Mining Electronic Health Records for Drugs Associated With 28-day Mortality in COVID-19: Pharmacopoeia-wide Association Study (PharmWAS)
et al., JMIR Medical Informatics, doi:10.2196/35190, Mar 2022
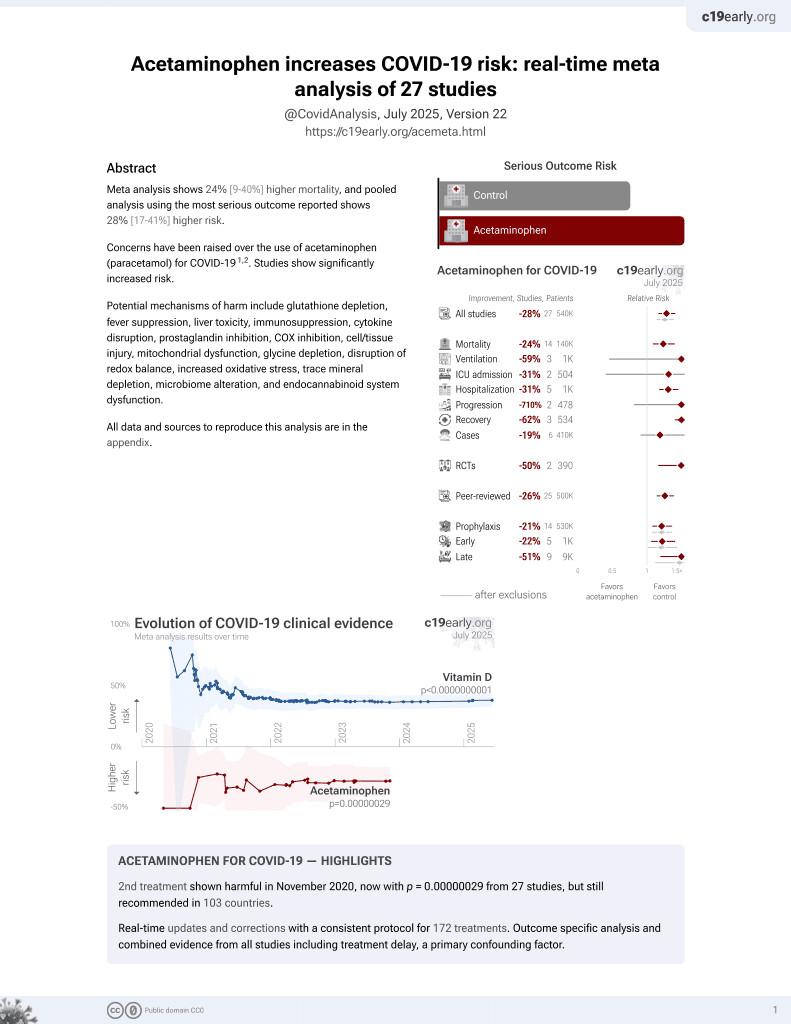
2nd treatment shown to increase risk in
November 2020, now with p = 0.00000029 from 27 studies, but still recommended in 103 countries.
6,400+ studies for
210+ treatments. c19early.org
|
Retrospective 5,783 hospitalized patients in France, showing higher mortality with paracetamol use, without statistical significance.
Paracetamol is also known as acetaminophen, Tylenol, Panadol, Calpol, Tempra, Calprofen, Doliprane, Efferalgan, Grippostad C, Dolo, Acamol, Fevadol, Crocin, and Perfalgan.
|
risk of death, 26.9% higher, RR 1.27, p = 0.10, odds ratio converted to relative risk, weighted and trimmed, day 28, control prevalance approximated with overall prevalence.
|
| Effect extraction follows pre-specified rules prioritizing more serious outcomes. Submit updates |
Lerner et al., 30 Mar 2022, retrospective, France, peer-reviewed, median age 69.2, 7 authors, study period 1 February, 2020 - 15 June, 2021.
Contact: antoine.neuraz@aphp.fr.
Mining Electronic Health Records for Drugs Associated With 28-day Mortality in COVID-19: Pharmacopoeia-wide Association Study (PharmWAS)
JMIR Medical Informatics, doi:10.2196/35190
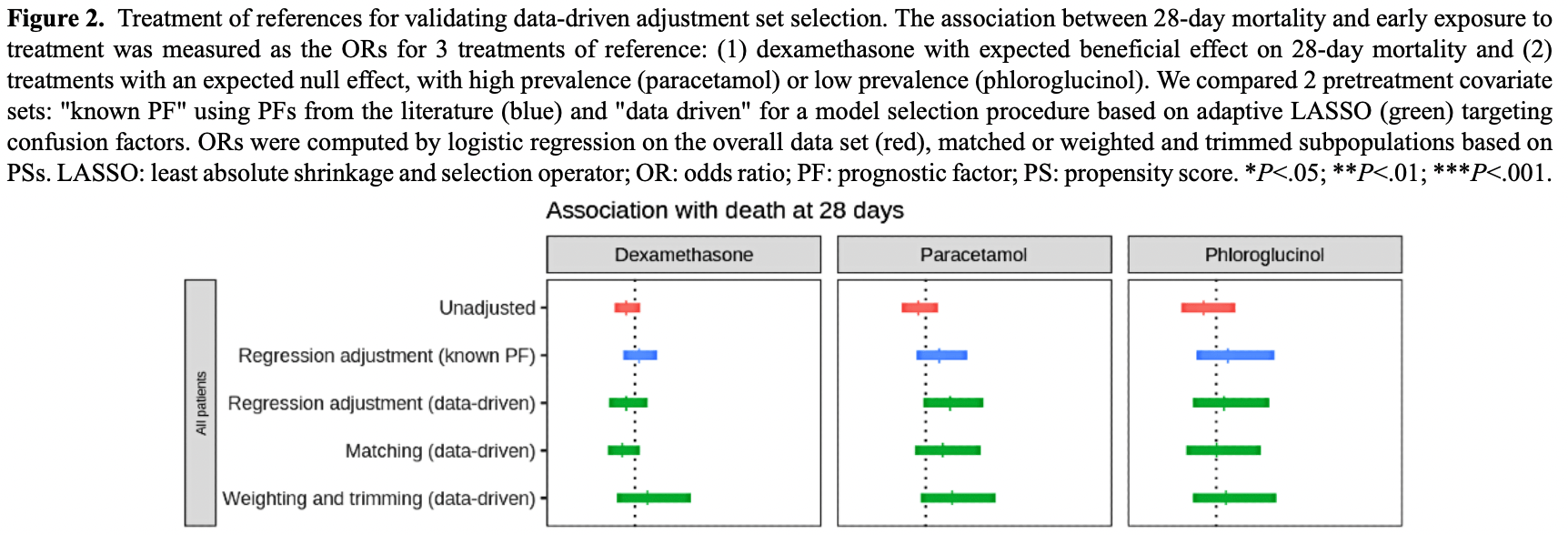
Background: Patients hospitalized for a given condition may be receiving other treatments for other contemporary conditions or comorbidities. The use of such observational clinical data for pharmacological hypothesis generation is appealing in the context of an emerging disease but particularly challenging due to the presence of drug indication bias. Objective: With this study, our main objective was the development and validation of a fully data-driven pipeline that would address this challenge. Our secondary objective was to generate pharmacological hypotheses in patients with COVID-19 and demonstrate the clinical relevance of the pipeline.
Methods: We developed a pharmacopeia-wide association study (PharmWAS) pipeline inspired from the PheWAS methodology, which systematically screens for associations between the whole pharmacopeia and a clinical phenotype. First, a fully data-driven procedure based on adaptive least absolute shrinkage and selection operator (LASSO) determined drug-specific adjustment sets. Second, we computed several measures of association, including robust methods based on propensity scores (PSs) to control indication bias. Finally, we applied the Benjamini and Hochberg procedure of the false discovery rate (FDR). We applied this method in a multicenter retrospective cohort study using electronic medical records from 16 university hospitals of the Greater Paris area. We included all adult patients between 18 and 95 years old hospitalized in conventional wards for COVID-19 between February 1, 2020, and June 15, 2021. We investigated the association between drug prescription within 48 hours from admission and 28-day mortality. We validated our data-driven pipeline against a knowledge-based pipeline on 3 treatments of reference, for which experts agreed on the expected association with mortality. We then demonstrated its clinical relevance by screening all drugs prescribed in more than 100 patients to generate pharmacological hypotheses. Results: A total of 5783 patients were included in the analysis. The median age at admission was 69.2 (IQR 56.7-81.1) years, and 3390 (58.62%) of the patients were male. The performance of our automated pipeline was comparable or better for controlling bias than the knowledge-based adjustment set for 3 reference drugs: dexamethasone, phloroglucinol, and paracetamol. After
Authors' Contributions IL contributed to conceptualization, data curation, formal analysis, methodology, software, and writing (original draft). ASL contributed to methodology and writing (reviewing and editing). LC contributed to conceptualization, formal analysis, and writing (reviewing and editing). BR and NG contributed to validation, and writing (reviewing and editing). AB contributed to project administration, resources, supervision, validation, and writing (reviewing and editing). AN contributed to conceptualization, data curation, formal analysis, software, resources, supervision, validation, writing (original draft), and writing (reviewing and editing).
Conflicts of Interest None declared.
Multimedia Appendix 1 Supplementary materials.
References
Austin, Grootendorst, Anderson, A comparison of the ability of different propensity score models to balance measured variables between treated and untreated subjects: a Monte Carlo study, Stat Med, doi:10.1002/sim.2580
Austin, Optimal caliper widths for propensity-score matching when estimating differences in means and differences in proportions in observational studies, Pharm Stat, doi:10.1002/pst.433
Bejan, Cahill, Staso, Choi, Peterson et al., DrugWAS: drug-wide association studies for COVID-19 drug repurposing, Clin Pharmacol Ther, doi:10.1002/cpt.2376
Benjamini, Hochberg, Controlling the false discovery rate: a practical and powerful approach to multiple testing, J R Stat Soc: Series B (Methodol, doi:10.1111/j.2517-6161.1995.tb02031.x
Boutron, Chaimani, Devane, Meerpohl, Tovey et al., Interventions for preventing and treating COVID-19: protocol for a living mapping of research and a living systematic review, Cochrane Database Syst Rev, doi:10.1002/14651858.CD013769
Brookhart, Schneeweiss, Rothman, Glynn, Avorn et al., Variable selection for propensity score models, Am J Epidemiol, doi:10.1093/aje/kwj149
Buuren, Groothuis-Oudshoorn, mice: Multivariate imputation by chained equations, J Stat Softw, doi:10.18637/jss.v045.i03
Caly, Druce, Catton, Jans, Wagstaff, The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro, Antiviral Res, doi:10.1016/j.antiviral.2020.104787
Castelnuovo, Bonaccio, Costanzo, Gialluisi, Antinori et al., Common cardiovascular risk factors and in-hospital mortality in 3,894 patients with COVID-19: survival analysis and machine learning-based findings from the multicentre Italian CORIST Study, Nutr Metab Cardiovasc Dis, doi:10.1016/j.numecd.2020.07.031
Choi, Carroll, Beck, Mosley, Roden et al., Evaluating statistical approaches to leverage large clinical datasets for uncovering therapeutic and adverse medication effects, Bioinformatics, doi:10.1093/bioinformatics/bty306
Chouchana, Beeker, Garcelon, Rance, Paris et al., Correction to: association of antihypertensive agents with the risk of in-hospital death in patients with Covid-19, Cardiovasc Drugs Ther, doi:10.1101/2020.11.23.20237362
Chouchana, Beeker, Garcelon, Rance, Paris et al., Universities/Inserm COVID-19 research collaboration‚ AP-HP Covid CDR Initiative‚"Entrepôt de Données de Santé" AP-HP Consortium, Cardiovasc Drugs Ther, doi:10.1007/s10557-021-07155-5
Cohen, Davidson, Gallus, Lassen, Prins et al., Efficacy and safety of fondaparinux for the prevention of venous thromboembolism in older acute medical patients: randomised placebo controlled trial, BMJ, doi:10.1136/bmj.38733.466748.7c
Core, R: A Language and Environment for Statistical Computing
Daniel, Serre, Orlova, Bréant, Paris et al., Initializing a hospital-wide data quality program. The AP-HP experience, Comput Methods Programs Biomed, doi:10.1016/j.cmpb.2018.10.016
Denny, Bastarache, Ritchie, Carroll, Zink et al., Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data, Nat Biotechnol, doi:10.3410/f.718185859.793488460
Denny, Ritchie, Basford, Pulley, Bastarache et al., PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations, Bioinformatics, doi:10.1093/bioinformatics/btq126
Duarte, Pelorosso, Nicolosi, Salgado, Vetulli et al., Telmisartan for treatment of Covid-19 patients: an open multicenter randomized clinical trial, EClinicalMedicine, doi:10.1016/j.eclinm.2021.100962
Ekström, Bornefalk-Hermansson, Abernethy, Currow, Safety of benzodiazepines and opioids in very severe respiratory disease: national prospective study, BMJ, doi:10.1136/bmj.g445
Friedman, Hastie, Tibshirani, Regularization paths for generalized linear models via coordinate descent, J Stat Softw, doi:10.18637/jss.v033.i01
Gue, Tennyson, Gao, Ren, Kanji et al., Development of a novel risk score to predict mortality in patients admitted to hospital with COVID-19, Sci Rep, doi:10.1038/s41598-020-78505-w
Gérard, Romani, Fresse, Viard, Parassol et al., French Network of Pharmacovigilance Centers. "Off-label" use of hydroxychloroquine, azithromycin, lopinavir-ritonavir and chloroquine in COVID-19: a survey of cardiac adverse drug reactions by the French Network of Pharmacovigilance Centers, Therapie, doi:10.1016/j.therap.2020.05.002
Hellwig, A COVID-19 prophylaxis? Lower incidence associated with prophylactic administration of ivermectin, Int J Antimicrob Agents, doi:10.1016/j.ijantimicag.2020.106248
Ho, Imai, King, Stuart, MatchIt: nonparametric preprocessing for parametric causal inference, J Stat Softw, doi:10.18637/jss.v042.i08
Kohane, Aronow, Avillach, Beaulieu-Jones, Bellazzi et al., What every reader should know about studies using electronic health record data but may be afraid to ask, J Med Internet Res, doi:10.2196/22219
Lerner, Pharmwas, GitLab
Li, Raghunathan, Db, Large-sample significance levels from multiply imputed data using moment-based statistics and an F reference distribution, J Am Stat Assoc, doi:10.2307/2290525
Murray, Deruyter, Harrison, Opioids and benzodiazepines, Crit Care Clin, doi:10.1016/s0749-0704(18)30042-3
Neuraz, Chouchana, Malamut, Beller, Roche et al., Phenome-wide association studies on a quantitative trait: application to TPMT enzyme activity and thiopurine therapy in pharmacogenomics, PLoS Comput Biol, doi:10.1371/journal.pcbi.1003405
Neuraz, Lerner, Digan, Paris, Tsopra et al., Natural language processing for rapid response to emergent diseases: case study of calcium channel blockers and hypertension in the COVID-19 pandemic, J Med Internet Res, doi:10.2196/20773
Nopp, Moik, Jilma, Pabinger, Ay, Risk of venous thromboembolism in patients with COVID-19: a systematic review and meta-analysis, Res Pract Thromb Haemost, doi:10.1002/rth2.12439
Patel, Ioannidis, Assessment of vibration of effects due to model specification can demonstrate the instability of observational associations, J Clin Epidemiol, doi:10.1016/j.jclinepi.2015.05.029
Patel, Ji, Sundquist, Ioannidis, Sundquist, Systematic assessment of pharmaceutical prescriptions in association with cancer risk: a method to conduct a population-wide medication-wide longitudinal study, Sci Rep, doi:10.1038/srep31308
Rothman, Lanes, Robins, Causal inference, Epidemiology, doi:10.1097/00001648-199311000-00013
Ryan, Madigan, Stang, Schuemie, Hripcsak, Medication-wide association studies, CPT Pharmacomet Syst Pharmacol, doi:10.1038/psp.2013.52
Samama, Cohen, Darmon, Desjardins, Eldor et al., A comparison of enoxaparin with placebo for the prevention of venous thromboembolism in acutely ill medical patients, N Engl J Med, doi:10.1056/nejm199909093411103
Siuka, Pfeifer, Pinter, Vitamin D supplementation during the COVID-19 pandemic, Mayo Clin Proc, doi:10.1016/j.mayocp.2020.05.036
Stuart, Lee, Leacy, Prognostic score-based balance measures can be a useful diagnostic for propensity score methods in comparative effectiveness research, J Clin Epidemiol, doi:10.1016/j.jclinepi.2013.01.013
Stürmer, Rothman, Avorn, Glynn, Treatment effects in the presence of unmeasured confounding: dealing with observations in the tails of the propensity score distribution; a simulation study, Am J Epidemiol, doi:10.1093/aje/kwq198
Stürmer, Webster-Clark, Lund, Ellis, Lunt, Propensity score weighting and trimming strategies for reducing variance and bias of treatment effect estimates: a simulation study, Am J Epidemiol, doi:10.1093/aje/kwab041
Van Buuren, Flexible Imputation of Missing Data, Second Edition
Walker, Patrick, Lauer, Hornbrook, Marin et al., A tool for assessing the feasibility of comparative effectiveness research, CER, doi:10.2147/cer.s40357
Wang, Cao, Zhang, Yang, Liu et al., Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro, Cell Res, doi:10.1038/s41422-020-0282-0
Williamson, Walker, Bhaskaran, Bacon, Bates et al., Factors associated with COVID-19-related death using OpenSAFELY, Nature, doi:10.1038/s41586-020-2521-4
Witte, Didelez, Covariate selection strategies for causal inference: classification and comparison, Biom J, doi:10.1002/bimj.201700294
Wong, Mackenna, Morton, Schultze, Walker et al., Use of non-steroidal anti-inflammatory drugs and risk of death from COVID-19: an OpenSAFELY cohort analysis based on two cohorts, Ann Rheum Dis, doi:10.1136/annrheumdis-2020-219517
Zheng, Peng, Xu, Zhao, Liu et al., None, J Infect, doi:10.1016/j.jinf.2020.04.021
Zhou, Tong, Li, Thomas, PSweight: An R Package for Propensity Score Weighting Analysis
Zou, The adaptive Lasso and its oracle properties, J Am Stat Assoc, doi:10.1198/016214506000000735
DOI record:
{
"DOI": "10.2196/35190",
"ISSN": [
"2291-9694"
],
"URL": "http://dx.doi.org/10.2196/35190",
"abstract": "<jats:sec>\n <jats:title>Background</jats:title>\n <jats:p>Patients hospitalized for a given condition may be receiving other treatments for other contemporary conditions or comorbidities. The use of such observational clinical data for pharmacological hypothesis generation is appealing in the context of an emerging disease but particularly challenging due to the presence of drug indication bias.</jats:p>\n </jats:sec>\n <jats:sec>\n <jats:title>Objective</jats:title>\n <jats:p>With this study, our main objective was the development and validation of a fully data-driven pipeline that would address this challenge. Our secondary objective was to generate pharmacological hypotheses in patients with COVID-19 and demonstrate the clinical relevance of the pipeline.</jats:p>\n </jats:sec>\n <jats:sec>\n <jats:title>Methods</jats:title>\n <jats:p>We developed a pharmacopeia-wide association study (PharmWAS) pipeline inspired from the PheWAS methodology, which systematically screens for associations between the whole pharmacopeia and a clinical phenotype. First, a fully data-driven procedure based on adaptive least absolute shrinkage and selection operator (LASSO) determined drug-specific adjustment sets. Second, we computed several measures of association, including robust methods based on propensity scores (PSs) to control indication bias. Finally, we applied the Benjamini and Hochberg procedure of the false discovery rate (FDR). We applied this method in a multicenter retrospective cohort study using electronic medical records from 16 university hospitals of the Greater Paris area. We included all adult patients between 18 and 95 years old hospitalized in conventional wards for COVID-19 between February 1, 2020, and June 15, 2021. We investigated the association between drug prescription within 48 hours from admission and 28-day mortality. We validated our data-driven pipeline against a knowledge-based pipeline on 3 treatments of reference, for which experts agreed on the expected association with mortality. We then demonstrated its clinical relevance by screening all drugs prescribed in more than 100 patients to generate pharmacological hypotheses.</jats:p>\n </jats:sec>\n <jats:sec>\n <jats:title>Results</jats:title>\n <jats:p>A total of 5783 patients were included in the analysis. The median age at admission was 69.2 (IQR 56.7-81.1) years, and 3390 (58.62%) of the patients were male. The performance of our automated pipeline was comparable or better for controlling bias than the knowledge-based adjustment set for 3 reference drugs: dexamethasone, phloroglucinol, and paracetamol. After correction for multiple testing, 4 drugs were associated with increased in-hospital mortality. Among these, diazepam and tramadol were the only ones not discarded by automated diagnostics, with adjusted odds ratios of 2.51 (95% CI 1.52-4.16, Q=.01) and 1.94 (95% CI 1.32-2.85, Q=.02), respectively.</jats:p>\n </jats:sec>\n <jats:sec>\n <jats:title>Conclusions</jats:title>\n <jats:p>Our innovative approach proved useful in generating pharmacological hypotheses in an outbreak setting, without requiring a priori knowledge of the disease. Our systematic analysis of early prescribed treatments from patients hospitalized for COVID-19 showed that diazepam and tramadol are associated with increased 28-day mortality. Whether these drugs could worsen COVID-19 needs to be further assessed.</jats:p>\n </jats:sec>",
"author": [
{
"ORCID": "http://orcid.org/0000-0002-5466-1707",
"affiliation": [],
"authenticated-orcid": false,
"family": "Lerner",
"given": "Ivan",
"sequence": "first"
},
{
"ORCID": "http://orcid.org/0000-0002-2642-2514",
"affiliation": [],
"authenticated-orcid": false,
"family": "Serret-Larmande",
"given": "Arnaud",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-4417-1197",
"affiliation": [],
"authenticated-orcid": false,
"family": "Rance",
"given": "Bastien",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-3326-2811",
"affiliation": [],
"authenticated-orcid": false,
"family": "Garcelon",
"given": "Nicolas",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-6855-4366",
"affiliation": [],
"authenticated-orcid": false,
"family": "Burgun",
"given": "Anita",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-9626-3571",
"affiliation": [],
"authenticated-orcid": false,
"family": "Chouchana",
"given": "Laurent",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-7142-6728",
"affiliation": [],
"authenticated-orcid": false,
"family": "Neuraz",
"given": "Antoine",
"sequence": "additional"
}
],
"container-title": "JMIR Medical Informatics",
"container-title-short": "JMIR Med Inform",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2022,
3,
11
]
],
"date-time": "2022-03-11T18:02:31Z",
"timestamp": 1647021751000
},
"deposited": {
"date-parts": [
[
2022,
4,
12
]
],
"date-time": "2022-04-12T18:21:52Z",
"timestamp": 1649787712000
},
"indexed": {
"date-parts": [
[
2022,
4,
12
]
],
"date-time": "2022-04-12T18:41:20Z",
"timestamp": 1649788880603
},
"is-referenced-by-count": 1,
"issue": "3",
"issued": {
"date-parts": [
[
2022,
3,
30
]
]
},
"journal-issue": {
"issue": "3",
"published-online": {
"date-parts": [
[
2022
]
]
}
},
"language": "en",
"member": "1010",
"original-title": [],
"page": "e35190",
"prefix": "10.2196",
"published": {
"date-parts": [
[
2022,
3,
30
]
]
},
"published-online": {
"date-parts": [
[
2022,
3,
30
]
]
},
"publisher": "JMIR Publications Inc.",
"reference": [
{
"key": "ref1",
"unstructured": "COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU)2021-08-27https://coronavirus.jhu.edu/map.html"
},
{
"DOI": "10.1002/14651858.CD013769",
"doi-asserted-by": "publisher",
"key": "ref2"
},
{
"DOI": "10.1056/nejmoa2021436",
"doi-asserted-by": "publisher",
"key": "ref3"
},
{
"DOI": "10.1016/j.therap.2020.05.002",
"doi-asserted-by": "publisher",
"key": "ref4"
},
{
"DOI": "10.1136/annrheumdis-2020-219517",
"doi-asserted-by": "publisher",
"key": "ref5"
},
{
"DOI": "10.1007/s10557-021-07155-5",
"doi-asserted-by": "publisher",
"key": "ref6"
},
{
"DOI": "10.1016/j.ijantimicag.2020.106248",
"doi-asserted-by": "publisher",
"key": "ref7"
},
{
"DOI": "10.1016/j.antiviral.2020.104787",
"doi-asserted-by": "publisher",
"key": "ref8"
},
{
"DOI": "10.1038/s41422-020-0282-0",
"doi-asserted-by": "publisher",
"key": "ref9"
},
{
"DOI": "10.1016/j.mayocp.2020.05.036",
"doi-asserted-by": "publisher",
"key": "ref10"
},
{
"DOI": "10.1002/rth2.12439",
"doi-asserted-by": "publisher",
"key": "ref11"
},
{
"DOI": "10.1136/bmj.38733.466748.7c",
"doi-asserted-by": "publisher",
"key": "ref12"
},
{
"DOI": "10.1056/nejm199909093411103",
"doi-asserted-by": "publisher",
"key": "ref13"
},
{
"DOI": "10.1093/bioinformatics/btq126",
"doi-asserted-by": "publisher",
"key": "ref14"
},
{
"DOI": "10.3410/f.718185859.793488460",
"doi-asserted-by": "publisher",
"key": "ref15"
},
{
"DOI": "10.1371/journal.pcbi.1003405",
"doi-asserted-by": "publisher",
"key": "ref16"
},
{
"DOI": "10.1038/psp.2013.52",
"doi-asserted-by": "publisher",
"key": "ref17"
},
{
"DOI": "10.1038/srep31308",
"doi-asserted-by": "publisher",
"key": "ref18"
},
{
"DOI": "10.1093/bioinformatics/bty306",
"doi-asserted-by": "publisher",
"key": "ref19"
},
{
"DOI": "10.1002/cpt.2376",
"doi-asserted-by": "publisher",
"key": "ref20"
},
{
"DOI": "10.1002/sim.2580",
"doi-asserted-by": "publisher",
"key": "ref21"
},
{
"DOI": "10.1093/aje/kwj149",
"doi-asserted-by": "publisher",
"key": "ref22"
},
{
"DOI": "10.1002/bimj.201700294",
"doi-asserted-by": "publisher",
"key": "ref23"
},
{
"DOI": "10.1093/aje/kwq198",
"doi-asserted-by": "publisher",
"key": "ref24"
},
{
"DOI": "10.1093/aje/kwab041",
"doi-asserted-by": "publisher",
"key": "ref25"
},
{
"DOI": "10.1097/00001648-199311000-00013",
"doi-asserted-by": "publisher",
"key": "ref26"
},
{
"DOI": "10.2147/cer.s40357",
"doi-asserted-by": "publisher",
"key": "ref27"
},
{
"DOI": "10.1016/j.cmpb.2018.10.016",
"doi-asserted-by": "publisher",
"key": "ref28"
},
{
"key": "ref29",
"unstructured": "Anatomical Therapeutic Chemical (ATC) Classification2021-05-02https://www.who.int/tools/atc-ddd-toolkit/atc-classification"
},
{
"DOI": "10.1198/016214506000000735",
"doi-asserted-by": "publisher",
"key": "ref30"
},
{
"DOI": "10.1002/pst.433",
"doi-asserted-by": "publisher",
"key": "ref31"
},
{
"DOI": "10.1016/j.jclinepi.2013.01.013",
"doi-asserted-by": "publisher",
"key": "ref32"
},
{
"DOI": "10.1111/j.2517-6161.1995.tb02031.x",
"doi-asserted-by": "publisher",
"key": "ref33"
},
{
"DOI": "10.1016/j.jinf.2020.04.021",
"doi-asserted-by": "publisher",
"key": "ref34"
},
{
"DOI": "10.1038/s41586-020-2521-4",
"doi-asserted-by": "publisher",
"key": "ref35"
},
{
"DOI": "10.1038/s41598-020-78505-w",
"doi-asserted-by": "publisher",
"key": "ref36"
},
{
"DOI": "10.1016/j.numecd.2020.07.031",
"doi-asserted-by": "publisher",
"key": "ref37"
},
{
"DOI": "10.1201/9780429492259",
"author": "van Buuren, S",
"doi-asserted-by": "crossref",
"journal-title": "Flexible Imputation of Missing Data, Second Edition",
"key": "ref38",
"year": "2018"
},
{
"DOI": "10.2307/2290525",
"doi-asserted-by": "publisher",
"key": "ref39"
},
{
"journal-title": "R: A Language and Environment for Statistical Computing",
"key": "ref40",
"year": "2018"
},
{
"DOI": "10.18637/jss.v045.i03",
"doi-asserted-by": "publisher",
"key": "ref41"
},
{
"DOI": "10.18637/jss.v033.i01",
"doi-asserted-by": "publisher",
"key": "ref42"
},
{
"DOI": "10.18637/jss.v042.i08",
"doi-asserted-by": "publisher",
"key": "ref43"
},
{
"key": "ref44",
"unstructured": "ZhouTTongGLiFThomasLPSweight: An R Package for Propensity Score Weighting Analysis20202021-08-27http://arxiv.org/abs/2010.08893"
},
{
"key": "ref45",
"unstructured": "pharmwas2021-11-29https://gitlab.com/lerner.ivan/pharmwas"
},
{
"DOI": "10.1136/bmj.g445",
"doi-asserted-by": "publisher",
"key": "ref46"
},
{
"DOI": "10.1016/s0749-0704(18)30042-3",
"doi-asserted-by": "publisher",
"key": "ref47"
},
{
"DOI": "10.1016/j.eclinm.2021.100962",
"doi-asserted-by": "publisher",
"key": "ref48"
},
{
"DOI": "10.1101/2020.11.23.20237362",
"doi-asserted-by": "publisher",
"key": "ref49"
},
{
"DOI": "10.2196/22219",
"doi-asserted-by": "publisher",
"key": "ref50"
},
{
"DOI": "10.1016/j.jclinepi.2015.05.029",
"doi-asserted-by": "publisher",
"key": "ref51"
},
{
"DOI": "10.2196/20773",
"doi-asserted-by": "publisher",
"key": "ref52"
},
{
"key": "ref53",
"unstructured": "LernerIpharmwasGitLab2021-11-29https://gitlab.com/lerner.ivan/pharmwas"
}
],
"reference-count": 53,
"references-count": 53,
"relation": {},
"resource": {
"primary": {
"URL": "https://medinform.jmir.org/2022/3/e35190"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"Health Information Management",
"Health Informatics"
],
"subtitle": [],
"title": "Mining Electronic Health Records for Drugs Associated With 28-day Mortality in COVID-19: Pharmacopoeia-wide Association Study (PharmWAS)",
"type": "journal-article",
"volume": "10"
}