Pharmacogenomic Study of SARS-CoV-2 Treatments: Identifying Polymorphisms Associated with Treatment Response in COVID-19 Patients
et al., Biomedicines, doi:10.3390/biomedicines13030553, Feb 2025
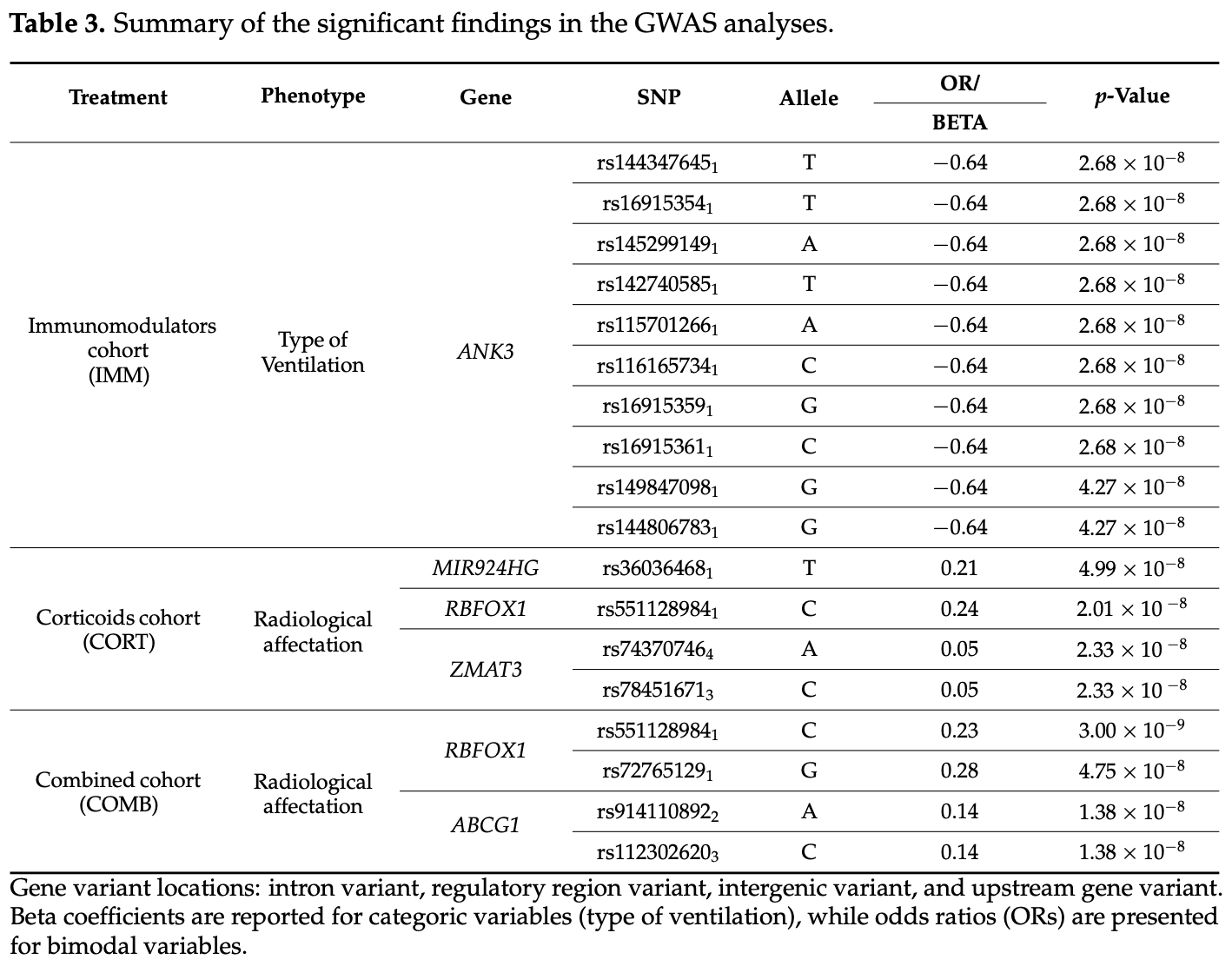
GWAS and candidate gene studies of 2,487 COVID-19 patients from Spain showing genetic variants associated with response to corticoid and immunomodulator treatments. Authors identified significant polymorphisms in 16 genes (including TLR1, TLR6, TLR10, CYP2C19, ACE2, UGT1A1, IL-1α, and others) that modulate treatment response across various outcomes including 90-day survival, ICU admission, radiological affectation, and ventilation requirements. Many identified genes interact with the NF-κB pathway, which regulates cytokine expression and is a target of corticoid therapy. Genetic variants in TLR family genes were associated with survival outcomes, while RBFOX1 variants were linked to lower probability of radiological affectation.
Serra-Llovich et al., 21 Feb 2025, Spain, peer-reviewed, mean age 66.0, 35 authors, study period March 2020 - December 2020.
Contact: alexserra@mutuaterrassa.cat (corresponding author), ddalmau@mutuaterrassa.cat, almudena.gil@usc.es, sheila.recarey.rama@usc.es, cflores@ull.edu.es, ncullell@mutuaterrassa.cat, mjarranz@mutuaterrassa.es.
Pharmacogenomic Study of SARS-CoV-2 Treatments: Identifying Polymorphisms Associated with Treatment Response in COVID-19 Patients
Biomedicines, doi:10.3390/biomedicines13030553
Background/Objectives: The COVID-19 pandemic resulted in 675 million cases and 6.9 million deaths by 2022. Despite substantial declines in case fatalities following widespread vaccination campaigns, the threat of future coronavirus outbreaks remains a concern. Current treatments for COVID-19 have been repurposed from existing therapies for other infectious and non-infectious diseases. Emerging evidence suggests a role for genetic factors in both susceptibility to SARS-CoV-2 infection and response to treatment. However, comprehensive studies correlating clinical outcomes with genetic variants are lacking. The main aim of our study is the identification of host genetic biomarkers that predict the clinical outcome of COVID-19 pharmacological treatments. Methods: In this study, we present findings from GWAS and candidate gene and pathway enrichment analyses leveraging diverse patient samples from the Spanish Coalition to Unlock Research of Host Genetics on COVID-19 (SCOURGE), representing patients treated with immunomodulators (n = 849), corticoids (n = 2202), and the combined cohort of both treatments (n = 2487) who developed different outcomes. We assessed various phenotypes as indicators of treatment response, including survival at 90 days, admission to the intensive care unit (ICU), radiological affectation, and type of ventilation. Results: We identified significant polymorphisms in 16 genes from the GWAS and candidate gene studies (TLR1, TLR6, TLR10, CYP2C19, ACE2, UGT1A1, IL-1α, ZMAT3, TLR4, MIR924HG, IFNG-AS1, ABCG1, RBFOX1, ABCB11, TLR5, and ANK3) that may modulate the response to corticoid and immunomodulator therapies in COVID-19 patients. Enrichment analyses revealed overrepresentation of genes involved in the innate immune system, drug ADME, viral infection, and the programmed cell death pathways associated with the response phenotypes. Conclusions: Our study provides an initial framework for understanding the genetic determinants of treatment response in COVID-19 patients, offering insights that could inform precision medicine approaches for future epidemics.
Conflicts of Interest: The authors declare no conflicts of interest.
References
Al-Eitan, Alahmad, Pharmacogenomics of Genetic Polymorphism within the Genes Responsible for SARS-CoV-2 Susceptibility and the Drug-Metabolising Genes Used in Treatment, Rev. Med. Virol, doi:10.1002/rmv.2194
Al-Taie, Büyük, Sardas, Considerations into Pharmacogenomics of COVID-19 Pharmacotherapy: Hope, Hype and Reality, Pulm. Pharmacol. Ther, doi:10.1016/j.pupt.2022.102172
Alors-Pérez, Pedraza-Arevalo, Blázquez-Encinas, García-Vioque, Agraz-Doblas et al., Altered CELF4 Splicing Factor Enhances Pancreatic Neuroendocrine Tumors Aggressiveness Influencing MTOR and Everolimus Response, Mol. Ther. Nucleic Acids, doi:10.1016/j.omtn.2023.102090
Badary, Pharmacogenomics and COVID-19: Clinical Implications of Human Genome Interactions with Repurposed Drugs, Pharmacogenomics J, doi:10.1038/s41397-021-00209-9
Bank, Skytt Andersen, Burisch, Pedersen, Roug et al., Polymorphisms in the Inflammatory Pathway Genes TLR2, TLR4, TLR9, LY96, NFKBIA, NFKB1, TNFA, TNFRSF1A, IL6R, IL10, IL23R, PTPN22, PLoS ONE, doi:10.1371/journal.pone.0098815
Battaglini, Lopes-Pacheco, Castro-Faria-Neto, Pelosi, Rocco, Laboratory Biomarkers for Diagnosis and Prognosis in COVID-19, Front. Immunol, doi:10.3389/fimmu.2022.857573
Biswas, Sawajan, Rungrotmongkol, Sanachai, Ershadian et al., Pharmacogenetics and Precision Medicine Aroaches for the Improvement of COVID-19 Therapies, Front. Pharmacol, doi:10.3389/fphar.2022.835136
Botton, Whirl-Carrillo, Del Tredici, Sangkuhl, Cavallari et al., PharmVar GeneFocus: CYP2C19, Clin. Pharmacol. Ther, doi:10.1002/cpt.1973
Brasileiro, Brazil, Programa de Pós-Graduação Profissional em Ensino de Biologia
Britt, Thompson, Sasse, Pabelick, Gerber et al., Th1 Cytokines TNF-and IFN-Promote Corticosteroid Resistance in Developing Human Airway Smooth Muscle, Am. J. Physiol. Lung. Cell. Mol. Physiol, doi:10.1152/ajplung.00547.2017
Cao, Zhang, Chen, Wang, Guo et al., The Critical Role of ABCG1 and PPARγ/LXRα Signaling in TLR4 Mediates Inflammatory Responses and Lipid Accumulation in Vascular Smooth Muscle Cells, Cell. Tissue Res, doi:10.1007/s00441-016-2518-3
Castilla Y León, None
Chakraborty, Sharma, Bhattacharya, Sharma, Lee et al., Consider TLR5 for New Therapeutic Development against COVID-19, J. Med. Virol, doi:10.1002/jmv.25997
Chavda, Vuu, Mishra, Kamaraj, Patel et al., Recent Review of COVID-19 Management: Diagnosis, Treatment and Vaccination, Pharmacol. Rep, doi:10.1007/s43440-022-00425-5
Creeden, Gordon, Stec, Hinds, Bilirubin as a Metabolic Hormone: The Physiological Relevance of Low Levels, Am. J. Physiol. -Endocrinol. Metab, doi:10.1152/ajpendo.00405.2020
Cruz, Diz-De Almeida, De Heredia, Quintela, Ceballos et al., A Novel Genes and Sex Differences in COVID-19 Severity, Hum. Mol. Genet, doi:10.1093/hmg/ddac132
De Niños, Gutierrez, None
De, None
Dery, Wong, Wei, Kupiec-Weglinski, Mechanistic Insights into Alternative Gene Splicing in Oxidative Stress and Tissue Injury, Antioxid. Redox. Signal, doi:10.1089/ars.2023.0437
Duggan, You, Cleaver, Larson, Garza et al., Synergistic Interactions of TLR2/6 and TLR9 Induce a High Level of Resistance to Lung Infection in Mice, J. Immunol, doi:10.4049/jimmunol.1002122
Díaz, None
El, Gerencia de Asistencia Sanitaria del Bierzo (GASBI)
El, Unidad Cuidados Intensivos
Engel, Van Der Made, Keur, Setiabudiawan, Röring et al., Dexamethasone Attenuates Interferon-Related Cytokine Hyperresponsiveness in COVID-19 Patients, Front. Immunol, doi:10.3389/fimmu.2023.1233318
Escola, Cicco, Natal, Brazil, None
Fan, Long, Ren, Chen, Li et al., Genome-Wide Association Study of SARS-CoV-2 Infection in Chinese Population, Eur. J. Clin. Microbiol. Infect. Dis, doi:10.1007/s10096-022-04478-5
Fania Hernandez 102, Hernandez-Ortega ; Moro 6, Hernández-Pérez 47, Hernández-Vaquero 104, Herraez 15 et al., María Lozano-Espinosa 112
Fardel, Payen, Courtois, Vernhet, Lecureur, Regulation of Biliary Drug Efflux Pump Expression by Hormones and Xenobiotics, Toxicology, doi:10.1016/S0300-483X(01)00456-5
Fatima, Ratnani, Husain, Surani, Radiological Findings in Patients with COVID-19, Cureus, doi:10.7759/cureus.7651
Fernández-Castañeda, Lu, Geraghty, Song, Lee et al., Mild Respiratory COVID Can Cause Multi-Lineage Neural Cell and Myelin Dysregulation, Cell, doi:10.1016/j.cell.2022.06.008
Finney, Glanville, Farne, Aniscenko, Fenwick et al., Inhaled Corticosteroids Downregulate the SARS-CoV-2 Receptor ACE2 in COPD through Suression of Type I Interferon, J. Allergy Clin. Immunol, doi:10.1016/j.jaci.2020.09.034
Fore, Indriputri, Mamutse, Nugraha, TLR10 and Its Unique Anti-Inflammatory Properties and Potential Use as a Target in Therapeutics, Immune Netw, doi:10.4110/in.2020.20.e21
Franczyk, Rysz, Miło Ński, Konecki, Rysz-Górzy Ńska et al., Will the Use of Pharmacogenetics Improve Treatment Efficiency in COVID-19?, Pharmaceuticals, doi:10.3390/ph15060739
Fricke-Galindo, Falfán-Valencia, Pharmacogenetics Aroach for the Improvement of COVID-19 Treatment, Viruses, doi:10.3390/v13030413
Fu, Xu, Wei, Why Tocilizumab Could Be an Effective Treatment for Severe COVID-19?, J. Transl. Med, doi:10.1186/s12967-020-02339-3
Gammal, Court, Haidar, Iwuchukwu, Gaur et al., Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for UGT1A1 and Atazanavir Prescribing, Clin. Med. Ther, doi:10.1002/cpt.269
Gao, Ren, Lee, Qiu, Chapski et al., RBFox1-Mediated RNA Splicing Regulates Cardiac Hypertrophy and Heart Failure, J. Clin. Investig, doi:10.1172/JCI84015
García-Menaya, Cordobés-Durán, García-Martín, Agúndez, Pharmacogenetic Factors Affecting Asthma Treatment Response. Potential Implications for Drug Therapy, Front. Pharmacol, doi:10.3389/fphar.2019.00520
Gil, Aguda, de La Comunitat En l'Adult: Etiologia i Biomarcadors Genètics de l'Hoste
Gong, Song, Hu, Che, Guo et al., Natural and Socio-Environmental Factors in the Transmission of COVID-19: A Comprehensive Analysis of Epidemiology and Mechanisms, BMC Public Health, doi:10.1186/s12889-024-19749-3
Graffelman, Moreno, The Mid P-Value in Exact Tests for Hardy-Weinberg Equilibrium, Stat. Al. Genet. Mol. Biol, doi:10.1515/sagmb-2012-0039
Hasanvand, COVID-19 and the Role of Cytokines in This Disease, Inflammopharmacology, doi:10.1007/s10787-022-00992-2
Hospitalarias, Complejo Hospitalario Universitario de A Coruña (CHUAC)
Hu, Huang, Yin, The Cytokine Storm and COVID-19, J. Med. Virol, doi:10.1002/jmv.26232
Inditex, Coruña, Spain, None
Jarczak, Nierhaus, Cytokine Storm-Definition, Causes, and Implications, Int. J. Mol. Sci, doi:10.3390/ijms231911740
Jeane, Medeiros 21, Medeiros, None, Francisco J. Medrano
Jiang, Rubin, Zhou, Zhang, Su et al., Pharmacological Therapies and Drug Development Targeting SARS-CoV-2 Infection, Cytokine Growth Factor Rev, doi:10.1016/j.cytogfr.2022.10.003
Kaushik, Bhandari, Kuhad, TLR4 as a Therapeutic Target for Respiratory and Neurological Complications of SARS-CoV-2, Expert Opin. Ther. Targets, doi:10.1080/14728222.2021.1918103
Kloth, Lozic, Tagoe, Hoffer, Van Der Ven et al., ANK3 Related Neurodevelopmental Disorders: Expanding the Spectrum of Heterozygous Loss-of-Function Variants, Neurogenetics, doi:10.1007/s10048-021-00655-4
Kotak, Khatri, Malik, Malik, Hassan et al., Use of Tocilizumab in COVID-19: A Systematic Review and Meta-Analysis of Current Evidence, Cureus, doi:10.7759/cureus.10869
Lee, Wang, Martin, Germer, Kenwright et al., Genetic Variation in UGT1A1 Typical of Gilbert Syndrome Is Associated with Unconjugated Hyperbilirubinemia in Patients Receiving Tocilizumab, Pharmacogenet. Genom, doi:10.1097/FPC.0b013e32834592fe
Matoulková, Pávek, Malý, Vlček, Cytochrome P450 Enzyme Regulation by Glucocorticoids and Consequences in Terms of Drug Interaction, Expert Opin. Drug Metab. Toxicol, doi:10.1517/17425255.2014.878703
Mcpeek, Malur, Tokarz, Lertpiriyapong, Gowdy et al., Alveolar Macrophage ABCG1 Deficiency Promotes Pulmonary Granulomatous Inflammation, Am. J. Respir. Cell Mol. Biol, doi:10.1165/rcmb.2018-0365OC
Mo, Fisher, A Review of Treatment Modalities for Middle East Respiratory Syndrome, J. Antimicrob. Chemother, doi:10.1093/jac/dkw338
Molina-Mora, González, Jiménez-Morgan, Cordero-Laurent, Brenes et al., Clinical Profiles at the Time of Diagnosis of SARS-CoV-2 Infection in Costa Rica During the Pre-Vaccination Period Using a Machine Learning Aroach, Phenomics, doi:10.1007/s43657-022-00058-x
Montazersaheb, Hosseiniyan Khatibi, Hejazi, Tarhriz, Farjami et al., COVID-19 Infection: An Overview on Cytokine Storm and Related Interventions, Virol. J, doi:10.1186/s12985-022-01814-1
Moreira, Jorge, Gomes, Kaupert, Filho et al., Pharmacogenetics of Glucocorticoid Replacement Could Optimize the Treatment of Congenital Adrenal Hyperplasia Due to 21-Hydroxylase Deficiency, Clinics, doi:10.1590/S1807-59322011000800009
Méndez-Echevarria, Joana, Nunes 23, Nuñez-Torres 15, Obrador-Hevia, Montserrat Robelo Pardo 104
Noureddine, Chakkour, El Roz, Reda, Al Sahily et al., The Emergence of SARS-CoV-2 Variant(s) and Its Impact on the Prevalence of COVID-19 Cases in the Nabatieh Region, Lebanon, Med. Sci, doi:10.3390/medsci9020040
O'beirne, Salit, Kaner, Crystal, Strulovici-Barel, Up-Regulation of ACE2, the SARS-CoV-2 Receptor, in Asthmatics on Maintenance Inhaled Corticosteroids, Respir. Res, doi:10.1186/s12931-021-01782-0
Oram, Vaughan, ATP-Binding Cassette Cholesterol Transporters and Cardiovascular Disease, Circ. Res, doi:10.1161/01.RES.0000250171.54048.5c
Otto Von, University, Departament of Microgravity and Translational Regenerative Medicine
Pino-Yanes, Corrales, Casula, Blanco, Muriel et al., Common Variants of TLR1 Associate with Organ Dysfunction and Sustained Pro-Inflammatory Responses during Sepsis, PLoS ONE, doi:10.1371/journal.pone.0013759
Purcell, Neale, Todd-Brown, Thomas, Ferreira et al., PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses, Am. J. Hum. Genet, doi:10.1086/519795
Quironprevención, None
Raghav, Mann, Ahluwalia, Rajalingam, Potential Treatments of COVID-19: Drug Repurposing and Therapeutic Interventions, J. Pharmacol. Sci, doi:10.1016/j.jphs.2023.02.004
Rendic, Guengerich, Metabolism and Interactions of Chloroquine and Hydroxychloroquine with Human Cytochrome P450 Enzymes and Drug Transporters, Curr. Drug Metab, doi:10.2174/1389200221999201208211537
Sant, De Deu, None
Scialo, Daniele, Amato, Pastore, Matera et al., ACE2: The Major Cell Entry Receptor for SARS-CoV-2, Lung, doi:10.1007/s00408-020-00408-4
Scott, Sangkuhl, Shuldiner, Hulot, Thorn et al., PharmGKB Summary: Very Important Pharmacogene Information for Cytochrome P450, Family 2, Subfamily C, Polypeptide 19, Pharmacogenet. Genom, doi:10.1097/FPC.0b013e32834d4962
Stein, Berhani, Schmiedel, Duev-Cohen, Seidel et al., IFNG-AS1 Enhances Interferon Gamma Production in Human Natural Killer Cells, iScience, doi:10.1016/j.isci.2018.12.034
Takahashi, Luzum, Nicol, Jacobson, Pharmacogenomics of COVID-19 Therapies, NPJ Genom. Med, doi:10.1038/s41525-020-00143-y
Taliun, Harris, Kessler, Carlson, Szpiech et al., Sequencing of 53,831 Diverse Genomes from the NHLBI TOPMed Program, Nature, doi:10.1038/s41586-021-03205-y
Tecnica De Saúde, Laboratorio de Vigilancia Molecular Aplicada
Thoeni, Waldherr, Scheuerer, Schmitteckert, Roeth et al., Expression Analysis of Atp-Binding Cassette Transporters Abcb11 and Abcb4 in Primary Sclerosing Cholangitis and Variety of Pediatric and Adult Cholestatic and Noncholestatic Liver Diseases, Can. J. Gastroenterol. Hepatol, doi:10.1155/2019/1085717
Tomlinson, Maggs, Park, Back, Dexamethasone Metabolism in Species Differences, J. Steroid. Biochem. Mol. Biol, doi:10.1016/S0960-0760(97)00038-1
Universitario, Espases, Unidad de Diagnóstico Molecular y Genética Clínica
Valenzuela, Ibáñez, Poli, Roessler, Aylwin et al., First Report of Tocilizumab Use in a Cohort of Latin American Patients Hospitalized for Severe COVID-19 Pneumonia, Front. Med, doi:10.3389/fmed.2020.596916
Vohra, Sharma, Satyamoorthy, Rai, Pharmacogenomic Considerations for Repurposing of Dexamethasone as a Potential Drug against SARS-CoV-2 Infection, Per. Med, doi:10.2217/pme-2020-0183
Wang, Cheng, Gao, Li, Ding et al., Investigation of the Shared Molecular Mechanisms and Hub Genes between Myocardial Infarction and Depression, Front. Cardiovasc. Med, doi:10.3389/fcvm.2023.1203168
Xose, Meijome, Patricia Moreira-Escriche 137
Yamasaki, Tanimoto, Kohno, Kadowaki, Takase et al., UGT1A1 *6 Polymorphism Predicts Outcome in Elderly Patients with Relapsed or Refractory Diffuse Large B-Cell Lymphoma Treated with Carboplatin, Dexamethasone, Etoposide and Irinotecan, Ann. Hematol, doi:10.1007/s00277-014-2170-5
Yang, Zhu, Hui, Sun, Effects of CACNA1C and ANK3 on Cognitive Function in Patients with Bipolar Disorder, Prog. Neuropsychopharmacol. Biol. Psychiatry, doi:10.1016/j.pnpbp.2024.111016
Yeh, Gaver, Patterson, Rezk, Baxter-Meheux et al., Lopinavir/Ritonavir Induces the Hepatic Activity of Cytochrome P450 Enzymes CYP2C9, CYP2C19, and CYP1A2 But Inhibits the Hepatic and Intestinal Activity of CYP3A as Measured by a Phenotyping Drug Cocktail in Healthy Volunteers, JAIDS J. Acquir. Immune Defic. Syndr, doi:10.1097/01.qai.0000219774.20174.64
Zaidat, Erwin Grüter, Hospital, Rafael Moscote-Salazar, Yan et al., A Rare Variant of ANK3 Is Associated With Intracranial Aneurysm, Front. Neurol, doi:10.3389/fneur.2021.672570
DOI record:
{
"DOI": "10.3390/biomedicines13030553",
"ISSN": [
"2227-9059"
],
"URL": "http://dx.doi.org/10.3390/biomedicines13030553",
"abstract": "<jats:p>Background/Objectives: The COVID-19 pandemic resulted in 675 million cases and 6.9 million deaths by 2022. Despite substantial declines in case fatalities following widespread vaccination campaigns, the threat of future coronavirus outbreaks remains a concern. Current treatments for COVID-19 have been repurposed from existing therapies for other infectious and non-infectious diseases. Emerging evidence suggests a role for genetic factors in both susceptibility to SARS-CoV-2 infection and response to treatment. However, comprehensive studies correlating clinical outcomes with genetic variants are lacking. The main aim of our study is the identification of host genetic biomarkers that predict the clinical outcome of COVID-19 pharmacological treatments. Methods: In this study, we present findings from GWAS and candidate gene and pathway enrichment analyses leveraging diverse patient samples from the Spanish Coalition to Unlock Research of Host Genetics on COVID-19 (SCOURGE), representing patients treated with immunomodulators (n = 849), corticoids (n = 2202), and the combined cohort of both treatments (n = 2487) who developed different outcomes. We assessed various phenotypes as indicators of treatment response, including survival at 90 days, admission to the intensive care unit (ICU), radiological affectation, and type of ventilation. Results: We identified significant polymorphisms in 16 genes from the GWAS and candidate gene studies (TLR1, TLR6, TLR10, CYP2C19, ACE2, UGT1A1, IL-1α, ZMAT3, TLR4, MIR924HG, IFNG-AS1, ABCG1, RBFOX1, ABCB11, TLR5, and ANK3) that may modulate the response to corticoid and immunomodulator therapies in COVID-19 patients. Enrichment analyses revealed overrepresentation of genes involved in the innate immune system, drug ADME, viral infection, and the programmed cell death pathways associated with the response phenotypes. Conclusions: Our study provides an initial framework for understanding the genetic determinants of treatment response in COVID-19 patients, offering insights that could inform precision medicine approaches for future epidemics.</jats:p>",
"alternative-id": [
"biomedicines13030553"
],
"author": [
{
"ORCID": "https://orcid.org/0000-0001-8318-851X",
"affiliation": [
{
"name": "Fundació Docència i Recerca Mutua Terrassa, 08221 Terrassa, Spain"
}
],
"authenticated-orcid": false,
"family": "Serra-Llovich",
"given": "Alexandre",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Fundació Docència i Recerca Mutua Terrassa, 08221 Terrassa, Spain"
},
{
"name": "Hospital Universitario Mutua Terrassa, 08221 Terrassa, Spain"
}
],
"family": "Cullell",
"given": "Natalia",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Fundación Pública Galega de Medicina Genómica (FPGMX), Centro Nacional de Genotipado (CEGEN), Servicio Gallego de Salud (SERGAS), 15706 Santiago de Compostela, Spain"
},
{
"name": "Grupo de Farmacogenómica y Descubrimiento de Medicamentos (GenDeM), Instituto de Investigación Sanitaria de Santiago de Compostela (IDIS), 15706 Santiago de Compostela, Spain"
},
{
"name": "Centre for Biomedical Network Research on Rare Diseases (CIBERER), Instituto de Salud Carlos III, 28029 Madrid, Spain"
}
],
"family": "Maroñas",
"given": "Olalla",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-6042-4185",
"affiliation": [
{
"name": "IIS La Fe, Plataforma de Farmacogenética, 43026 Valencia, Spain"
},
{
"name": "Departamento de Farmacología, Universidad de Valencia, 46010 Valencia, Spain"
}
],
"authenticated-orcid": false,
"family": "José Herrero",
"given": "María",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-6964-8898",
"affiliation": [
{
"name": "Centre for Biomedical Network Research on Rare Diseases (CIBERER), Instituto de Salud Carlos III, 28029 Madrid, Spain"
},
{
"name": "Centro Nacional de Genotipado (CEGEN), Universidad de Santiago de Compostela, 15706 Santiago de Compostela, Spain"
},
{
"name": "Instituto de Investigación Sanitaria de Santiago (IDIS), 15706 Santiago de Compostela, Spain"
},
{
"name": "Centro Singular de Investigación en Medicina Molecular y Enfermedades Crónicas (CIMUS), Universidade de Santiago de Compostela, 15782 Santiago de Compostela, Spain"
}
],
"authenticated-orcid": false,
"family": "Cruz",
"given": "Raquel",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Centre for Biomedical Network Research on Rare Diseases (CIBERER), Instituto de Salud Carlos III, 28029 Madrid, Spain"
},
{
"name": "Department of Genetics and Genomics, Instituto de Investigación Sanitaria-Fundación Jiménez Díaz University Hospital-Universidad Autónoma de Madrid (IIS-FJD, UAM), 28040 Madrid, Spain"
}
],
"family": "Almoguera",
"given": "Berta",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-9242-7065",
"affiliation": [
{
"name": "Centre for Biomedical Network Research on Rare Diseases (CIBERER), Instituto de Salud Carlos III, 28029 Madrid, Spain"
},
{
"name": "Department of Genetics and Genomics, Instituto de Investigación Sanitaria-Fundación Jiménez Díaz University Hospital-Universidad Autónoma de Madrid (IIS-FJD, UAM), 28040 Madrid, Spain"
}
],
"authenticated-orcid": false,
"family": "Ayuso",
"given": "Carmen",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-4713-0633",
"affiliation": [
{
"name": "Centre for Biomedical Network Research on Rare Diseases (CIBERER), Instituto de Salud Carlos III, 28029 Madrid, Spain"
},
{
"name": "Department of Genetics and Genomics, Instituto de Investigación Sanitaria-Fundación Jiménez Díaz University Hospital-Universidad Autónoma de Madrid (IIS-FJD, UAM), 28040 Madrid, Spain"
}
],
"authenticated-orcid": false,
"family": "López-Rodríguez",
"given": "Rosario",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Unidad Diagnóstico Molecular, Fundación Rioja Salud, 26006 La Rioja, Spain"
}
],
"family": "Domínguez-Garrido",
"given": "Elena",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Tecnologico de Monterrey, Escuela de Medicina y Ciencias de la Salud and Hospital San Jose TecSalud, Monterrey 64718, Mexico"
}
],
"family": "Ortiz-Lopez",
"given": "Rocio",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-4728-8174",
"affiliation": [
{
"name": "Instituto Murciano de Investigación Biosanitaria (IMIB-Arrixaca), 30120 Murcia, Spain"
},
{
"name": "Departamento de Ciencias de la Salud, Universidad Católica San Antonio de Murcia (UCAM), 30120 Murcia, Spain"
}
],
"authenticated-orcid": false,
"family": "Barreda-Sánchez",
"given": "María",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Centre for Biomedical Network Research on Rare Diseases (CIBERER), Instituto de Salud Carlos III, 28029 Madrid, Spain"
},
{
"name": "Department of Genetics and Genomics, Instituto de Investigación Sanitaria-Fundación Jiménez Díaz University Hospital-Universidad Autónoma de Madrid (IIS-FJD, UAM), 28040 Madrid, Spain"
}
],
"family": "Corton",
"given": "Marta",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Fundació Docència i Recerca Mutua Terrassa, 08221 Terrassa, Spain"
},
{
"name": "Hospital Universitario Mutua Terrassa, 08221 Terrassa, Spain"
}
],
"family": "Dalmau",
"given": "David",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Hospital Universitario Mutua Terrassa, 08221 Terrassa, Spain"
},
{
"name": "Faculty of Medicine and Health Sciences, Universitat Internacional de Catalunya, 08017 Barcelona, Spain"
}
],
"family": "Calbo",
"given": "Esther",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Hospital Universitario Mutua Terrassa, 08221 Terrassa, Spain"
}
],
"family": "Boix-Palop",
"given": "Lucía",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-6011-6986",
"affiliation": [
{
"name": "Hospital Universitario Mutua Terrassa, 08221 Terrassa, Spain"
}
],
"authenticated-orcid": false,
"family": "Dietl",
"given": "Beatriz",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Hospital Universitario Mutua Terrassa, 08221 Terrassa, Spain"
}
],
"family": "Sangil",
"given": "Anna",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-7637-6915",
"affiliation": [
{
"name": "Grupo de Farmacogenómica y Descubrimiento de Medicamentos (GenDeM), Instituto de Investigación Sanitaria de Santiago de Compostela (IDIS), 15706 Santiago de Compostela, Spain"
},
{
"name": "Grupo de Medicina Genómica, CIMUS, Universidad de Santiago de Compostela, 15782 Santiago de Compostela, Spain"
}
],
"authenticated-orcid": false,
"family": "Gil-Rodriguez",
"given": "Almudena",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Centre for Biomedical Network Research on Rare Diseases (CIBERER), Instituto de Salud Carlos III, 28029 Madrid, Spain"
},
{
"name": "Instituto Murciano de Investigación Biosanitaria (IMIB-Arrixaca), 30120 Murcia, Spain"
},
{
"name": "Sección Genética Médica-Servicio de Pediatría, Hospital Clínico Universitario Virgen de la Arrixaca, Servicio Murciano de Salud, 30120 Murcia, Spain"
},
{
"name": "Departamento Cirugía, Pediatría, Obstetricia y Ginecología, Facultad de Medicina, Universidad de Murcia (UMU), 30120 Murcia, Spain"
}
],
"family": "Guillén-Navarro",
"given": "Encarna",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-1535-9062",
"affiliation": [
{
"name": "Department of Immunology, Hospital Universitario 12 de Octubre, 28041 Madrid, Spain"
},
{
"name": "Transplant Immunology and Immunodeficiencies Group, Instituto de Investigación Sanitaria Hospital 12 de Octubre (imas12), 28041 Madrid, Spain"
}
],
"authenticated-orcid": false,
"family": "Mancebo",
"given": "Esther",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Laboratorios MICROLAB Vios, Tegucigalpa 11101, Honduras"
}
],
"family": "Lira-Albarrán",
"given": "Saúl",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-4099-9421",
"affiliation": [
{
"name": "Centre for Biomedical Network Research on Rare Diseases (CIBERER), Instituto de Salud Carlos III, 28029 Madrid, Spain"
},
{
"name": "Department of Genetics and Genomics, Instituto de Investigación Sanitaria-Fundación Jiménez Díaz University Hospital-Universidad Autónoma de Madrid (IIS-FJD, UAM), 28040 Madrid, Spain"
}
],
"authenticated-orcid": false,
"family": "Minguez",
"given": "Pablo",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Immunology, Hospital Universitario 12 de Octubre, 28041 Madrid, Spain"
},
{
"name": "Transplant Immunology and Immunodeficiencies Group, Instituto de Investigación Sanitaria Hospital 12 de Octubre (imas12), 28041 Madrid, Spain"
},
{
"name": "Department of Immunology, Ophthalmology and ENT, Universidad Complutense de Madrid, 28040 Madrid, Spain"
},
{
"name": "Centro de Investigación Biomédica en Red de Enfermedades Infecciosas (CIBERINFEC), Instituto de Salud Carlos III, 28029 Madrid, Spain"
}
],
"family": "Paz-Artal",
"given": "Estela",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-7698-2122",
"affiliation": [
{
"name": "IIS La Fe, Plataforma de Farmacogenética, 43026 Valencia, Spain"
},
{
"name": "Departamento de Farmacología, Universidad de Valencia, 46010 Valencia, Spain"
}
],
"authenticated-orcid": false,
"family": "Olivera",
"given": "Gladys G.",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0009-0003-1623-1110",
"affiliation": [
{
"name": "Grupo de Farmacogenómica y Descubrimiento de Medicamentos (GenDeM), Instituto de Investigación Sanitaria de Santiago de Compostela (IDIS), 15706 Santiago de Compostela, Spain"
},
{
"name": "Grupo de Medicina Genómica, CIMUS, Universidad de Santiago de Compostela, 15782 Santiago de Compostela, Spain"
}
],
"authenticated-orcid": false,
"family": "Recarey-Rama",
"given": "Sheila",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-3176-9763",
"affiliation": [
{
"name": "IIS La Fe, Plataforma de Farmacogenética, 43026 Valencia, Spain"
},
{
"name": "Departamento de Farmacología, Universidad de Valencia, 46010 Valencia, Spain"
}
],
"authenticated-orcid": false,
"family": "Sendra",
"given": "Luis",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "IIS La Fe, Plataforma de Farmacogenética, 43026 Valencia, Spain"
},
{
"name": "Departamento de Farmacología, Universidad de Valencia, 46010 Valencia, Spain"
}
],
"family": "Zucchet",
"given": "Enrique G.",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-2826-9565",
"affiliation": [
{
"name": "Centre for Biomedical Network Research on Rare Diseases (CIBERER), Instituto de Salud Carlos III, 28029 Madrid, Spain"
}
],
"authenticated-orcid": false,
"family": "López de Heredia",
"given": "Miguel",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-5352-069X",
"affiliation": [
{
"name": "Genomics Division, Instituto Tecnológico y de Energías Renovables, 38600 Santa Cruz de Tenerife, Spain"
},
{
"name": "Research Unit, Hospital Universitario Nuestra Señora de Candelaria, Instituto de Investigación Sanitaria de Canarias, 38010 Santa Cruz de Tenerife, Spain"
},
{
"name": "Centre for Biomedical Network Research on Respiratory Diseases (CIBERES), Instituto de Salud Carlos III, 28029 Madrid, Spain"
},
{
"name": "Facultad de Ciencias de la Salud, Universidad Fernando Pessoa Canarias, 35450 Las Palmas de Gran Canaria, Spain"
}
],
"authenticated-orcid": false,
"family": "Flores",
"given": "Carlos",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-0691-8755",
"affiliation": [
{
"name": "Centre for Biomedical Network Research on Rare Diseases (CIBERER), Instituto de Salud Carlos III, 28029 Madrid, Spain"
},
{
"name": "Servicio de Medicina Interna, Hospital U.M. Valdecilla, Universidad de Cantabria, IDIVAL, 39008 Santander, Spain"
}
],
"authenticated-orcid": false,
"family": "Riancho",
"given": "José A.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Tecnologico de Monterrey, Escuela de Medicina y Ciencias de la Salud and Hospital San Jose TecSalud, Monterrey 64718, Mexico"
}
],
"family": "Rojas-Martinez",
"given": "Augusto",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-6324-4825",
"affiliation": [
{
"name": "Fundación Pública Galega de Medicina Genómica (FPGMX), Centro Nacional de Genotipado (CEGEN), Servicio Gallego de Salud (SERGAS), 15706 Santiago de Compostela, Spain"
},
{
"name": "Centre for Biomedical Network Research on Rare Diseases (CIBERER), Instituto de Salud Carlos III, 28029 Madrid, Spain"
},
{
"name": "Instituto de Investigación Sanitaria de Santiago (IDIS), 15706 Santiago de Compostela, Spain"
}
],
"authenticated-orcid": false,
"family": "Lapunzina",
"given": "Pablo",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-1085-8986",
"affiliation": [
{
"name": "Fundación Pública Galega de Medicina Genómica (FPGMX), Centro Nacional de Genotipado (CEGEN), Servicio Gallego de Salud (SERGAS), 15706 Santiago de Compostela, Spain"
},
{
"name": "Grupo de Medicina Genómica, CIMUS, Universidad de Santiago de Compostela, 15782 Santiago de Compostela, Spain"
},
{
"name": "Grupo de Genética, Instituto de Investigación Sanitaria de Santiago (IDIS), 15706 Santiago de Compostela, Spain"
},
{
"name": "Centro de Investigación Biomédica en Red de Enfermedades Raras, Instituto de Salud Carlos III, 28029 Madrid, Spain"
}
],
"authenticated-orcid": false,
"family": "Carracedo",
"given": "Ángel",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-6757-9198",
"affiliation": [
{
"name": "Fundació Docència i Recerca Mutua Terrassa, 08221 Terrassa, Spain"
}
],
"authenticated-orcid": false,
"family": "Arranz",
"given": "María J.",
"sequence": "additional"
},
{
"affiliation": [],
"name": "SCOURGE COHORT GROUP",
"sequence": "additional"
}
],
"container-title": "Biomedicines",
"container-title-short": "Biomedicines",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2025,
2,
21
]
],
"date-time": "2025-02-21T12:53:06Z",
"timestamp": 1740142386000
},
"deposited": {
"date-parts": [
[
2025,
3,
4
]
],
"date-time": "2025-03-04T11:26:30Z",
"timestamp": 1741087590000
},
"funder": [
{
"DOI": "10.13039/501100004587",
"award": [
"COV20_00622",
"PI20/00876",
"COV20/00181"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100004587",
"id-type": "DOI"
}
],
"name": "Instituto de Salud Carlos III"
},
{
"award": [
"PMP22/00056"
],
"name": "BioFRAM project"
},
{
"name": "European Union (ERDF) ‘A way of making Europe’"
},
{
"name": "Fundación Amancio Ortega, Banco de Santander"
},
{
"DOI": "10.13039/100007650",
"award": [
"PIFIISC23/05"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/100007650",
"id-type": "DOI"
}
],
"name": "Fundación Canaria Instituto de Investigación Sanitaria de Canarias"
},
{
"award": [
"JTC_2021",
"AC21_2/00039"
],
"name": "ERA PerMed"
},
{
"DOI": "10.13039/501100010801",
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/501100010801",
"id-type": "DOI"
}
],
"name": "Xunta de Galicia"
},
{
"DOI": "10.13039/100031478",
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/100031478",
"id-type": "DOI"
}
],
"name": "Next Generation EU"
},
{
"name": "Recovery Mechanism and Resilience"
},
{
"DOI": "10.13039/501100004587",
"award": [
"PT17/0019"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100004587",
"id-type": "DOI"
}
],
"name": "CEGEN-PRB3-ISCIII"
},
{
"DOI": "10.13039/501100004587",
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/501100004587",
"id-type": "DOI"
}
],
"name": "ISCIII"
},
{
"DOI": "10.13039/501100008530",
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/501100008530",
"id-type": "DOI"
}
],
"name": "ERDF"
}
],
"indexed": {
"date-parts": [
[
2025,
3,
5
]
],
"date-time": "2025-03-05T05:35:25Z",
"timestamp": 1741152925725,
"version": "3.38.0"
},
"is-referenced-by-count": 0,
"issue": "3",
"issued": {
"date-parts": [
[
2025,
2,
21
]
]
},
"journal-issue": {
"issue": "3",
"published-online": {
"date-parts": [
[
2025,
3
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
2,
21
]
],
"date-time": "2025-02-21T00:00:00Z",
"timestamp": 1740096000000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/2227-9059/13/3/553/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "553",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2025,
2,
21
]
]
},
"published-online": {
"date-parts": [
[
2025,
2,
21
]
]
},
"publisher": "MDPI AG",
"reference": [
{
"DOI": "10.1007/s43657-022-00058-x",
"article-title": "Clinical Profiles at the Time of Diagnosis of SARS-CoV-2 Infection in Costa Rica During the Pre-Vaccination Period Using a Machine Learning Aroach",
"author": "Brenes",
"doi-asserted-by": "crossref",
"first-page": "312",
"journal-title": "Phenomics",
"key": "ref_1",
"volume": "2",
"year": "2022"
},
{
"DOI": "10.1016/j.jphs.2023.02.004",
"article-title": "Potential Treatments of COVID-19: Drug Repurposing and Therapeutic Interventions",
"author": "Raghav",
"doi-asserted-by": "crossref",
"first-page": "1",
"journal-title": "J. Pharmacol. Sci.",
"key": "ref_2",
"volume": "152",
"year": "2023"
},
{
"key": "ref_3",
"unstructured": "(2024, February 26). COVID-19 Vaccinations Have Saved More than 1.4 Million Lives in the WHO European Region, a New Study Finds. Available online: https://www.who.int/europe/news-room/16-01-2024-covid-19-vaccinations-have-saved-more-than-1.4-million-lives-in-the-who-european-region--a-new-study-finds."
},
{
"DOI": "10.1101/2021.04.08.21255005",
"doi-asserted-by": "crossref",
"key": "ref_4",
"unstructured": "Noureddine, F.Y., Chakkour, M., El Roz, A., Reda, J., Al Sahily, R., Assi, A., Joma, M., Salami, H., Hashem, S.J., and Harb, B. (2021). The Emergence of SARS-CoV-2 Variant(s) and Its Impact on the Prevalence of COVID-19 Cases in the Nabatieh Region, Lebanon. Med. Sci., 9."
},
{
"DOI": "10.1186/s12889-024-19749-3",
"doi-asserted-by": "crossref",
"key": "ref_5",
"unstructured": "Gong, Z., Song, T., Hu, M., Che, Q., Guo, J., Zhang, H., Li, H., Wang, Y., Liu, B., and Shi, N. (2024). Natural and Socio-Environmental Factors in the Transmission of COVID-19: A Comprehensive Analysis of Epidemiology and Mechanisms. BMC Public Health, 24."
},
{
"DOI": "10.1093/hmg/ddac132",
"article-title": "A Novel Genes and Sex Differences in COVID-19 Severity",
"author": "Cruz",
"doi-asserted-by": "crossref",
"first-page": "3789",
"journal-title": "Hum. Mol. Genet.",
"key": "ref_6",
"volume": "31",
"year": "2022"
},
{
"DOI": "10.1093/jac/dkw338",
"article-title": "A Review of Treatment Modalities for Middle East Respiratory Syndrome",
"author": "Mo",
"doi-asserted-by": "crossref",
"first-page": "3340",
"journal-title": "J. Antimicrob. Chemother.",
"key": "ref_7",
"volume": "71",
"year": "2016"
},
{
"DOI": "10.1007/s43440-022-00425-5",
"article-title": "Recent Review of COVID-19 Management: Diagnosis, Treatment and Vaccination",
"author": "Chavda",
"doi-asserted-by": "crossref",
"first-page": "1120",
"journal-title": "Pharmacol. Rep.",
"key": "ref_8",
"volume": "74",
"year": "2022"
},
{
"DOI": "10.1002/jmv.26232",
"article-title": "The Cytokine Storm and COVID-19",
"author": "Hu",
"doi-asserted-by": "crossref",
"first-page": "250",
"journal-title": "J. Med. Virol.",
"key": "ref_9",
"volume": "93",
"year": "2020"
},
{
"DOI": "10.3389/fmed.2020.596916",
"doi-asserted-by": "crossref",
"key": "ref_10",
"unstructured": "Valenzuela, O., Ibáñez, S., Poli, M.C., Roessler, P., Aylwin, M., Roizen, G., Iruretagoyena, M., Agar, V., Donoso, J., and Fierro, M. (2020). First Report of Tocilizumab Use in a Cohort of Latin American Patients Hospitalized for Severe COVID-19 Pneumonia. Front. Med., 7."
},
{
"DOI": "10.1186/s12967-020-02339-3",
"article-title": "Why Tocilizumab Could Be an Effective Treatment for Severe COVID-19?",
"author": "Fu",
"doi-asserted-by": "crossref",
"first-page": "164",
"journal-title": "J. Transl. Med.",
"key": "ref_11",
"volume": "18",
"year": "2020"
},
{
"article-title": "Use of Tocilizumab in COVID-19: A Systematic Review and Meta-Analysis of Current Evidence",
"author": "Kotak",
"first-page": "e10869",
"journal-title": "Cureus",
"key": "ref_12",
"volume": "12",
"year": "2020"
},
{
"DOI": "10.3389/fphar.2022.835136",
"doi-asserted-by": "crossref",
"key": "ref_13",
"unstructured": "Biswas, M., Sawajan, N., Rungrotmongkol, T., Sanachai, K., Ershadian, M., and Sukasem, C. (2022). Pharmacogenetics and Precision Medicine Aroaches for the Improvement of COVID-19 Therapies. Front. Pharmacol., 13."
},
{
"DOI": "10.1097/01.qai.0000219774.20174.64",
"article-title": "Lopinavir/Ritonavir Induces the Hepatic Activity of Cytochrome P450 Enzymes CYP2C9, CYP2C19, and CYP1A2 But Inhibits the Hepatic and Intestinal Activity of CYP3A as Measured by a Phenotyping Drug Cocktail in Healthy Volunteers",
"author": "Yeh",
"doi-asserted-by": "crossref",
"first-page": "52",
"journal-title": "JAIDS J. Acquir. Immune Defic. Syndr.",
"key": "ref_14",
"volume": "42",
"year": "2006"
},
{
"article-title": "Metabolism and Interactions of Chloroquine and Hydroxychloroquine with Human Cytochrome P450 Enzymes and Drug Transporters",
"author": "Rendic",
"first-page": "1127",
"journal-title": "Curr. Drug Metab.",
"key": "ref_15",
"volume": "21",
"year": "2020"
},
{
"DOI": "10.1016/S0960-0760(97)00038-1",
"article-title": "Dexamethasone Metabolism in Species Differences",
"author": "Tomlinson",
"doi-asserted-by": "crossref",
"first-page": "345",
"journal-title": "J. Steroid. Biochem. Mol. Biol.",
"key": "ref_16",
"volume": "62",
"year": "1997"
},
{
"DOI": "10.3389/fphar.2019.00520",
"doi-asserted-by": "crossref",
"key": "ref_17",
"unstructured": "García-Menaya, J.M., Cordobés-Durán, C., García-Martín, E., and Agúndez, J.A.G. (2019). Pharmacogenetic Factors Affecting Asthma Treatment Response. Potential Implications for Drug Therapy. Front. Pharmacol., 10."
},
{
"DOI": "10.1038/s41397-021-00209-9",
"article-title": "Pharmacogenomics and COVID-19: Clinical Implications of Human Genome Interactions with Repurposed Drugs",
"author": "Badary",
"doi-asserted-by": "crossref",
"first-page": "275",
"journal-title": "Pharmacogenomics J.",
"key": "ref_18",
"volume": "21",
"year": "2021"
},
{
"DOI": "10.3390/v13030413",
"doi-asserted-by": "crossref",
"key": "ref_19",
"unstructured": "Fricke-Galindo, I., and Falfán-Valencia, R. (2021). Pharmacogenetics Aroach for the Improvement of COVID-19 Treatment. Viruses, 13."
},
{
"DOI": "10.1371/journal.pone.0013759",
"doi-asserted-by": "crossref",
"key": "ref_20",
"unstructured": "Pino-Yanes, M., Corrales, A., Casula, M., Blanco, J., Muriel, A., Espinosa, E., García-Bello, M., Torres, A., Ferrer, M., and Zavala, E. (2010). Common Variants of TLR1 Associate with Organ Dysfunction and Sustained Pro-Inflammatory Responses during Sepsis. PLoS ONE, 5."
},
{
"key": "ref_21",
"unstructured": "San Gil, A. (2017). Pneumònia Aguda de La Comunitat En l’Adult: Etiologia i Biomarcadors Genètics de l’Hoste. [Ph.D. Thesis, Universitat Internacional de Catalunya]."
},
{
"DOI": "10.1002/rmv.2194",
"article-title": "Pharmacogenomics of Genetic Polymorphism within the Genes Responsible for SARS-CoV-2 Susceptibility and the Drug-Metabolising Genes Used in Treatment",
"author": "Alahmad",
"doi-asserted-by": "crossref",
"first-page": "e2194",
"journal-title": "Rev. Med. Virol.",
"key": "ref_22",
"volume": "31",
"year": "2021"
},
{
"DOI": "10.3390/ph15060739",
"doi-asserted-by": "crossref",
"key": "ref_23",
"unstructured": "Franczyk, B., Rysz, J., Miłoński, J., Konecki, T., Rysz-Górzyńska, M., and Gluba-Brzózka, A. (2022). Will the Use of Pharmacogenetics Improve Treatment Efficiency in COVID-19?. Pharmaceuticals, 15."
},
{
"article-title": "Radiological Findings in Patients with COVID-19",
"author": "Fatima",
"first-page": "e7651",
"journal-title": "Cureus",
"key": "ref_24",
"volume": "12",
"year": "2020"
},
{
"article-title": "The Mid P-Value in Exact Tests for Hardy-Weinberg Equilibrium",
"author": "Graffelman",
"first-page": "433",
"journal-title": "Stat. Al. Genet. Mol. Biol.",
"key": "ref_25",
"volume": "12",
"year": "2013"
},
{
"DOI": "10.1038/s41586-021-03205-y",
"article-title": "Sequencing of 53,831 Diverse Genomes from the NHLBI TOPMed Program",
"author": "Taliun",
"doi-asserted-by": "crossref",
"first-page": "290",
"journal-title": "Nature",
"key": "ref_26",
"volume": "590",
"year": "2021"
},
{
"DOI": "10.1016/j.pupt.2022.102172",
"article-title": "Considerations into Pharmacogenomics of COVID-19 Pharmacotherapy: Hope, Hype and Reality",
"author": "Sardas",
"doi-asserted-by": "crossref",
"first-page": "102172",
"journal-title": "Pulm. Pharmacol. Ther.",
"key": "ref_27",
"volume": "77",
"year": "2022"
},
{
"DOI": "10.1590/S1807-59322011000800009",
"article-title": "Pharmacogenetics of Glucocorticoid Replacement Could Optimize the Treatment of Congenital Adrenal Hyperplasia Due to 21-Hydroxylase Deficiency",
"author": "Moreira",
"doi-asserted-by": "crossref",
"first-page": "1361",
"journal-title": "Clinics",
"key": "ref_28",
"volume": "66",
"year": "2011"
},
{
"DOI": "10.1186/s12985-022-01814-1",
"article-title": "COVID-19 Infection: An Overview on Cytokine Storm and Related Interventions",
"author": "Montazersaheb",
"doi-asserted-by": "crossref",
"first-page": "92",
"journal-title": "Virol. J.",
"key": "ref_29",
"volume": "19",
"year": "2022"
},
{
"DOI": "10.2217/pme-2020-0183",
"article-title": "Pharmacogenomic Considerations for Repurposing of Dexamethasone as a Potential Drug against SARS-CoV-2 Infection",
"author": "Vohra",
"doi-asserted-by": "crossref",
"first-page": "389",
"journal-title": "Per. Med.",
"key": "ref_30",
"volume": "18",
"year": "2021"
},
{
"DOI": "10.1097/FPC.0b013e32834592fe",
"article-title": "Genetic Variation in UGT1A1 Typical of Gilbert Syndrome Is Associated with Unconjugated Hyperbilirubinemia in Patients Receiving Tocilizumab",
"author": "Lee",
"doi-asserted-by": "crossref",
"first-page": "365",
"journal-title": "Pharmacogenet. Genom.",
"key": "ref_31",
"volume": "21",
"year": "2011"
},
{
"DOI": "10.1016/S0300-483X(01)00456-5",
"article-title": "Regulation of Biliary Drug Efflux Pump Expression by Hormones and Xenobiotics",
"author": "Fardel",
"doi-asserted-by": "crossref",
"first-page": "37",
"journal-title": "Toxicology",
"key": "ref_32",
"volume": "167",
"year": "2001"
},
{
"DOI": "10.1016/j.cytogfr.2022.10.003",
"article-title": "Pharmacological Therapies and Drug Development Targeting SARS-CoV-2 Infection",
"author": "Jiang",
"doi-asserted-by": "crossref",
"first-page": "13",
"journal-title": "Cytokine Growth Factor Rev.",
"key": "ref_33",
"volume": "68",
"year": "2022"
},
{
"DOI": "10.3390/ijms231911740",
"doi-asserted-by": "crossref",
"key": "ref_34",
"unstructured": "Jarczak, D., and Nierhaus, A. (2022). Cytokine Storm—Definition, Causes, and Implications. Int. J. Mol. Sci., 23."
},
{
"DOI": "10.1007/s10787-022-00992-2",
"doi-asserted-by": "crossref",
"key": "ref_35",
"unstructured": "Hasanvand, A. (2022). COVID-19 and the Role of Cytokines in This Disease. Inflammopharmacology, 789–798."
},
{
"DOI": "10.1152/ajplung.00547.2017",
"article-title": "Th1 Cytokines TNF-and IFN-Promote Corticosteroid Resistance in Developing Human Airway Smooth Muscle",
"author": "Britt",
"doi-asserted-by": "crossref",
"first-page": "71",
"journal-title": "Am. J. Physiol. Lung. Cell. Mol. Physiol.",
"key": "ref_36",
"volume": "316",
"year": "2019"
},
{
"DOI": "10.1086/519795",
"article-title": "PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses",
"author": "Purcell",
"doi-asserted-by": "crossref",
"first-page": "559",
"journal-title": "Am. J. Hum. Genet.",
"key": "ref_37",
"volume": "81",
"year": "2007"
},
{
"DOI": "10.1038/s41525-020-00143-y",
"article-title": "Pharmacogenomics of COVID-19 Therapies",
"author": "Takahashi",
"doi-asserted-by": "crossref",
"first-page": "35",
"journal-title": "NPJ Genom. Med.",
"key": "ref_38",
"volume": "5",
"year": "2020"
},
{
"key": "ref_39",
"unstructured": "Zaidat, O.O., Erwin Grüter, B., Cantonal Hospital, A., Luis Rafael Moscote-Salazar, S., Yan, J., Jiang, W., Liu, J., Liao, X., Zhou, J., and Li, B. (2021). A Rare Variant of ANK3 Is Associated With Intracranial Aneurysm. Front. Neurol., 1."
},
{
"DOI": "10.1016/j.pnpbp.2024.111016",
"doi-asserted-by": "crossref",
"key": "ref_40",
"unstructured": "Yang, Y., Zhu, Z., Hui, L., and Sun, P. (2024). Effects of CACNA1C and ANK3 on Cognitive Function in Patients with Bipolar Disorder. Prog. Neuropsychopharmacol. Biol. Psychiatry, 133."
},
{
"DOI": "10.1007/s10048-021-00655-4",
"article-title": "ANK3 Related Neurodevelopmental Disorders: Expanding the Spectrum of Heterozygous Loss-of-Function Variants",
"author": "Kloth",
"doi-asserted-by": "crossref",
"first-page": "263",
"journal-title": "Neurogenetics",
"key": "ref_41",
"volume": "22",
"year": "2021"
},
{
"article-title": "Expression Analysis of Atp-Binding Cassette Transporters Abcb11 and Abcb4 in Primary Sclerosing Cholangitis and Variety of Pediatric and Adult Cholestatic and Noncholestatic Liver Diseases",
"author": "Thoeni",
"first-page": "1085717",
"journal-title": "Can. J. Gastroenterol. Hepatol.",
"key": "ref_42",
"volume": "2019",
"year": "2019"
},
{
"DOI": "10.4110/in.2020.20.e21",
"article-title": "TLR10 and Its Unique Anti-Inflammatory Properties and Potential Use as a Target in Therapeutics",
"author": "Fore",
"doi-asserted-by": "crossref",
"first-page": "e21",
"journal-title": "Immune Netw.",
"key": "ref_43",
"volume": "20",
"year": "2020"
},
{
"DOI": "10.1007/s00441-016-2518-3",
"article-title": "The Critical Role of ABCG1 and PPARγ/LXRα Signaling in TLR4 Mediates Inflammatory Responses and Lipid Accumulation in Vascular Smooth Muscle Cells",
"author": "Cao",
"doi-asserted-by": "crossref",
"first-page": "145",
"journal-title": "Cell. Tissue Res.",
"key": "ref_44",
"volume": "368",
"year": "2017"
},
{
"DOI": "10.1371/journal.pone.0098815",
"doi-asserted-by": "crossref",
"key": "ref_45",
"unstructured": "Bank, S., Skytt Andersen, P., Burisch, J., Pedersen, N., Roug, S., Galsgaard, J., Ydegaard Turino, S., Broder Brodersen, J., Rashid, S., and Kaiser Rasmussen, B. (2014). Polymorphisms in the Inflammatory Pathway Genes TLR2, TLR4, TLR9, LY96, NFKBIA, NFKB1, TNFA, TNFRSF1A, IL6R, IL10, IL23R, PTPN22, and PPARG Are Associated with Susceptibility of Inflammatory Bowel Disease in a Danish Cohort. PLoS ONE, 9."
},
{
"DOI": "10.4049/jimmunol.1002122",
"article-title": "Synergistic Interactions of TLR2/6 and TLR9 Induce a High Level of Resistance to Lung Infection in Mice",
"author": "Duggan",
"doi-asserted-by": "crossref",
"first-page": "5916",
"journal-title": "J. Immunol.",
"key": "ref_46",
"volume": "186",
"year": "2011"
},
{
"DOI": "10.1002/jmv.25997",
"doi-asserted-by": "crossref",
"key": "ref_47",
"unstructured": "Chakraborty, C., Sharma, A.R., Bhattacharya, M., Sharma, G., Lee, S.S., and Agoramoorthy, G. (2020). Consider TLR5 for New Therapeutic Development against COVID-19. J. Med. Virol., 2314–2315."
},
{
"DOI": "10.1080/14728222.2021.1918103",
"doi-asserted-by": "crossref",
"key": "ref_48",
"unstructured": "Kaushik, D., Bhandari, R., and Kuhad, A. (2021). TLR4 as a Therapeutic Target for Respiratory and Neurological Complications of SARS-CoV-2. Expert Opin. Ther. Targets, 491–508."
},
{
"DOI": "10.1172/JCI84015",
"article-title": "RBFox1-Mediated RNA Splicing Regulates Cardiac Hypertrophy and Heart Failure",
"author": "Gao",
"doi-asserted-by": "crossref",
"first-page": "195",
"journal-title": "J. Clin. Investig.",
"key": "ref_49",
"volume": "126",
"year": "2016"
},
{
"DOI": "10.1089/ars.2023.0437",
"article-title": "Mechanistic Insights into Alternative Gene Splicing in Oxidative Stress and Tissue Injury",
"author": "Dery",
"doi-asserted-by": "crossref",
"first-page": "890",
"journal-title": "Antioxid. Redox. Signal.",
"key": "ref_50",
"volume": "41",
"year": "2023"
},
{
"DOI": "10.3389/fcvm.2023.1203168",
"doi-asserted-by": "crossref",
"key": "ref_51",
"unstructured": "Wang, M., Cheng, L., Gao, Z., Li, J., Ding, Y., Shi, R., Xiang, Q., and Chen, X. (2023). Investigation of the Shared Molecular Mechanisms and Hub Genes between Myocardial Infarction and Depression. Front. Cardiovasc. Med., 10."
},
{
"article-title": "Genome-Wide Association Study of SARS-CoV-2 Infection in Chinese Population",
"author": "Fan",
"first-page": "1155",
"journal-title": "Eur. J. Clin. Microbiol. Infect. Dis.",
"key": "ref_52",
"volume": "41",
"year": "2022"
},
{
"article-title": "Altered CELF4 Splicing Factor Enhances Pancreatic Neuroendocrine Tumors Aggressiveness Influencing MTOR and Everolimus Response",
"first-page": "102090",
"journal-title": "Mol. Ther. Nucleic Acids",
"key": "ref_53",
"volume": "35",
"year": "2023"
},
{
"DOI": "10.3389/fimmu.2023.1233318",
"doi-asserted-by": "crossref",
"key": "ref_54",
"unstructured": "Engel, J.J., van der Made, C.I., Keur, N., Setiabudiawan, T., Röring, R.J., Damoraki, G., Dijkstra, H., Lemmers, H., Ioannou, S., and Poulakou, G. (2023). Dexamethasone Attenuates Interferon-Related Cytokine Hyperresponsiveness in COVID-19 Patients. Front. Immunol., 14."
},
{
"DOI": "10.1002/cpt.1973",
"article-title": "PharmVar GeneFocus: CYP2C19",
"author": "Botton",
"doi-asserted-by": "crossref",
"first-page": "352",
"journal-title": "Clin. Pharmacol. Ther.",
"key": "ref_55",
"volume": "109",
"year": "2020"
},
{
"DOI": "10.1517/17425255.2014.878703",
"article-title": "Cytochrome P450 Enzyme Regulation by Glucocorticoids and Consequences in Terms of Drug Interaction",
"doi-asserted-by": "crossref",
"first-page": "425",
"journal-title": "Expert Opin. Drug Metab. Toxicol.",
"key": "ref_56",
"volume": "10",
"year": "2014"
},
{
"DOI": "10.1097/FPC.0b013e32834d4962",
"article-title": "PharmGKB Summary: Very Important Pharmacogene Information for Cytochrome P450, Family 2, Subfamily C, Polypeptide 19",
"author": "Scott",
"doi-asserted-by": "crossref",
"first-page": "159",
"journal-title": "Pharmacogenet. Genom.",
"key": "ref_57",
"volume": "22",
"year": "2012"
},
{
"DOI": "10.1152/ajpendo.00405.2020",
"article-title": "Bilirubin as a Metabolic Hormone: The Physiological Relevance of Low Levels",
"author": "Creeden",
"doi-asserted-by": "crossref",
"first-page": "191",
"journal-title": "Am. J. Physiol. -Endocrinol. Metab.",
"key": "ref_58",
"volume": "320",
"year": "2021"
},
{
"article-title": "Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for UGT1A1 and Atazanavir Prescribing",
"author": "Gammal",
"first-page": "363",
"journal-title": "Clin. Med. Ther.",
"key": "ref_59",
"volume": "99",
"year": "2016"
},
{
"DOI": "10.1007/s00277-014-2170-5",
"article-title": "UGT1A1 *6 Polymorphism Predicts Outcome in Elderly Patients with Relapsed or Refractory Diffuse Large B-Cell Lymphoma Treated with Carboplatin, Dexamethasone, Etoposide and Irinotecan",
"author": "Yamasaki",
"doi-asserted-by": "crossref",
"first-page": "65",
"journal-title": "Ann. Hematol.",
"key": "ref_60",
"volume": "94",
"year": "2015"
},
{
"DOI": "10.1007/s00408-020-00408-4",
"doi-asserted-by": "crossref",
"key": "ref_61",
"unstructured": "Scialo, F., Daniele, A., Amato, F., Pastore, L., Matera, M.G., Cazzola, M., Castaldo, G., and Bianco, A. (2020). ACE2: The Major Cell Entry Receptor for SARS-CoV-2. Lung, 867–877."
},
{
"DOI": "10.1016/j.jaci.2020.09.034",
"article-title": "Inhaled Corticosteroids Downregulate the SARS-CoV-2 Receptor ACE2 in COPD through Suression of Type I Interferon",
"author": "Finney",
"doi-asserted-by": "crossref",
"first-page": "510",
"journal-title": "J. Allergy Clin. Immunol.",
"key": "ref_62",
"volume": "147",
"year": "2021"
},
{
"DOI": "10.1186/s12931-021-01782-0",
"article-title": "Up-Regulation of ACE2, the SARS-CoV-2 Receptor, in Asthmatics on Maintenance Inhaled Corticosteroids",
"author": "Salit",
"doi-asserted-by": "crossref",
"first-page": "200",
"journal-title": "Respir. Res.",
"key": "ref_63",
"volume": "22",
"year": "2021"
},
{
"DOI": "10.1016/j.isci.2018.12.034",
"article-title": "IFNG-AS1 Enhances Interferon Gamma Production in Human Natural Killer Cells",
"author": "Stein",
"doi-asserted-by": "crossref",
"first-page": "466",
"journal-title": "iScience",
"key": "ref_64",
"volume": "11",
"year": "2019"
},
{
"DOI": "10.1161/01.RES.0000250171.54048.5c",
"article-title": "ATP-Binding Cassette Cholesterol Transporters and Cardiovascular Disease",
"author": "Oram",
"doi-asserted-by": "crossref",
"first-page": "1031",
"journal-title": "Circ. Res.",
"key": "ref_65",
"volume": "99",
"year": "2006"
},
{
"DOI": "10.1165/rcmb.2018-0365OC",
"article-title": "Alveolar Macrophage ABCG1 Deficiency Promotes Pulmonary Granulomatous Inflammation",
"author": "McPeek",
"doi-asserted-by": "crossref",
"first-page": "332",
"journal-title": "Am. J. Respir. Cell Mol. Biol.",
"key": "ref_66",
"volume": "61",
"year": "2019"
},
{
"DOI": "10.1016/j.cell.2022.06.008",
"article-title": "Mild Respiratory COVID Can Cause Multi-Lineage Neural Cell and Myelin Dysregulation",
"author": "Lu",
"doi-asserted-by": "crossref",
"first-page": "2452",
"journal-title": "Cell",
"key": "ref_67",
"volume": "185",
"year": "2022"
},
{
"DOI": "10.3389/fimmu.2022.857573",
"doi-asserted-by": "crossref",
"key": "ref_68",
"unstructured": "Battaglini, D., Lopes-Pacheco, M., Castro-Faria-Neto, H.C., Pelosi, P., and Rocco, P.R.M. (2022). Laboratory Biomarkers for Diagnosis and Prognosis in COVID-19. Front. Immunol., 13."
}
],
"reference-count": 68,
"references-count": 68,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.mdpi.com/2227-9059/13/3/553"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Pharmacogenomic Study of SARS-CoV-2 Treatments: Identifying Polymorphisms Associated with Treatment Response in COVID-19 Patients",
"type": "journal-article",
"volume": "13"
}