep. 2021;70:1291.",
"volume": "70",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2021.06.020",
"author": "C Liu",
"doi-asserted-by": "publisher",
"first-page": "4220",
"journal-title": "Cell",
"key": "2223_CR101",
"unstructured": "Liu C, Ginn HM, Dejnirattisai W, Supasa P, Wang B, Tuekprakhon A, et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell. 2021;184:4220-4236.e13.",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1038/s41423-021-00772-y",
"author": "P Arora",
"doi-asserted-by": "publisher",
"first-page": "2557",
"journal-title": "Cell Mol Immunol",
"key": "2223_CR102",
"unstructured": "Arora P, Kempf A, Nehlmeier I, Graichen L, Sidarovich A, Winkler MS, et al. Delta variant (B.1.617.2) sublineages do not show increased neutralization resistance. Cell Mol Immunol. 2021;18:2557–9.",
"volume": "18",
"year": "2021"
},
{
"DOI": "10.1126/science.abf9302",
"author": "TN Starr",
"doi-asserted-by": "publisher",
"first-page": "850",
"journal-title": "Science (-80)",
"key": "2223_CR103",
"unstructured": "Starr TN, Greaney AJ, Addetia A, Hannon WW, Choudhary MC, Dingens AS, et al. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science (-80). 2021;371:850–4.",
"volume": "371",
"year": "2021"
},
{
"DOI": "10.1056/NEJMc2119358",
"author": "I Nemet",
"doi-asserted-by": "publisher",
"first-page": "492",
"issue": "5",
"journal-title": "N Engl J Med.",
"key": "2223_CR104",
"unstructured": "Nemet I, Kliker L, Lustig Y, Zuckerman N, Erster O, et al. Third BNT162b2 vaccination neutralization of SARS-CoV-2 Omicron infection. N Engl J Med. 2022;386(5):492–4. https://doi.org/10.1056/NEJMc2119358.",
"volume": "386",
"year": "2022"
},
{
"key": "2223_CR105",
"unstructured": "Administration. USF and D. Coronavirus (COVID-19) Update: FDA Limits Use of Certain Monoclonal Antibodies to Treat COVID-19 Due to the Omicron Variant [Internet]. 2022. Available from: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-limits-use-certain-monoclonal-antibodies-treat-covid-19-due-omicron"
},
{
"DOI": "10.3390/v14061334",
"author": "H Zhou",
"doi-asserted-by": "publisher",
"first-page": "1334",
"journal-title": "Viruses",
"key": "2223_CR106",
"unstructured": "Zhou H, Dcosta BM, Landau NR, Tada T. Resistance of SARS-CoV-2 Omicron BA 1 and BA 2 Variants to Vaccine-Elicited Sera and Therapeutic Monoclonal Antibodies. Viruses. 2022;14:1334.",
"volume": "14",
"year": "2022"
},
{
"key": "2223_CR107",
"unstructured": "Administration USF and D. FDA authorizes revisions to Evusheld dosing [Internet]. 2022. Available from: https://www.fda.gov/drugs/drug-safety-and-availability/fda-authorizes-revisions-evusheld-dosing"
},
{
"DOI": "10.1038/s41591-021-01678-y",
"author": "LA VanBlargan",
"doi-asserted-by": "publisher",
"first-page": "490",
"journal-title": "Nat Med.",
"key": "2223_CR108",
"unstructured": "VanBlargan LA, Errico JM, Halfmann PJ, Zost SJ, Crowe JE Jr, et al. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat Med. 2022;28:490–5. https://doi.org/10.1038/s41591-021-01678-y.",
"volume": "28",
"year": "2022"
},
{
"DOI": "10.1038/s41586-022-04594-4",
"author": "S Iketani",
"doi-asserted-by": "publisher",
"first-page": "553",
"issue": "7906",
"journal-title": "Nature",
"key": "2223_CR109",
"unstructured": "Iketani S, Liu L, Guo Y, Liu L, Chan JF-W, Huang Y, et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature. 2022;604(7906):553–6.",
"volume": "604",
"year": "2022"
},
{
"DOI": "10.1038/s41467-022-30681-1",
"author": "C Kurhade",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Nat Commun",
"key": "2223_CR110",
"unstructured": "Kurhade C, Zou J, Xia H, Cai H, Yang Q, Cutler M, et al. Neutralization of Omicron BA. 1, BA. 2, and BA. 3 SARS-CoV-2 by 3 doses of BNT162b2 vaccine. Nat Commun. 2022;13:1–4.",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.1002/mco2.130",
"author": "Q Li",
"doi-asserted-by": "publisher",
"journal-title": "MedComm",
"key": "2223_CR111",
"unstructured": "Li Q, Zhang M, Liang Z, Zhang L, Wu X, Yang C, et al. Antigenicity comparison of SARS-CoV-2 Omicron sublineages with other variants contained multiple mutations in RBD. MedComm. 2022;3: e130.",
"volume": "3",
"year": "2022"
},
{
"DOI": "10.1016/S1473-3099(22)00580-1",
"author": "H Gruell",
"doi-asserted-by": "publisher",
"first-page": "1422",
"journal-title": "Lancet Infect Dis",
"key": "2223_CR112",
"unstructured": "Gruell H, Vanshylla K, Tober-Lau P, Hillus D, Sander LE, Kurth F, et al. Neutralisation sensitivity of the SARS-CoV-2 omicron BA.2.75 sublineage. Lancet Infect Dis. 2022;22:1422–3.",
"volume": "22",
"year": "2022"
},
{
"DOI": "10.1016/S1473-3099(22)00816-7",
"author": "R Uraki",
"doi-asserted-by": "publisher",
"first-page": "30",
"journal-title": "Lancet Infect Dis",
"key": "2223_CR113",
"unstructured": "Uraki R, Ito M, Furusawa Y, Yamayoshi S, Iwatsuki-Horimoto K, Adachi E, et al. Humoral immune evasion of the omicron subvariants BQ. 1.1 and XBB. Lancet Infect Dis. 2023;23:30–2.",
"volume": "23",
"year": "2023"
},
{
"DOI": "10.1056/NEJMc2214293",
"author": "ME Davis-Gardner",
"doi-asserted-by": "publisher",
"first-page": "183",
"journal-title": "N Engl J Med",
"key": "2223_CR114",
"unstructured": "Davis-Gardner ME, Lai L, Wali B, Samaha H, Solis D, Lee M, et al. Neutralization against BA. 2.75. 2, BQ.1.1, and XBB from mRNA Bivalent Booster. N Engl J Med. 2023;388:183–5.",
"volume": "388",
"year": "2023"
}
],
"reference-count": 114,
"references-count": 114,
"relation": {},
"resource": {
"primary": {
"URL": "https://link.springer.com/10.1007/s15010-024-02223-y"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "SARS-CoV-2 journey: from alpha variant to omicron and its sub-variants",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1007/springer_crossmark_policy",
"volume": "52"
}
hattab