Molecular Aspects of Viral Pathogenesis in Emerging SARS-CoV-2 Variants: Evolving Mechanisms of Infection and Host Response
et al., International Journal of Molecular Sciences, doi:10.3390/ijms27020891, Jan 2026
Review of molecular mechanisms underlying SARS-CoV-2 pathogenesis in emerging variants, particularly late Omicron sublineages including BA.2.86, JN.1, and KP descendants.
Muntean et al., 15 Jan 2026, Romania, peer-reviewed, 8 authors.
Contact: andreea-raluca.szoke@umfst.ro (corresponding author), sofia.harsan@umfst.ro, andreea-catalina.tinca@umfst.ro, silviu.vultur@yahoo.com, nemes.mara.1997@gmail.com.
Molecular Aspects of Viral Pathogenesis in Emerging SARS-CoV-2 Variants: Evolving Mechanisms of Infection and Host Response
International Journal of Molecular Sciences, doi:10.3390/ijms27020891
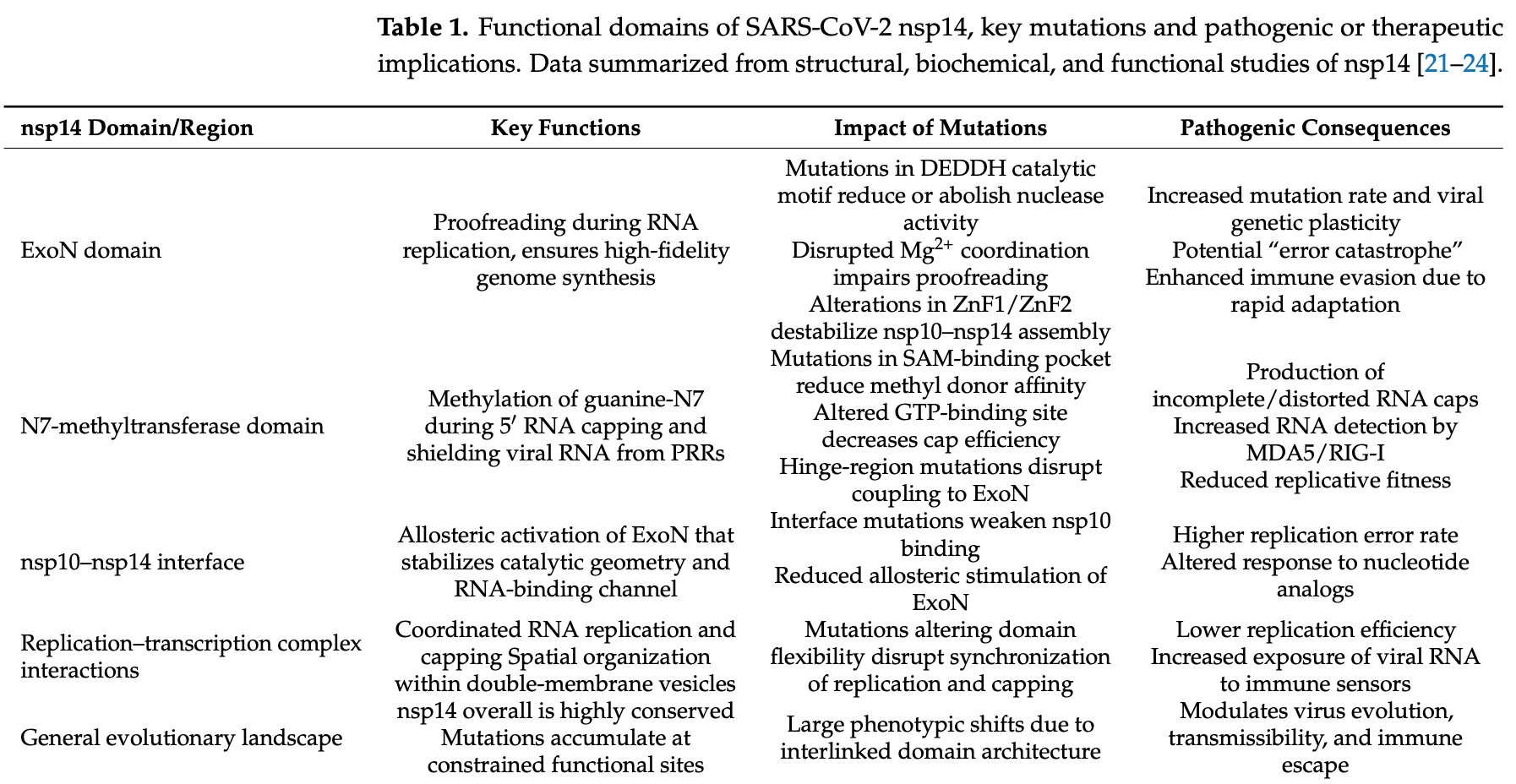
Although the SARS-CoV-2 pandemic no longer poses a global emergency, the virus continues to diversify and acquire immunoevasive properties. Understanding the molecular pathways that shape SARS-CoV-2 pathogenesis has become essential. In this paper, we summarize the most recent current evidence on how the spike protein structurally evolves, on changes in key non-structural proteins, such as nsp14, and on host factors, such as TMPRSS2 and neuropilin-1. These changes, together, shape viral entry, replication fidelity and interferon antagonism. Given the emerging Omicron variants of SARS-CoV-2, recent articles in the literature, cryo-EM analyses, and artificial intelligence-assisted mutational modeling were analyzed to infer and contextualize mutation-driven mechanisms. It is through these changes that the virus adapts and evolves, such as optimizing angiotensinconverting enzyme binding, modifying antigenic surfaces, and accumulating mutations that affect CD8 + T-cell recognition. Multi-omics data studies further support SARS-CoV-2 pathogenesis through convergent evidence linking viral adaptation to host immune and metabolic reprogramming, as occurs in myocarditis, liver injury, and acute kidney injury. By integrating proteomic, transcriptomic, and structural findings, this work presents how the virus persists and dictates disease severity through interferon antagonism (ORF6, ORF9b, and nsp1), adaptive immune evasion, and metabolic rewiring. All these insights underscore the need for next-generation interventions that provide a multidimensional framework for understanding the evolution of SARS-CoV-2 and guiding future antiviral strategies.
Conflicts of Interest: The authors declare no conflicts of interest.
Abbreviations
References
Agu, Afiukwa, Orji, Ezeh, Ofoke et al., Molecular Docking as a Tool for the Discovery of Molecular Targets of Nutraceuticals in Disease Management, Sci. Rep, doi:10.1038/s41598-023-40160-2
Albalawi, Thomas, Mughal, Kotsiri, Roper et al., SARS-CoV-2 S, M, and E Structural Glycoproteins Differentially Modulate Endoplasmic Reticulum Stress Responses, Int. J. Mol. Sci, doi:10.3390/ijms26031047
Altamirano, Huang, Tsai, Tzeng, Teruel et al., Coronavirus nsp14 Drives Internal m7G Modification to Rewire Host Splicing and Promote Viral Replication, bioRxiv, doi:10.1101/2025.09.03.673802
Augustus, Radhakrishnan, Bhaskar, Ramamurthi, Shunmugiah, Tannic Acid Modulates SARS-CoV-2 Pathogenesis by Curbing Key Host Receptors and Oxidative Stress, Toxicol. Vitr, doi:10.1016/j.tiv.2024.105971
Babačić, Christ, Araújo, Mermelekas, Sharma et al., Comprehensive Proteomics and Meta-Analysis of COVID-19 Host Response, Nat. Commun, doi:10.1038/s41467-023-41159-z
Bolland, Marechal, Petiot, Porrot, Guivel-Benhassine et al., SARS-CoV-2 Entry and Fusion Are Independent of ACE2 Localization to Lipid Rafts, J. Virol, doi:10.1128/jvi.01823-24
Branda, Ciccozzi, Scarpa, Features of the SARS-CoV-2 KP.3 Variant Mutations, Infect. Dis, doi:10.1080/23744235.2024.2385500
Budin, Cocuz, Enache, Rent, Cazacu et al., Neutrophil-to-Lymphocyte and Platelet-to-Lymphocyte Ratio: Side by Side with Molecular Mutations in Patients with Non-Small Cell Lung Cancer-The INOLUNG Study, Cancers, doi:10.3390/cancers16162903
Budin, Nemes, Râjnoveanu, Nemes, Rajnoveanu et al., The Inflammatory Profile Correlates with COVID-19 Severity and Mortality in Cancer Patients, J. Pers. Med, doi:10.3390/jpm13081235
Chen, Jeng, Lee, Wang, Tsai et al., Viral Mitochondriopathy in COVID-19, Redox Biol, doi:10.1016/j.redox.2025.103766
Dette, Moers, Mayr, Stillfried, Bernhardt et al., Differential Expression of Viral Entry Protein Neuropilin 1 (NRP1) and Neuropilin 2 (NRP2) in Fatal COVID-19, J. Virol, doi:10.1128/jvi.01384-25
Feng, Huang, Baboo, Diedrich, Bangaru et al., Structural and Functional Insights into the Evolution of SARS-CoV-2 KP.3.1.1 Spike Protein, Cell Rep, doi:10.1016/j.celrep.2025.115941
Holmes, Colaneri, Palomba, Gori, Exploring Post-Sepsis and Post-COVID-19 Syndromes: Crossovers from Pathophysiology to Therapeutic Approach, Front. Med, doi:10.3389/fmed.2023.1280951
Honda, Toyama, Matsumoto, Sanada, Yasui et al., Intranasally Inoculated SARS-CoV-2 Spike Protein Combined with Mucoadhesive Polymer Induces Broad and Long-Lasting Immunity, Vaccines, doi:10.3390/vaccines12070794
Isazadeh, Heris, Shahabi, Mohammadinasab, Shomali et al., Pattern-Recognition Receptors (PRRs) in SARS-CoV-2, Life Sci, doi:10.1016/j.lfs.2023.121940
Jadhav, Liang, Ansari, Tan, Tan et al., Design of Quinoline SARS-CoV-2 Papain-Like Protease Inhibitors as Oral Antiviral Drug Candidates, Nat. Commun, doi:10.1038/s41467-025-56902-x
Jayaraman, Rajagopal, Paranjpe, Suarez-Farinas, Liharska et al., Peripheral Transcriptomics in Acute and Long-Term Kidney Dysfunction in SARS-CoV-2 Infection, Kidney, doi:10.34067/KID.0000000727
Kim, Bae, Myoung, Domain-Specific Impacts of Spike Protein Mutations on Infectivity and Antibody Escape in SARS-CoV-2 Omicron BA.1, J. Microbiol. Biotechnol, doi:10.4014/jmb.2507.07040
Lee, Kwon, Kim, Woo, Oh, Electrochemical Pan-Variant Detection of SARS-CoV-2 through Host Cell Receptor-Mimicking Molecular Recognition, Biosens. Bioelectron, doi:10.1016/j.bios.2025.117311
Li, Faraone, Hsu, Chamblee, Liu et al., Neutralization and Spike Stability of JN.1-Derived LB.1, KP.2.3, KP.3, doi:10.1128/mbio.00464-25
Li, Wu, Pattern Recognition Receptors in Health and Diseases, Signal Transduct. Target. Ther, doi:10.1038/s41392-021-00687-0
Liu, Wang, Zheng, Du, Maarouf et al., Roles of circRNAs in Viral Pathogenesis, Front. Cell. Infect. Microbiol, doi:10.3389/fcimb.2025.1564258
Luo, Mo, Wang, Wei, Chen et al., SARS-CoV-2 nsp14 Inhibitor Exhibits Potent Antiviral Activity and Reverses nsp14-Driven Host Modulation, Nat. Commun, doi:10.1038/s41467-025-64674-7
Maddaloni, Bugani, Fracella, Bitossi, Aloisi et al., Pattern Recognition Receptors (PRRs) Expression and Activation in COVID-19 and Long COVID, Microorganisms, doi:10.3390/microorganisms13092176
Mahdi, Kiarie, Mótyán, Hoffka, Al-Muffti et al., Receptor Binding for the Entry Mechanisms of SARS-CoV-2: Insights from the Original Strain and Emerging Variants, Viruses, doi:10.3390/v17050691
Mambelli, De Araujo, Farias, De Andrade, Ferreira et al., An Update on Anti-COVID-19 Vaccines and the Challenges to Protect Against New SARS-CoV-2 Variants, Pathogens, doi:10.3390/pathogens14010023
Mcmahan, Wegmann, Aid, Sciacca, Liu et al., Mucosal Boosting Enhances Vaccine Protection against SARS-CoV-2 in Macaques, Nature, doi:10.1038/s41586-023-06951-3
Meyer, Garzia, Miller, Huggins, Myers et al., Small-Molecule Inhibition of SARS-CoV-2 nsp14 RNA Cap Methyltransferase, Nature, doi:10.1038/s41586-024-08320-0
Noureldeen, Aziz, Shouman, Mohamed, Attia et al., Molecular Design, Spectroscopic, DFT, Pharmacological, and Molecular Docking Studies of Novel Ruthenium(III)-Schiff Base Complex: An Inhibitor of Progression in HepG2 Cells, Int. J. Environ. Res. Public Health, doi:10.3390/ijerph192013624
Pollett, Chenoweth, Dalgard, Epsi, Richard et al., Proteomic and Transcriptomic Signatures of SARS-CoV-2-Associated Myocarditis, Open Forum Infect. Dis, doi:10.1093/ofid/ofae631.2094
Rau, Schicht, Zahn, Ali, Coroneo et al., Detection of SARS-CoV-2 Binding Receptors and Miscellaneous Targets as well as Mucosal Surface Area of the Human Lacrimal Drainage System, Sci. Direct
Sanches De Carvalho, Blanco Capassi Santos, Shiroma Graziano, Peres, Sousa Silva et al., Expression of ACE2 and TMPRSS2 and the Severity of COVID-19, Biomed. Rep, doi:10.3892/br.2025.2069
Siddiqui, Musharaf, Gulumbe, The, 1 Variant of COVID-19: Immune Evasion, Transmissibility, and Implications for Global Health, Ther. Adv. Infect. Dis, doi:10.1177/20499361251314763
Silva-Ríos, Mora-Ornelas, Flores-Medina, Muñoz-Valle, Díaz-Palomera et al., Beyond Processing: Furin as a Central Hub in Viral Pathogenesis and Genetic Susceptibility, Biomolecules, doi:10.3390/biom15111530
Tian, Shang, Zhang, Guo, Li et al., T Cell Immune Evasion by SARS-CoV-2 JN.1 Escapees Targeting Two Cytotoxic T Cell Epitope Hotspots, Nat. Immunol, doi:10.1038/s41590-024-02051-0
Uzun, Pant, Bartoszek, Gueguen, Frei et al., Analysis of Hyperexpanded T Cell Clones in SARS-CoV-2 Vaccine-Associated Liver Injury by Spatial Proteomics and Transcriptomics, Liver Int, doi:10.1111/liv.70172
Wang, Guo, Mellis, Wu, Mohri et al., Antibody Evasiveness of SARS-CoV-2 Subvariants KP.3.1.1 and XEC, Cell Rep, doi:10.1016/j.celrep.2025.115543
Wang, Hao, Ding, Rong, Ning et al., Characterization of Private Mutations in the Spike Protein of SARS-CoV-2 Correlates with Viral Prevalence, BMC Infect. Dis, doi:10.1186/s12879-025-11414-3
Wang, Li, Du, Chen, Sun et al., Ancestral SARS-CoV-2 Immune Imprinting Persists on RBD but Not NTD after Sequential Omicron Infections, iScience, doi:10.1016/j.isci.2024.111557
Wang, Liu, Lou, Targeting Coronaviral nsp14: A Bifunctional Enzyme for RNA Capping and Proofreading, Biophys. Rep, doi:10.52601/bpr.2025.250036
Wee, Wei, Rapid Response to Fast Viral Evolution Using AlphaFold 3-Assisted Topological Deep Learning, Virus Evol, doi:10.1093/ve/veaf026
Wu, Wu, Hu, Zheng, Chang et al., Immune Evasion of Omicron Variants JN.1, KP.2, and KP.3 to Polyclonal and Monoclonal Antibodies, Antiviral Res, doi:10.1016/j.antiviral.2025.106092
Zhang, Fong, Dang, Fintzi, Chen et al., Neutralizing Antibody Immune Correlates in COVAIL Trial Recipients of an mRNA Second COVID-19 Vaccine Boost, Nat. Commun, doi:10.1038/s41467-025-55931-w
Zhang, Kennedy, Jorge, Xing, Reid et al., SARS-CoV-2 Remodels the Golgi Apparatus to Facilitate Viral Assembly and Secretion, PLoS Pathog, doi:10.1371/journal.ppat.1013295
Zodda, Pons, Demoya-Valenzuela, Calvo-González, Benítez-Rodríguez et al., Induction of the Inflammasome by the SARS-CoV-2 Accessory Protein ORF9b, J. Med Virol, doi:10.1002/jmv.70145
DOI record:
{
"DOI": "10.3390/ijms27020891",
"ISSN": [
"1422-0067"
],
"URL": "http://dx.doi.org/10.3390/ijms27020891",
"abstract": "<jats:p>Although the SARS-CoV-2 pandemic no longer poses a global emergency, the virus continues to diversify and acquire immunoevasive properties. Understanding the molecular pathways that shape SARS-CoV-2 pathogenesis has become essential. In this paper, we summarize the most recent current evidence on how the spike protein structurally evolves, on changes in key non-structural proteins, such as nsp14, and on host factors, such as TMPRSS2 and neuropilin-1. These changes, together, shape viral entry, replication fidelity and interferon antagonism. Given the emerging Omicron variants of SARS-CoV-2, recent articles in the literature, cryo-EM analyses, and artificial intelligence-assisted mutational modeling were analyzed to infer and contextualize mutation-driven mechanisms. It is through these changes that the virus adapts and evolves, such as optimizing angiotensin-converting enzyme binding, modifying antigenic surfaces, and accumulating mutations that affect CD8+ T-cell recognition. Multi-omics data studies further support SARS-CoV-2 pathogenesis through convergent evidence linking viral adaptation to host immune and metabolic reprogramming, as occurs in myocarditis, liver injury, and acute kidney injury. By integrating proteomic, transcriptomic, and structural findings, this work presents how the virus persists and dictates disease severity through interferon antagonism (ORF6, ORF9b, and nsp1), adaptive immune evasion, and metabolic rewiring. All these insights underscore the need for next-generation interventions that provide a multidimensional framework for understanding the evolution of SARS-CoV-2 and guiding future antiviral strategies.</jats:p>",
"alternative-id": [
"ijms27020891"
],
"author": [
{
"ORCID": "https://orcid.org/0009-0007-7066-7424",
"affiliation": [
{
"name": "Doctoral School of Medicine and Pharmacy, George Emil Palade University of Medicine, Pharmacy, Science, and Technology of Târgu Mures, 540139 Targu Mures, Romania"
},
{
"name": "Department of Pathophysiology, George Emil Palade University of Medicine, Pharmacy, Science, and Technology of Târgu Mures, 540139 Targu Mures, Romania"
},
{
"name": "Pneumology Department, Clinical County Hospital of Mureș, 540142 Targu Mures, Romania"
}
],
"authenticated-orcid": false,
"family": "Muntean",
"given": "Sofia Teodora",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Department of Pathophysiology, George Emil Palade University of Medicine, Pharmacy, Science, and Technology of Târgu Mures, 540139 Targu Mures, Romania"
},
{
"name": "Pathology Department, Clinical County Hospital of Mureș, 540011 Targu Mures, Romania"
}
],
"family": "Cozac-Szoke",
"given": "Andreea-Raluca",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-9603-6256",
"affiliation": [
{
"name": "Department of Pathophysiology, George Emil Palade University of Medicine, Pharmacy, Science, and Technology of Târgu Mures, 540139 Targu Mures, Romania"
},
{
"name": "Pathology Department, Clinical County Hospital of Mureș, 540011 Targu Mures, Romania"
}
],
"authenticated-orcid": false,
"family": "Tinca",
"given": "Andreea Cătălina",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-1172-8262",
"affiliation": [
{
"name": "Department of Pathophysiology, George Emil Palade University of Medicine, Pharmacy, Science, and Technology of Târgu Mures, 540139 Targu Mures, Romania"
},
{
"name": "Clinical Laboratory Department, Clinical County Hospital of Mureș, 540072 Targu Mures, Romania"
}
],
"authenticated-orcid": false,
"family": "Kosovski",
"given": "Irina Bianca",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Plastic, Aesthetic and Reconstructive Microsurgery Department, County Emergency Clinical Hospital of Târgu Mureș, 540136 Targu Mures, Romania"
}
],
"family": "Vultur",
"given": "Silviu",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Plastic, Aesthetic and Reconstructive Microsurgery Department, County Emergency Clinical Hospital of Târgu Mureș, 540136 Targu Mures, Romania"
}
],
"family": "Vultur",
"given": "Mara",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Pathophysiology, George Emil Palade University of Medicine, Pharmacy, Science, and Technology of Târgu Mures, 540139 Targu Mures, Romania"
},
{
"name": "Pathology Department, Clinical County Hospital of Mureș, 540011 Targu Mures, Romania"
}
],
"family": "Cotoi",
"given": "Ovidiu Simion",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Genetics and Cellular and Molecular Biology, George Emil Palade University of Medicine, Pharmacy, Science, and Technology of Târgu Mures, 540139 Targu Mures, Romania"
}
],
"family": "Sin",
"given": "Anca Ileana",
"sequence": "additional"
}
],
"container-title": "International Journal of Molecular Sciences",
"container-title-short": "IJMS",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2026,
1,
15
]
],
"date-time": "2026-01-15T16:21:13Z",
"timestamp": 1768494073000
},
"deposited": {
"date-parts": [
[
2026,
1,
15
]
],
"date-time": "2026-01-15T16:25:30Z",
"timestamp": 1768494330000
},
"indexed": {
"date-parts": [
[
2026,
1,
16
]
],
"date-time": "2026-01-16T10:09:53Z",
"timestamp": 1768558193521,
"version": "3.49.0"
},
"is-referenced-by-count": 0,
"issue": "2",
"issued": {
"date-parts": [
[
2026,
1,
15
]
]
},
"journal-issue": {
"issue": "2",
"published-online": {
"date-parts": [
[
2026,
1
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2026,
1,
15
]
],
"date-time": "2026-01-15T00:00:00Z",
"timestamp": 1768435200000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/1422-0067/27/2/891/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "891",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2026,
1,
15
]
]
},
"published-online": {
"date-parts": [
[
2026,
1,
15
]
]
},
"publisher": "MDPI AG",
"reference": [
{
"DOI": "10.3390/jpm13081235",
"doi-asserted-by": "crossref",
"key": "ref_1",
"unstructured": "Budin, C.E., Nemeș, A.F., Râjnoveanu, R.-M., Nemeș, R.M., Rajnoveanu, A.G., Sabău, A.H., Cocuz, I.G., Mareș, R.G., Oniga, V.I., and Pătrîntașu, D.E. (2023). The Inflammatory Profile Correlates with COVID-19 Severity and Mortality in Cancer Patients. J. Pers. Med., 13."
},
{
"DOI": "10.20944/preprints202407.0487.v1",
"doi-asserted-by": "crossref",
"key": "ref_2",
"unstructured": "Budin, C.E., Cocuz, I.G., Enache, L.S., Rența, I.A., Cazacu, C., Pătrîntașu, D.E., Olteanu, M., Râjnoveanu, R.-M., Ianoși, E.S., and Râjnoveanu, A. (2024). Neutrophil-to-Lymphocyte and Platelet-to-Lymphocyte Ratio: Side by Side with Molecular Mutations in Patients with Non-Small Cell Lung Cancer—The INOLUNG Study. Cancers, 16."
},
{
"DOI": "10.1016/j.tiv.2024.105971",
"article-title": "Tannic Acid Modulates SARS-CoV-2 Pathogenesis by Curbing Key Host Receptors and Oxidative Stress",
"author": "Augustus",
"doi-asserted-by": "crossref",
"first-page": "105971",
"journal-title": "Toxicol. Vitr.",
"key": "ref_3",
"volume": "103",
"year": "2025"
},
{
"DOI": "10.1186/s12879-025-11414-3",
"doi-asserted-by": "crossref",
"key": "ref_4",
"unstructured": "Wang, S., Hao, Z., Ding, Y., Rong, H., Ning, S., Qiao, Q., Zhu, X., Wu, T., Zhao, K., and Zhu, L. (2025). Characterization of Private Mutations in the Spike Protein of SARS-CoV-2 Correlates with Viral Prevalence. BMC Infect. Dis., 25."
},
{
"DOI": "10.1038/s41598-023-40160-2",
"article-title": "Molecular Docking as a Tool for the Discovery of Molecular Targets of Nutraceuticals in Disease Management",
"author": "Agu",
"doi-asserted-by": "crossref",
"first-page": "13398",
"journal-title": "Sci. Rep.",
"key": "ref_5",
"volume": "13",
"year": "2023"
},
{
"DOI": "10.3390/ijerph192013624",
"doi-asserted-by": "crossref",
"key": "ref_6",
"unstructured": "Noureldeen, A.F.H., Aziz, S.W., Shouman, S.A., Mohamed, M.M., Attia, Y.M., Ramadan, R.M., and Elhady, M.M. (2022). Molecular Design, Spectroscopic, DFT, Pharmacological, and Molecular Docking Studies of Novel Ruthenium(III)-Schiff Base Complex: An Inhibitor of Progression in HepG2 Cells. Int. J. Environ. Res. Public Health, 19."
},
{
"DOI": "10.1128/mbio.00464-25",
"article-title": "Neutralization and Spike Stability of JN.1-Derived LB.1, KP.2.3, KP.3, and KP.3.1.1 Subvariants",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "e00464-25",
"journal-title": "mBio",
"key": "ref_7",
"volume": "16",
"year": "2025"
},
{
"DOI": "10.1016/j.celrep.2025.115941",
"article-title": "Structural and Functional Insights into the Evolution of SARS-CoV-2 KP.3.1.1 Spike Protein",
"author": "Feng",
"doi-asserted-by": "crossref",
"first-page": "115941",
"journal-title": "Cell Rep.",
"key": "ref_8",
"volume": "44",
"year": "2025"
},
{
"DOI": "10.1016/j.celrep.2025.115543",
"article-title": "Antibody Evasiveness of SARS-CoV-2 Subvariants KP.3.1.1 and XEC",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "115543",
"journal-title": "Cell Rep.",
"key": "ref_9",
"volume": "44",
"year": "2025"
},
{
"DOI": "10.1080/23744235.2024.2385500",
"article-title": "Features of the SARS-CoV-2 KP.3 Variant Mutations",
"author": "Branda",
"doi-asserted-by": "crossref",
"first-page": "894",
"journal-title": "Infect. Dis.",
"key": "ref_10",
"volume": "56",
"year": "2024"
},
{
"DOI": "10.1016/j.bios.2025.117311",
"article-title": "Electrochemical Pan-Variant Detection of SARS-CoV-2 through Host Cell Receptor-Mimicking Molecular Recognition",
"author": "Lee",
"doi-asserted-by": "crossref",
"first-page": "117311",
"journal-title": "Biosens. Bioelectron.",
"key": "ref_11",
"volume": "278",
"year": "2025"
},
{
"DOI": "10.1093/ve/veaf026",
"article-title": "Rapid Response to Fast Viral Evolution Using AlphaFold 3-Assisted Topological Deep Learning",
"author": "Wee",
"doi-asserted-by": "crossref",
"first-page": "veaf026",
"journal-title": "Virus Evol.",
"key": "ref_12",
"volume": "11",
"year": "2025"
},
{
"article-title": "Detection of SARS-CoV-2 Binding Receptors and Miscellaneous Targets as well as Mucosal Surface Area of the Human Lacrimal Drainage System",
"author": "Rau",
"first-page": "296",
"journal-title": "Sci. Direct",
"key": "ref_13",
"volume": "34",
"year": "2024"
},
{
"DOI": "10.3892/br.2025.2069",
"article-title": "Expression of ACE2 and TMPRSS2 and the Severity of COVID-19",
"author": "Peres",
"doi-asserted-by": "crossref",
"first-page": "1",
"journal-title": "Biomed. Rep.",
"key": "ref_14",
"volume": "23",
"year": "2025"
},
{
"DOI": "10.1128/jvi.01384-25",
"article-title": "Differential Expression of Viral Entry Protein Neuropilin 1 (NRP1) and Neuropilin 2 (NRP2) in Fatal COVID-19",
"author": "Dette",
"doi-asserted-by": "crossref",
"first-page": "e01384-25",
"journal-title": "J. Virol.",
"key": "ref_15",
"volume": "99",
"year": "2025"
},
{
"DOI": "10.3390/v17050691",
"doi-asserted-by": "crossref",
"key": "ref_16",
"unstructured": "Mahdi, M., Kiarie, I.W., Mótyán, J.A., Hoffka, G., Al-Muffti, A.S., Tóth, A., and Tőzsér, J. (2025). Receptor Binding for the Entry Mechanisms of SARS-CoV-2: Insights from the Original Strain and Emerging Variants. Viruses, 17."
},
{
"DOI": "10.1038/s41467-025-56902-x",
"article-title": "Design of Quinoline SARS-CoV-2 Papain-Like Protease Inhibitors as Oral Antiviral Drug Candidates",
"author": "Jadhav",
"doi-asserted-by": "crossref",
"first-page": "1604",
"journal-title": "Nat. Commun.",
"key": "ref_17",
"volume": "16",
"year": "2025"
},
{
"DOI": "10.1128/jvi.01823-24",
"article-title": "SARS-CoV-2 Entry and Fusion Are Independent of ACE2 Localization to Lipid Rafts",
"author": "Bolland",
"doi-asserted-by": "crossref",
"first-page": "e01823-24",
"journal-title": "J. Virol.",
"key": "ref_18",
"volume": "99",
"year": "2024"
},
{
"DOI": "10.15252/rc.2025229520",
"doi-asserted-by": "crossref",
"key": "ref_19",
"unstructured": "Zhang, J., Kennedy, A., Jorge, D.M.d.M., Xing, L., Reid, W., Bui, S., Joppich, J., Rose, M., Ercan, S., and Tang, Q. (2025). SARS-CoV-2 Remodels the Golgi Apparatus to Facilitate Viral Assembly and Secretion. PLoS Pathog., 21."
},
{
"DOI": "10.3390/biom15111530",
"doi-asserted-by": "crossref",
"key": "ref_20",
"unstructured": "Silva-Ríos, A.A., Mora-Ornelas, C.E., Flores-Medina, L.G., Muñoz-Valle, J.F., Díaz-Palomera, C.D., García-Chagollan, M., Vizcaíno-Quirarte, A.M., and Viera-Segura, O. (2025). Beyond Processing: Furin as a Central Hub in Viral Pathogenesis and Genetic Susceptibility. Biomolecules, 15."
},
{
"article-title": "Targeting Coronaviral nsp14: A Bifunctional Enzyme for RNA Capping and Proofreading",
"author": "Wang",
"first-page": "1",
"journal-title": "Biophys. Rep.",
"key": "ref_21",
"volume": "11",
"year": "2025"
},
{
"key": "ref_22",
"unstructured": "Altamirano, E.E.S., Huang, C., Tsai, Y., Tzeng, T.J., Teruel, N.F.B., Pandey, I., Oswal, N., Chen, Y.G., Chang, C., and Rajsbaum, R. (2025). Coronavirus nsp14 Drives Internal m7G Modification to Rewire Host Splicing and Promote Viral Replication. bioRxiv."
},
{
"DOI": "10.1038/s41586-024-08320-0",
"article-title": "Small-Molecule Inhibition of SARS-CoV-2 nsp14 RNA Cap Methyltransferase",
"author": "Meyer",
"doi-asserted-by": "crossref",
"first-page": "1178",
"journal-title": "Nature",
"key": "ref_23",
"volume": "637",
"year": "2025"
},
{
"DOI": "10.1038/s41467-025-64674-7",
"article-title": "SARS-CoV-2 nsp14 Inhibitor Exhibits Potent Antiviral Activity and Reverses nsp14-Driven Host Modulation",
"author": "Luo",
"doi-asserted-by": "crossref",
"first-page": "9671",
"journal-title": "Nat. Commun.",
"key": "ref_24",
"volume": "16",
"year": "2025"
},
{
"DOI": "10.1038/s41467-023-41159-z",
"article-title": "Comprehensive Proteomics and Meta-Analysis of COVID-19 Host Response",
"author": "Christ",
"doi-asserted-by": "crossref",
"first-page": "5921",
"journal-title": "Nat. Commun.",
"key": "ref_25",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.1093/ofid/ofae631.2094",
"article-title": "Proteomic and Transcriptomic Signatures of SARS-CoV-2-Associated Myocarditis",
"author": "Pollett",
"doi-asserted-by": "crossref",
"first-page": "ofae631.2094",
"journal-title": "Open Forum Infect. Dis.",
"key": "ref_26",
"volume": "12",
"year": "2025"
},
{
"DOI": "10.1111/liv.70172",
"article-title": "Analysis of Hyperexpanded T Cell Clones in SARS-CoV-2 Vaccine-Associated Liver Injury by Spatial Proteomics and Transcriptomics",
"author": "Uzun",
"doi-asserted-by": "crossref",
"first-page": "e70172",
"journal-title": "Liver Int.",
"key": "ref_27",
"volume": "45",
"year": "2025"
},
{
"DOI": "10.34067/KID.0000000727",
"article-title": "Peripheral Transcriptomics in Acute and Long-Term Kidney Dysfunction in SARS-CoV-2 Infection",
"author": "Jayaraman",
"doi-asserted-by": "crossref",
"first-page": "921",
"journal-title": "Kidney360",
"key": "ref_28",
"volume": "6",
"year": "2025"
},
{
"DOI": "10.3390/ijms26031047",
"doi-asserted-by": "crossref",
"key": "ref_29",
"unstructured": "Albalawi, W., Thomas, J., Mughal, F., Kotsiri, A., Roper, K.J., Alshehri, A., Kelbrick, M., Pollakis, G., and Paxton, W.A. (2025). SARS-CoV-2 S, M, and E Structural Glycoproteins Differentially Modulate Endoplasmic Reticulum Stress Responses. Int. J. Mol. Sci., 26."
},
{
"DOI": "10.3389/fcimb.2025.1564258",
"doi-asserted-by": "crossref",
"key": "ref_30",
"unstructured": "Liu, J., Wang, Y., Zheng, M., Du, J., Maarouf, M., and Chen, J.-L. (2025). Roles of circRNAs in Viral Pathogenesis. Front. Cell. Infect. Microbiol., 15."
},
{
"DOI": "10.3389/fmed.2023.1280951",
"doi-asserted-by": "crossref",
"key": "ref_31",
"unstructured": "Holmes, D., Colaneri, M., Palomba, E., and Gori, A. (2024). Exploring Post-Sepsis and Post-COVID-19 Syndromes: Crossovers from Pathophysiology to Therapeutic Approach. Front. Med., 10."
},
{
"DOI": "10.1038/s41392-021-00687-0",
"article-title": "Pattern Recognition Receptors in Health and Diseases",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "291",
"journal-title": "Signal Transduct. Target. Ther.",
"key": "ref_32",
"volume": "6",
"year": "2021"
},
{
"DOI": "10.1016/j.lfs.2023.121940",
"article-title": "Pattern-Recognition Receptors (PRRs) in SARS-CoV-2",
"author": "Isazadeh",
"doi-asserted-by": "crossref",
"first-page": "121940",
"journal-title": "Life Sci.",
"key": "ref_33",
"volume": "329",
"year": "2023"
},
{
"DOI": "10.3390/microorganisms13092176",
"doi-asserted-by": "crossref",
"key": "ref_34",
"unstructured": "Maddaloni, L., Bugani, G., Fracella, M., Bitossi, C., D’auria, A., Aloisi, F., Azri, A., Santinelli, L., Ben M’hadheb, M., and Pierangeli, A. (2025). Pattern Recognition Receptors (PRRs) Expression and Activation in COVID-19 and Long COVID. Microorganisms, 13."
},
{
"DOI": "10.1002/jmv.70145",
"article-title": "Induction of the Inflammasome by the SARS-CoV-2 Accessory Protein ORF9b",
"author": "Zodda",
"doi-asserted-by": "crossref",
"first-page": "e70145",
"journal-title": "J. Med Virol.",
"key": "ref_35",
"volume": "97",
"year": "2025"
},
{
"DOI": "10.4014/jmb.2507.07040",
"article-title": "Domain-Specific Impacts of Spike Protein Mutations on Infectivity and Antibody Escape in SARS-CoV-2 Omicron BA.1",
"author": "Kim",
"doi-asserted-by": "crossref",
"first-page": "e2507040",
"journal-title": "J. Microbiol. Biotechnol.",
"key": "ref_36",
"volume": "35",
"year": "2025"
},
{
"DOI": "10.1016/j.isci.2024.111557",
"article-title": "Ancestral SARS-CoV-2 Immune Imprinting Persists on RBD but Not NTD after Sequential Omicron Infections",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "111557",
"journal-title": "iScience",
"key": "ref_37",
"volume": "28",
"year": "2025"
},
{
"DOI": "10.1038/s41590-024-02051-0",
"article-title": "T Cell Immune Evasion by SARS-CoV-2 JN.1 Escapees Targeting Two Cytotoxic T Cell Epitope Hotspots",
"author": "Tian",
"doi-asserted-by": "crossref",
"first-page": "265",
"journal-title": "Nat. Immunol.",
"key": "ref_38",
"volume": "26",
"year": "2025"
},
{
"article-title": "The JN.1 Variant of COVID-19: Immune Evasion, Transmissibility, and Implications for Global Health",
"author": "Siddiqui",
"first-page": "20499361251314763",
"journal-title": "Ther. Adv. Infect. Dis.",
"key": "ref_39",
"volume": "12",
"year": "2025"
},
{
"DOI": "10.1016/j.redox.2025.103766",
"article-title": "Viral Mitochondriopathy in COVID-19",
"author": "Chen",
"doi-asserted-by": "crossref",
"first-page": "103766",
"journal-title": "Redox Biol.",
"key": "ref_40",
"volume": "85",
"year": "2025"
},
{
"DOI": "10.1016/j.antiviral.2025.106092",
"article-title": "Immune Evasion of Omicron Variants JN.1, KP.2, and KP.3 to Polyclonal and Monoclonal Antibodies",
"author": "Wu",
"doi-asserted-by": "crossref",
"first-page": "106092",
"journal-title": "Antiviral Res.",
"key": "ref_41",
"volume": "235",
"year": "2025"
},
{
"DOI": "10.3390/pathogens14010023",
"doi-asserted-by": "crossref",
"key": "ref_42",
"unstructured": "Mambelli, F., de Araujo, A.C.V.S.C., Farias, J.P., de Andrade, K.Q., Ferreira, L.C.S., Minoprio, P., Leite, L.C.C., and Oliveira, S.C. (2025). An Update on Anti-COVID-19 Vaccines and the Challenges to Protect Against New SARS-CoV-2 Variants. Pathogens, 14."
},
{
"DOI": "10.3390/vaccines12070794",
"doi-asserted-by": "crossref",
"key": "ref_43",
"unstructured": "Honda, T., Toyama, S., Matsumoto, Y., Sanada, T., Yasui, F., Koseki, A., Kono, R., Yamamoto, N., Kamishita, T., and Kodake, N. (2024). Intranasally Inoculated SARS-CoV-2 Spike Protein Combined with Mucoadhesive Polymer Induces Broad and Long-Lasting Immunity. Vaccines, 12."
},
{
"DOI": "10.1038/s41586-023-06951-3",
"article-title": "Mucosal Boosting Enhances Vaccine Protection against SARS-CoV-2 in Macaques",
"author": "McMahan",
"doi-asserted-by": "crossref",
"first-page": "385",
"journal-title": "Nature",
"key": "ref_44",
"volume": "626",
"year": "2024"
},
{
"DOI": "10.1038/s41467-025-55931-w",
"article-title": "Neutralizing Antibody Immune Correlates in COVAIL Trial Recipients of an mRNA Second COVID-19 Vaccine Boost",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "759",
"journal-title": "Nat. Commun.",
"key": "ref_45",
"volume": "16",
"year": "2025"
}
],
"reference-count": 45,
"references-count": 45,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.mdpi.com/1422-0067/27/2/891"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Molecular Aspects of Viral Pathogenesis in Emerging SARS-CoV-2 Variants: Evolving Mechanisms of Infection and Host Response",
"type": "journal-article",
"update-policy": "https://doi.org/10.3390/mdpi_crossmark_policy",
"volume": "27"
}