Multi-omics and machine learning identify novel biomarkers and therapeutic targets of COVID-19
et al., Frontiers in Immunology, doi:10.3389/fimmu.2025.1671936, Oct 2025
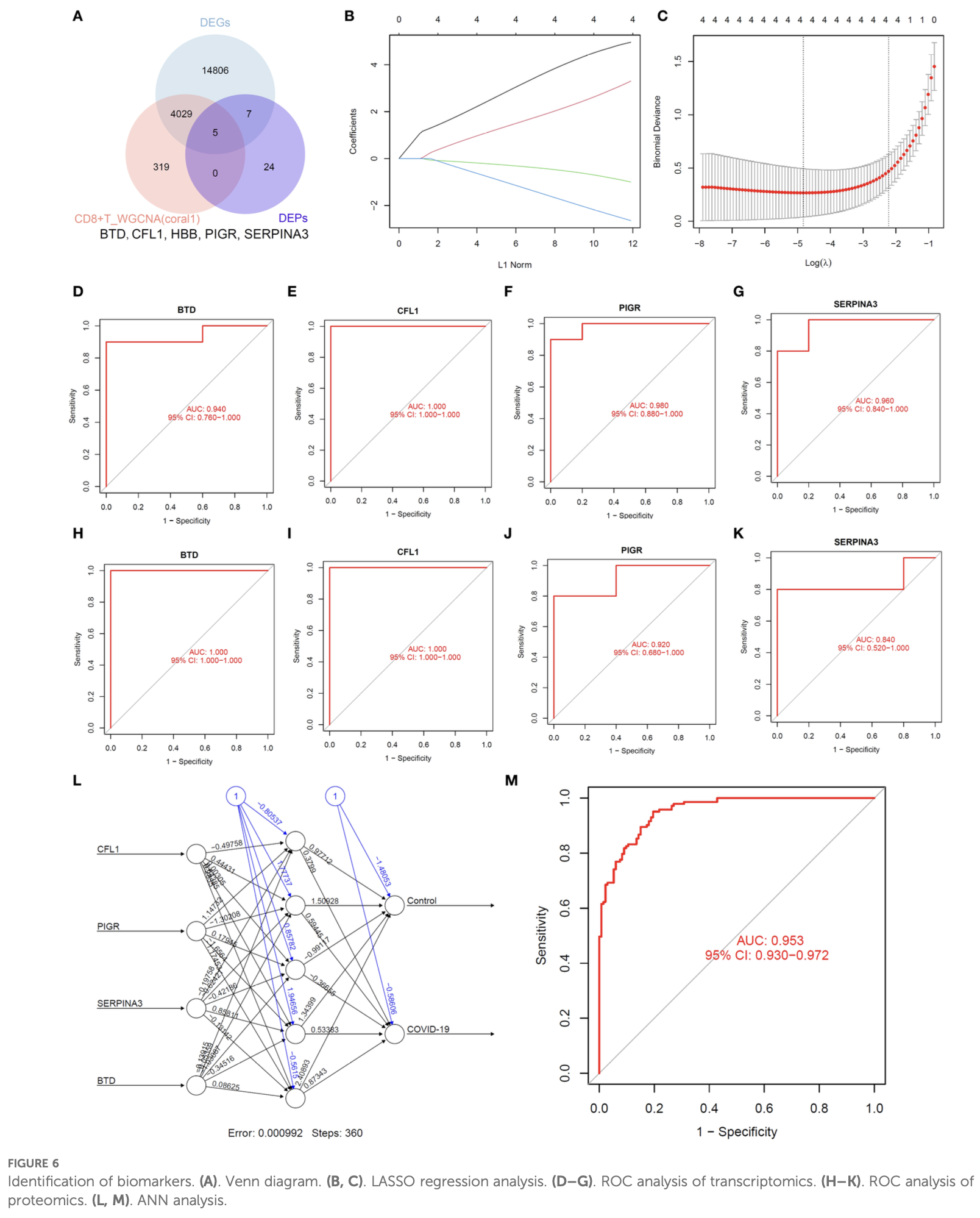
In silico study showing potential therapeutic targets BTD, CFL1, PIGR, and SERPINA3 for COVID-19 through integrated analysis of clinical data, single-cell RNA sequencing, bulk RNA sequencing, and proteomics.
Zhou et al., 1 Oct 2025, peer-reviewed, 7 authors, study period January 2023 - June 2023.
Contact: doctorwang2009@126.com, wangqi710@126.com.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Multi-omics and machine learning identify novel biomarkers and therapeutic targets of COVID-19
Frontiers in Immunology, doi:10.3389/fimmu.2025.1671936
Introduction: COVID-19 has caused over 7 million deaths worldwide since its onset in 2019, and the virus remains a significant health threat. Identifying sensitive and specific biomarkers, along with elucidating immune-mediated mechanisms, is essential for improving the diagnosis, treatment, and prevention of COVID-19. To predict key molecular markers of COVID-19 using an established multi-omics framework combined with machine learning models. Methods: We conducted an integrated analysis of single-cell RNA sequencing (scRNA-seq), bulk RNA sequencing, and proteomics data to identify critical biomarkers associated with COVID-19. The multi-omics approach enabled the characterization of gene expression dynamics and alterations in immune cell subsets in COVID-19 patients. Machine learning techniques and molecular docking analyses were employed to identify biomarkers and therapeutic targets within the disease's pathophysiological network. Results: Principal component analysis effectively grouped samples based on clinical characteristics. Using random forest and SVM-RFE models, we identified clinical indicators capable of accurately distinguishing COVID-19 patients. Transcriptomic analysis, including scRNA-seq, highlighted the pivotal role of CD8 + T cells, and WGCNA identified related module genes. Proteomic analysis, integrated with machine learning, revealed 36 DEPs. Further investigation identified several genes associated with monocyte proportions. Correlation analysis showed that BTD, CFL1, PIGR, and SERPINA3 were strongly linked to CD8 + T cell abundance in COVID-19 patients. ROC curve analysis demonstrated that these genes could effectively distinguish between COVID-19 patients and healthy individuals. Concordant findings from both transcriptomic and proteomic levels support BTD, CFL1, PIGR, and SERPINA3 as potential auxiliary diagnostic markers. Finally, AlphaFold-based molecular docking analysis suggested these biomarkers may also serve as candidate therapeutic targets.
Ethics statement The studies involving humans were approved by the Ethics Committee of the Second Affiliated Hospital of Mudanjiang Medical College (Approval No. 202328 ). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
Conflict of interest The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement The author(s) declare that no Generative AI was used in the creation of this manuscript. Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material The Supplementary Material for this article can be found online at: ..
References
Amrute, Perry, Anand, Cruchaga, Hock et al., Cell specific peripheral immune responses predict survival in critical COVID-19 patients, Nat Commun, doi:10.1038/s41467-022-28505-3
Athaya, Ripan, Li, Hu, Multimodal deep learning approaches for singlecell multi-omics data integration, Briefings Bioinf, doi:10.1093/bib/bbad313
Bilich, Nelde, Heitmann, Maringer, Roerden et al., T cell and antibody kinetics delineate SARS-CoV-2 peptides mediating long-term immune responses in COVID-19 convalescent individuals, Sci Transl Med, doi:10.1126/scitranslmed.abf7517
Bo, Fan, Wang, Zou, Yu et al., Suppressed T cellmediated immunity in patients with COVID-19: a clinical retrospective study in Wuhan, China, J Infection, doi:10.1016/j.jinf.2020.04.012
Cai, Gao, Adamo, Rivera-Ballesteros, Hansson et al., SARS-CoV-2 vaccination enhances the effector qualities of spike-specific T cells induced by COVID-19, Sci Immunol, doi:10.1126/sciimmunol.adh0687
Chen, Huang, Zhang, Song, Zhao et al., Dynamic changes and future trend predictions of the global burden of anxiety disorders: analysis of 204 countries and regions from 1990 to 2021 and the impact of the COVID-19 pandemic, eClinicalMedicine, doi:10.1016/j.eclinm.2024.103014
Choe, Farzan, How SARS-CoV-2 first adapted in humans, Science, doi:10.1126/science.abi4711
Gao, Zhang, Wheelock, Xin, Cai et al., Immunomics in one health: understanding the human, animal, and environmental aspects of COVID-19, Front Immunol, doi:10.3389/fimmu.2024.1450380
Gozzi-Silva, Oliveira, Alberca, Pereira, Yoshikawa et al., Generation of cytotoxic T cells and dysfunctional CD8 T cells in severe COVID-19 patients, Cells, doi:10.3390/cells11213359
Huang, Wang, Li, Ren, Zhao et al., Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China, Lancet, doi:10.1016/S0140-6736(20)30183-5
Jiang, Zhang, Ru, Yang, Vu et al., Systematic investigation of cytokine signaling activity at the tissue and single-cell levels, Nat Methods, doi:10.1038/s41592-021-01274-5
Kahraman, Yıldız, Çıkı, Erdal, Akar et al., COVID-19 in inherited metabolic disorders: Clinical features and risk factors for disease severity, Mol Genet Metab, doi:10.1016/j.ymgme.2023.107607
Karl, Hofmann, Thimme, Role of antiviral CD8+ T cell immunity to SARS-CoV-2 infection and vaccination, J Virol, doi:10.1128/jvi.01350-24
Kovarik, Bileck, Hagn, Meier-Menches, Frey et al., A multi-omics based anti-inflammatory immune signature characterizes long COVID-19 syndrome, iScience, doi:10.1016/j.isci.2022.105717
Kwon, Safer, Nguyen, Hoksza, May et al., Genomics 2 Proteins portal: a resource and discovery tool for linking genetic screening outputs to protein sequences and structures, Nat Methods, doi:10.1038/s41592-024-02409-0
Laprise, Arduini, Duguay, Pan, Liang, SARS-CoV-2 accessory protein ORF8 targets the dimeric IgA receptor pIgR, Viruses, doi:10.3390/v16071008
Li, Hilgenfeld, Whitley, Clercq, Therapeutic strategies for COVID-19: progress and lessons learned, Nat Rev Drug Discovery, doi:10.1038/s41573-023-00672-y
Li, Zaslavsky, Su, Guo, Sikora et al., KIR + CD8 + T cells suppress pathogenic T cells and are active in autoimmune diseases and COVID-19, Science, doi:10.1126/science.abi9591
Liu, Yang, Gan, Chen, Xiao et al., CB-Dock2: improved proteinligand blind docking by integrating cavity detection, docking and homologous template fitting, Nucleic Acids Res, doi:10.1093/nar/gkac394
Mohr, Ortega-Santos, Whisner, Klein-Seetharaman, Jasbi, Navigating challenges and opportunities in multi-omics integration for personalized healthcare, Biomedicines, doi:10.3390/biomedicines12071496
Najjar-Debbiny, Gronich, Weber, Khoury, Amar et al., Effectiveness of paxlovid in reducing severe coronavirus disease 2019 and mortality in high-risk patients, Clin Infect Diseases, doi:10.1093/cid/ciac443
Nguema, Picard, Hajj, Dupaty, Fenwick et al., Subunit protein CD40.SARS.CoV2 vaccine induces SARS-CoV-2-specific stem cell-like memory CD8+ T cells, EBioMedicine, doi:10.1016/j.ebiom.2024.105479
Nguyen, Rowntree, Petersen, Chua, Hensen et al., CD8+ T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope display high naive precursor frequency and TCR promiscuity, Immunity, doi:10.1016/j.immuni.2021.04.009
Parkins, Lee, Acosta, Bautista, Hubert et al., Wastewater-based surveillance as a tool for public health action: SARS-CoV-2 and beyond, Clin Microbiol Rev, doi:10.1128/cmr.00103-22
Rabaan, Sh, Muhammad, Khan, Sule et al., Role of inflammatory cytokines in COVID-19 patients: a review on molecular mechanisms, immune functions, immunopathology and immunomodulatory drugs to counter cytokine storm, Vaccines, doi:10.3390/vaccines9050436
Rha, Shin, Activation or exhaustion of CD8+ T cells in patients with COVID-19, Cell Mol Immunol, doi:10.1038/s41423-021-00750-4
Santos, Humbard, Lambrou, Lin, Padilla et al., The SARS-CoV-2 test scale-up in the USA: an analysis of the number of tests produced and used over time and their modelled impact on the COVID-19 pandemic, Lancet Public Health, doi:10.1016/S2468-2667(24)00279-2
Steiner, Kratzel, Barut, Lang, Moreira et al., SARS-CoV-2 biology and host interactions, Nat Rev Microbiol, doi:10.1038/s41579-023-01003-z
Swinnerton, Fillmore, Vo, La, Elbers et al., Leveraging near-real-time patient and population data to incorporate fluctuating risk of severe COVID-19: development and prospective validation of a personalised risk prediction tool, eClinicalMedicine, doi:10.1016/j.eclinm.2025.103114
Tan, Wang, Zhang, Ding, Huang et al., Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study, Signal Transduction Targeted Ther, doi:10.1038/s41392-020-0148-4
Tang, Lao, Surani, Development and applications of single-cell transcriptome analysis, Nat Methods, doi:10.1038/nmeth.1557
Trott, Olson, AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J Comput Chem, doi:10.1002/jcc.21334
Wang, Wang, Cao, Wu, Yuan et al., Regulatory T cells in COVID-19, Aging disease, doi:10.14336/AD.2021.0709
Weiskopf, Schmitz, Raadsen, Grifoni, Okba et al., Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome, Sci Immunol, doi:10.1126/sciimmunol.abd2071
Wørzner, Schmidt, Zimmermann, Polacek, Fernandez-Antunez, Intranasal recombinant protein subunit vaccine targeting TLR3 induces respiratory tract IgA and CD8 T cell responses and protects against respiratory virus infection, EBioMedicine, doi:10.1016/j.ebiom.2025.105615
Xing, Wang, Peng, Li, Niu et al., The role of actin cytoskeleton CFL1 and ADF/cofilin superfamily in inflammatory response, Front Mol Biosci, doi:10.3389/fmolb.2024.1408287
Yang, Huang, Jiang, Advancements in serine protease inhibitors: from mechanistic insights to clinical applications, Catalysts, doi:10.3390/catal14110787
Ying, Wu, Jia, Yang, Liu et al., Single-cell transcriptome-wide Mendelian randomization and colocalization reveals immune-mediated regulatory mechanisms and drug targets for COVID-19, EBioMedicine, doi:10.1016/j.ebiom.2025.105596
Zhang, Han, Li, Cui, Yuan et al., Clinical outcomes of hospitalized immunocompromised patients with COVID-19 and the impact of hyperinflammation: A retrospective cohort study, J Inflammation Res, doi:10.2147/JIR.S482940
Zhang, Meng, Wang, Zhang, Sheng, Inflammation and antiviral immune response associated with severe progression of COVID-19, Front Immunol, doi:10.3389/fimmu.2021.631226
Zhou, Wu, Zhu, Lu, Zhang et al., Mucosal immune response in biology, disease prevention and treatment, Signal Transduction Targeted Ther, doi:10.1038/s41392-024-02043-4
DOI record:
{
"DOI": "10.3389/fimmu.2025.1671936",
"ISSN": [
"1664-3224"
],
"URL": "http://dx.doi.org/10.3389/fimmu.2025.1671936",
"abstract": "<jats:sec><jats:title>Introduction</jats:title><jats:p>COVID-19 has caused over 7 million deaths worldwide since its onset in 2019, and the virus remains a significant health threat. Identifying sensitive and specific biomarkers, along with elucidating immune-mediated mechanisms, is essential for improving the diagnosis, treatment, and prevention of COVID-19. To predict key molecular markers of COVID-19 using an established multi-omics framework combined with machine learning models.</jats:p></jats:sec><jats:sec><jats:title>Methods</jats:title><jats:p>We conducted an integrated analysis of single-cell RNA sequencing (scRNA-seq), bulk RNA sequencing, and proteomics data to identify critical biomarkers associated with COVID-19. The multi-omics approach enabled the characterization of gene expression dynamics and alterations in immune cell subsets in COVID-19 patients. Machine learning techniques and molecular docking analyses were employed to identify biomarkers and therapeutic targets within the disease’s pathophysiological network.</jats:p></jats:sec><jats:sec><jats:title>Results</jats:title><jats:p>Principal component analysis effectively grouped samples based on clinical characteristics. Using random forest and SVM-RFE models, we identified clinical indicators capable of accurately distinguishing COVID-19 patients. Transcriptomic analysis, including scRNA-seq, highlighted the pivotal role of CD8<jats:sup>+</jats:sup> T cells, and WGCNA identified related module genes. Proteomic analysis, integrated with machine learning, revealed 36 DEPs. Further investigation identified several genes associated with monocyte proportions. Correlation analysis showed that BTD, CFL1, PIGR, and SERPINA3 were strongly linked to CD8<jats:sup>+</jats:sup> T cell abundance in COVID-19 patients. ROC curve analysis demonstrated that these genes could effectively distinguish between COVID-19 patients and healthy individuals. Concordant findings from both transcriptomic and proteomic levels support BTD, CFL1, PIGR, and SERPINA3 as potential auxiliary diagnostic markers. Finally, AlphaFold-based molecular docking analysis suggested these biomarkers may also serve as candidate therapeutic targets.</jats:p></jats:sec><jats:sec><jats:title>Discussion</jats:title><jats:p>Preliminary findings indicate that BTD, CFL1, PIGR, and SERPINA3 are vital molecular biomarkers related of CD8+ T cell, providing new insights into the molecular mechanisms and long-term prevention of COVID-19.</jats:p></jats:sec>",
"alternative-id": [
"10.3389/fimmu.2025.1671936"
],
"article-number": "1671936",
"author": [
{
"affiliation": [],
"family": "Zhou",
"given": "Yumei",
"sequence": "first"
},
{
"affiliation": [],
"family": "Fan",
"given": "Pengbei",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Zhang",
"given": "Haiyun",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Han",
"given": "Shuai",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Bai",
"given": "Minghua",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Wang",
"given": "Ji",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Wang",
"given": "Qi",
"sequence": "additional"
}
],
"container-title": "Frontiers in Immunology",
"container-title-short": "Front. Immunol.",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"frontiersin.org"
]
},
"created": {
"date-parts": [
[
2025,
10,
2
]
],
"date-time": "2025-10-02T05:35:27Z",
"timestamp": 1759383327000
},
"deposited": {
"date-parts": [
[
2025,
10,
2
]
],
"date-time": "2025-10-02T05:35:28Z",
"timestamp": 1759383328000
},
"indexed": {
"date-parts": [
[
2025,
10,
2
]
],
"date-time": "2025-10-02T06:15:26Z",
"timestamp": 1759385726096,
"version": "build-2065373602"
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2025,
10,
2
]
]
},
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
10,
2
]
],
"date-time": "2025-10-02T00:00:00Z",
"timestamp": 1759363200000
}
}
],
"link": [
{
"URL": "https://www.frontiersin.org/articles/10.3389/fimmu.2025.1671936/full",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1965",
"original-title": [],
"prefix": "10.3389",
"published": {
"date-parts": [
[
2025,
10,
2
]
]
},
"published-online": {
"date-parts": [
[
2025,
10,
2
]
]
},
"publisher": "Frontiers Media SA",
"reference": [
{
"DOI": "10.1016/j.ebiom.2025.105596",
"article-title": "Single-cell transcriptome-wide Mendelian randomization and colocalization reveals immune-mediated regulatory mechanisms and drug targets for COVID-19",
"author": "Ying",
"doi-asserted-by": "publisher",
"first-page": "105596",
"journal-title": "EBioMedicine",
"key": "B1",
"volume": "113",
"year": "2025"
},
{
"DOI": "10.1016/j.eclinm.2024.103014",
"article-title": "Dynamic changes and future trend predictions of the global burden of anxiety disorders: analysis of 204 countries and regions from 1990 to 2021 and the impact of the COVID-19 pandemic",
"author": "Chen",
"doi-asserted-by": "publisher",
"first-page": "103014",
"journal-title": "eClinicalMedicine",
"key": "B2",
"volume": "79",
"year": "2025"
},
{
"DOI": "10.1126/science.abi4711",
"article-title": "How SARS-CoV-2 first adapted in humans",
"author": "Choe",
"doi-asserted-by": "publisher",
"journal-title": "Science",
"key": "B3",
"volume": "372",
"year": "2021"
},
{
"DOI": "10.1038/s41392-020-0148-4",
"article-title": "Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study",
"author": "Tan",
"doi-asserted-by": "publisher",
"first-page": "33",
"journal-title": "Signal Transduction Targeted Ther",
"key": "B4",
"volume": "5",
"year": "2020"
},
{
"DOI": "10.1038/s41573-023-00672-y",
"article-title": "Therapeutic strategies for COVID-19: progress and lessons learned",
"author": "Li",
"doi-asserted-by": "publisher",
"journal-title": "Nat Rev Drug Discovery",
"key": "B5",
"volume": "22",
"year": "2023"
},
{
"DOI": "10.1016/S0140-6736(20)30183-5",
"article-title": "Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China",
"author": "Huang",
"doi-asserted-by": "publisher",
"first-page": "497",
"journal-title": "Lancet",
"key": "B6",
"volume": "395",
"year": "2020"
},
{
"DOI": "10.2147/JIR.S482940",
"article-title": "Clinical outcomes of hospitalized immunocompromised patients with COVID-19 and the impact of hyperinflammation: A retrospective cohort study",
"author": "Zhang",
"doi-asserted-by": "publisher",
"journal-title": "J Inflammation Res",
"key": "B7",
"volume": "18",
"year": "2025"
},
{
"DOI": "10.1093/bib/bbad313",
"article-title": "Multimodal deep learning approaches for single-cell multi-omics data integration",
"author": "Athaya",
"doi-asserted-by": "publisher",
"first-page": "bbad313",
"journal-title": "Briefings Bioinf",
"key": "B8",
"volume": "24",
"year": "2023"
},
{
"DOI": "10.1038/nmeth.1557",
"article-title": "Development and applications of single-cell transcriptome analysis",
"author": "Tang",
"doi-asserted-by": "publisher",
"first-page": "S6",
"journal-title": "Nat Methods",
"key": "B9",
"volume": "8",
"year": "2011"
},
{
"DOI": "10.1038/s41467-022-28505-3",
"article-title": "Cell specific peripheral immune responses predict survival in critical COVID-19 patients",
"author": "Amrute",
"doi-asserted-by": "publisher",
"first-page": "882",
"journal-title": "Nat Commun",
"key": "B10",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.1038/s41592-021-01274-5",
"article-title": "Systematic investigation of cytokine signaling activity at the tissue and single-cell levels",
"author": "Jiang",
"doi-asserted-by": "publisher",
"journal-title": "Nat Methods",
"key": "B11",
"volume": "18",
"year": "2021"
},
{
"DOI": "10.1038/s41592-024-02409-0",
"article-title": "Genomics 2 Proteins portal: a resource and discovery tool for linking genetic screening outputs to protein sequences and structures",
"author": "Kwon",
"doi-asserted-by": "publisher",
"journal-title": "Nat Methods",
"key": "B12",
"volume": "21",
"year": "2024"
},
{
"DOI": "10.3390/vaccines9050436",
"article-title": "Role of inflammatory cytokines in COVID-19 patients: a review on molecular mechanisms, immune functions, immunopathology and immunomodulatory drugs to counter cytokine storm",
"author": "Rabaan",
"doi-asserted-by": "publisher",
"first-page": "436",
"journal-title": "Vaccines",
"key": "B13",
"volume": "9",
"year": "2021"
},
{
"DOI": "10.1016/j.jinf.2020.04.012",
"article-title": "Suppressed T cell-mediated immunity in patients with COVID-19: a clinical retrospective study in Wuhan, China",
"author": "Bo",
"doi-asserted-by": "publisher",
"journal-title": "J Infection",
"key": "B14",
"volume": "81",
"year": "2020"
},
{
"DOI": "10.1128/jvi.01350-24",
"article-title": "Role of antiviral CD8+ T cell immunity to SARS-CoV-2 infection and vaccination",
"author": "Karl",
"doi-asserted-by": "publisher",
"journal-title": "J Virol",
"key": "B15",
"volume": "99",
"year": "2025"
},
{
"DOI": "10.3390/cells11213359",
"article-title": "Generation of cytotoxic T cells and dysfunctional CD8 T cells in severe COVID-19 patients",
"author": "Gozzi-Silva",
"doi-asserted-by": "publisher",
"first-page": "3359",
"journal-title": "Cells",
"key": "B16",
"volume": "11",
"year": "2022"
},
{
"DOI": "10.1038/s41423-021-00750-4",
"article-title": "Activation or exhaustion of CD8+ T cells in patients with COVID-19",
"author": "Rha",
"doi-asserted-by": "publisher",
"journal-title": "Cell Mol Immunol",
"key": "B17",
"volume": "18",
"year": "2021"
},
{
"DOI": "10.14336/AD.2021.0709",
"article-title": "Regulatory T cells in COVID-19",
"author": "Wang",
"doi-asserted-by": "publisher",
"first-page": "1545",
"journal-title": "Aging disease",
"key": "B18",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1126/sciimmunol.abd2071",
"article-title": "Phenotype and kinetics of SARS-CoV-2–specific T cells in COVID-19 patients with acute respiratory distress syndrome",
"author": "Weiskopf",
"doi-asserted-by": "publisher",
"journal-title": "Sci Immunol",
"key": "B19",
"volume": "5",
"year": "2020"
},
{
"DOI": "10.3389/fimmu.2021.631226",
"article-title": "Inflammation and antiviral immune response associated with severe progression of COVID-19",
"author": "Zhang",
"doi-asserted-by": "publisher",
"journal-title": "Front Immunol",
"key": "B20",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1093/cid/ciac443",
"article-title": "Effectiveness of paxlovid in reducing severe coronavirus disease 2019 and mortality in high-risk patients",
"author": "Najjar-Debbiny",
"doi-asserted-by": "publisher",
"journal-title": "Clin Infect Diseases",
"key": "B21",
"volume": "76",
"year": "2022"
},
{
"DOI": "10.1093/nar/gkac394",
"article-title": "CB-Dock2: improved protein–ligand blind docking by integrating cavity detection, docking and homologous template fitting",
"author": "Liu",
"doi-asserted-by": "publisher",
"journal-title": "Nucleic Acids Res",
"key": "B22",
"volume": "50",
"year": "2022"
},
{
"DOI": "10.1002/jcc.21334",
"article-title": "AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading",
"author": "Trott",
"doi-asserted-by": "publisher",
"journal-title": "J Comput Chem",
"key": "B23",
"volume": "31",
"year": "2010"
},
{
"DOI": "10.1016/S2468-2667(24)00279-2",
"article-title": "The SARS-CoV-2 test scale-up in the USA: an analysis of the number of tests produced and used over time and their modelled impact on the COVID-19 pandemic",
"author": "Santos",
"doi-asserted-by": "publisher",
"journal-title": "Lancet Public Health",
"key": "B24",
"volume": "10",
"year": "2025"
},
{
"DOI": "10.1128/cmr.00103-22",
"article-title": "Wastewater-based surveillance as a tool for public health action: SARS-CoV-2 and beyond",
"author": "Parkins",
"doi-asserted-by": "publisher",
"journal-title": "Clin Microbiol Rev",
"key": "B25",
"volume": "37",
"year": "2024"
},
{
"DOI": "10.1038/s41579-023-01003-z",
"article-title": "SARS-CoV-2 biology and host interactions",
"author": "Steiner",
"doi-asserted-by": "publisher",
"journal-title": "Nat Rev Microbiol",
"key": "B26",
"volume": "22",
"year": "2024"
},
{
"DOI": "10.3389/fimmu.2024.1450380",
"article-title": "Immunomics in one health: understanding the human, animal, and environmental aspects of COVID-19",
"author": "Gao",
"doi-asserted-by": "publisher",
"journal-title": "Front Immunol",
"key": "B27",
"volume": "15",
"year": "2024"
},
{
"DOI": "10.1016/j.ebiom.2025.105615",
"article-title": "Intranasal recombinant protein subunit vaccine targeting TLR3 induces respiratory tract IgA and CD8 T cell responses and protects against respiratory virus infection",
"author": "Wørzner",
"doi-asserted-by": "publisher",
"first-page": "105615",
"journal-title": "EBioMedicine",
"key": "B28",
"volume": "113",
"year": "2025"
},
{
"DOI": "10.1016/j.ebiom.2024.105479",
"article-title": "Subunit protein CD40.SARS.CoV2 vaccine induces SARS-CoV-2-specific stem cell-like memory CD8+ T cells",
"author": "Nguema",
"doi-asserted-by": "publisher",
"first-page": "105479",
"journal-title": "EBioMedicine",
"key": "B29",
"volume": "111",
"year": "2025"
},
{
"DOI": "10.1126/scitranslmed.abf7517",
"article-title": "T cell and antibody kinetics delineate SARS-CoV-2 peptides mediating long-term immune responses in COVID-19 convalescent individuals",
"author": "Bilich",
"doi-asserted-by": "publisher",
"journal-title": "Sci Transl Med",
"key": "B30",
"volume": "13",
"year": "2021"
},
{
"DOI": "10.1126/sciimmunol.adh0687",
"article-title": "SARS-CoV-2 vaccination enhances the effector qualities of spike-specific T cells induced by COVID-19",
"author": "Cai",
"doi-asserted-by": "publisher",
"journal-title": "Sci Immunol",
"key": "B31",
"volume": "8",
"year": "2023"
},
{
"DOI": "10.1016/j.immuni.2021.04.009",
"article-title": "CD8+ T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope display high naive precursor frequency and TCR promiscuity",
"author": "Nguyen",
"doi-asserted-by": "publisher",
"first-page": "1066",
"journal-title": "Immunity",
"key": "B32",
"volume": "54",
"year": "2021"
},
{
"DOI": "10.1126/science.abi9591",
"article-title": "KIR+CD8+ T cells suppress pathogenic T cells and are active in autoimmune diseases and COVID-19",
"author": "Li",
"doi-asserted-by": "publisher",
"journal-title": "Science",
"key": "B33",
"volume": "376",
"year": "2022"
},
{
"DOI": "10.1016/j.isci.2022.105717",
"article-title": "A multi-omics based anti-inflammatory immune signature characterizes long COVID-19 syndrome",
"author": "Kovarik",
"doi-asserted-by": "publisher",
"first-page": "105717",
"journal-title": "iScience",
"key": "B34",
"volume": "26",
"year": "2023"
},
{
"DOI": "10.1016/j.ymgme.2023.107607",
"article-title": "COVID-19 in inherited metabolic disorders: Clinical features and risk factors for disease severity",
"author": "Kahraman",
"doi-asserted-by": "publisher",
"first-page": "107607",
"journal-title": "Mol Genet Metab",
"key": "B35",
"volume": "139",
"year": "2023"
},
{
"DOI": "10.3389/fmolb.2024.1408287",
"article-title": "The role of actin cytoskeleton CFL1 and ADF/cofilin superfamily in inflammatory response",
"author": "Xing",
"doi-asserted-by": "publisher",
"journal-title": "Front Mol Biosci",
"key": "B36",
"volume": "11",
"year": "2024"
},
{
"DOI": "10.1038/s41392-024-02043-4",
"article-title": "Mucosal immune response in biology, disease prevention and treatment",
"author": "Zhou",
"doi-asserted-by": "publisher",
"first-page": "7",
"journal-title": "Signal Transduction Targeted Ther",
"key": "B37",
"volume": "10",
"year": "2025"
},
{
"DOI": "10.3390/v16071008",
"article-title": "SARS-CoV-2 accessory protein ORF8 targets the dimeric IgA receptor pIgR",
"author": "Laprise",
"doi-asserted-by": "publisher",
"first-page": "1008",
"journal-title": "Viruses",
"key": "B38",
"volume": "16",
"year": "2024"
},
{
"DOI": "10.3390/catal14110787",
"article-title": "Advancements in serine protease inhibitors: from mechanistic insights to clinical applications",
"author": "Yang",
"doi-asserted-by": "publisher",
"first-page": "787",
"journal-title": "Catalysts",
"key": "B39",
"volume": "14",
"year": "2024"
},
{
"DOI": "10.3390/biomedicines12071496",
"article-title": "Navigating challenges and opportunities in multi-omics integration for personalized healthcare",
"author": "Mohr",
"doi-asserted-by": "publisher",
"first-page": "1496",
"journal-title": "Biomedicines",
"key": "B40",
"volume": "12",
"year": "2024"
},
{
"DOI": "10.1016/j.eclinm.2025.103114",
"article-title": "Leveraging near-real-time patient and population data to incorporate fluctuating risk of severe COVID-19: development and prospective validation of a personalised risk prediction tool",
"author": "Swinnerton",
"doi-asserted-by": "publisher",
"first-page": "103114",
"journal-title": "eClinicalMedicine",
"key": "B41",
"volume": "81",
"year": "2025"
}
],
"reference-count": 41,
"references-count": 41,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.frontiersin.org/articles/10.3389/fimmu.2025.1671936/full"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Multi-omics and machine learning identify novel biomarkers and therapeutic targets of COVID-19",
"type": "journal-article",
"update-policy": "https://doi.org/10.3389/crossmark-policy",
"volume": "16"
}