Temporal TCR dynamics and epitope diversity mark recovery in severe COVID-19 patients
et al., Frontiers in Immunology, doi:10.3389/fimmu.2025.1582949, CTRI/2020/05/025209, Jul 2025
Longitudinal study of 36 ICU-admitted severe COVID-19 patients showing dynamic T-cell receptor (TCR) repertoire changes during recovery.
Khare et al., 10 Jul 2025, India, peer-reviewed, 6 authors, study period 31 May, 2020 - 12 October, 2020, trial CTRI/2020/05/025209.
Contact: rajeshp@igib.in, rajesh.p@igib.res.in.
Temporal TCR dynamics and epitope diversity mark recovery in severe COVID-19 patients
Frontiers in Immunology, doi:10.3389/fimmu.2025.1582949
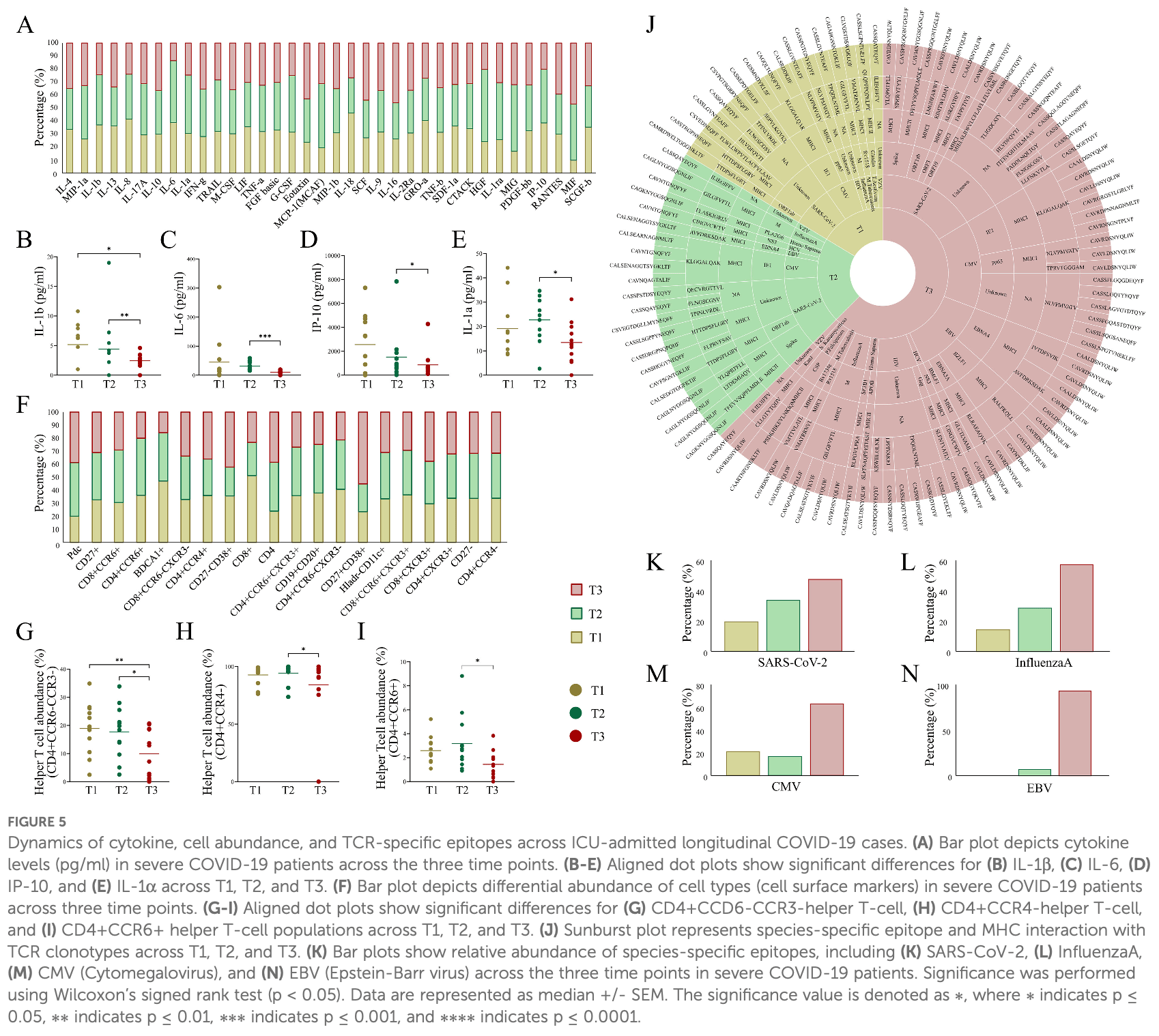
Introduction: Severe COVID-19 is characterized by immune dysregulation, with T cells playing a central role in disease progression and recovery. However, the longitudinal dynamics of the T cell receptor (TCR) repertoire during the course of severe illness remain unclear. Methods: To investigate temporal changes in adaptive immunity, we analyzed peripheral blood samples from the ICU-admitted severe COVID-19 patients (n = 36) collected at three time points: Day 1 (T1), Day 4 (T2), and Day 7 (T3). Bulk RNAsequencing was performed to extract TCR repertoires, and cytokine profiles were assessed in parallel. TCR clonotypes were annotated using VDJdb and TCRex to infer potential epitope specificities. Results: By T3, we observed a 2.3-fold expansion in TCR clonotypes along with increased TCR-b (TRB) chain usage, indicating the emergence of a broad polyclonal T cell response. In contrast, TCR-g (TRG) chain prevalence declined. Pro-inflammatory cytokines, including IL-1b and IL-6, were reduced over time, marking a shift toward immune resolution. Changes in CDR3 motifs and preferential TRBV gene segment usage were detected, suggesting repertoire adaptation. Additionally, annotated TCR clonotypes at T3 mapped to SARS-CoV-2 and other pathogen-associated epitopes (e.g., CMV, Plasmodium), reflecting possible cross-reactivity or memory T cell recruitment. Discussion: These findings suggest a coordinated transition from immune dysfunction to recovery in severe COVID-19, marked by expanding TCR diversity, reduced inflammation, and predicted broadening of antigen recognition. The integrated analysis of TCR repertoire dynamics and cytokine profiles provides insights into the adaptive immune mechanisms underlying viral clearance and immune stabilization.
Ethics statement The studies involving humans were approved by Institutional Review Boards of CSIR-Indian Institute of Chemical Biology, Kolkata. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
Conflict of interest The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher's note All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1582949/ full#supplementary-material
References
Alhamlan, Aa, Pro-inflammatory and antiinflammatory interleukins in infectious diseases: A comprehensive review, Trop Med Infect Dis, doi:10.3390/tropicalmed9010013
Attaf, Legut, Cole, Sewell, The T cell antigen receptor: the Swiss army knife of the immune system, Clin Exp Immunol, doi:10.1111/cei.12622
Blanco, Garcıá, Garcıá-Mariscal, Hernańdez-Munain, Control of V (D)J recombination through transcriptional elongation and changes in locus chromatin structure and nuclear organization, Genet Res Int, doi:10.4061/2011/970968
Boechat, Chora, Morais, Delgado, The immune response to SARS-CoV-2 and COVID-19 immunopathology -Current perspectives, Pulmonology, doi:10.1016/j.pulmoe.2021.03.008
Bolger, Lohse, Usadel, Trimmomatic: A flexible trimmer for Illumina sequence data, Bioinformatics, doi:10.1093/bioinformatics/btu170
Bolotin, Poslavsky, Mitrophanov, Shugay, Mamedov et al., MiXCR: software for comprehensive adaptive immunity profiling, Nat Methods, doi:10.1038/nmeth.3364
Burrows, Silins, Moss, Khanna, Misko et al., T cell receptor repertoire for a viral epitope in humans is diversified by tolerance to a background major histocompatibility complex antigen, J Exp Med, doi:10.1084/jem.182.6.1703
Carter, Notter, COVID-19 Disease: a critical care perspective, Clinics Integrated Care, doi:10.1016/j.intcar.2020.100003
Chen, Yang, Ko, Sun, Gao et al., Sequence and structural analyses reveal distinct and highly diverse human CD8+ TCR repertoires to immunodominant viral antigens, Cell Rep, doi:10.1016/j.celrep.2017.03.072
Chi, Pepper, Thomas, Principles and therapeutic applications of adaptive immunity, Cell, doi:10.1016/j.cell.2024.03.037
Clark, Gil, Thapa, Aslan, Ghersi et al., Cross-reactivity influences changes in human influenza A virus and Epstein Barr virus specific CD8 memory T cell receptor alpha and beta repertoires between young and old, Front Immunol, doi:10.3389/fimmu.2022.1011935
Connors, Kovacs, Krevat, Gea-Banacloche, Sneller et al., HIV infection induces changes in CD4+ T-cell phenotype and depletions within the CD4+ T-cell repertoire that are not immediately restored by antiviral or immunebased therapies, Nat Med, doi:10.1038/nm0597-533
Conti, Ronconi, Caraffa, Gallenga, Ross et al., Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies, J Biol Regul Homeost Agents, doi:10.23812/CONTI-E
Crooks, Hon, Chandonia, Brenner, WebLogo: a sequence logo generator, Genome Res, doi:10.1101/gr.849004
Dhawan, Rabaan, Fawarah, Almuthree, Alsubki et al., Updated Insights into the T Cell-Mediated Immune Response against SARS-CoV-2: A Step towards Efficient and Reliable Vaccines, Vaccines, doi:10.3390/vaccines11010101
Dutta, Zhao, Love, New insights into TCR b-selection, Trends Immunol, doi:10.1016/j.it.2021.06.005
Gielis, Moris, Bittremieux, Neuter, Ogunjimi et al., Detection of enriched T cell epitope specificity in full T cell receptor sequence repertoires, Front Immunol, doi:10.3389/fimmu.2019.02820
Gittelman, Lavezzo, Snyder, Zahid, Carty et al., Longitudinal analysis of T cell receptor repertoires reveals shared patterns of antigenspecific response to SARS-CoV-2 infection, JCI Insight, doi:10.1172/jci.insight.151849
Gras, Chen, Miles, Liu, Bell et al., Allelic polymorphism in the T cell receptor and its impact on immune responses, J Exp Med, doi:10.1084/jem.20100603
Grifoni, Sidney, Vita, Peters, Crotty et al., SARS-CoV-2 human T cell epitopes: Adaptive immune response against COVID-19, Cell Host Microbe, doi:10.1016/j.chom.2021.05.010
Gruta, Gras, Daley, Thomas, Rossjohn, Understanding the drivers of MHC restriction of T cell receptors, Nat Rev Immunol, doi:10.1038/s41577-018-0007-5
Gutierrez, Beckford, Alachkar, Deciphering the TCR repertoire to solve the COVID-19 mystery, Trends Pharmacol Sci, doi:10.1016/j.tips.2020.06.001
Hou, Wang, Chen, Mo, Wang, T-cell receptor repertoires as potential diagnostic markers for patients with COVID-19, Int J Infect Dis, doi:10.1016/j.ijid.2021.10.033
Hu, Guo, Zhou, Shi, Characteristics of SARS-CoV-2 and COVID-19, Nat Rev Microbiol, doi:10.1038/s41579-020-00459-7
Kervevan, Chakrabarti, Role of CD4+ T cells in the control of viral infections: recent advances and open questions, Int J Mol Sci, doi:10.3390/ijms22020523
Khare, Yadav, Tarai, Budhiraja, Pandey, TCR repertoire dynamics and their responses underscores dengue severity, iScience, doi:10.1016/j.isci.2024.110983
Lamers, Haagmans, SARS-CoV-2 pathogenesis, Nat Rev Microbiol, doi:10.1038/s41579-022-00713-0
Leary, Scott, Gupta, Waite, Skokos et al., Designing meaningful continuous representations of T cell receptor sequences with deep generative models, Nat Commun, doi:10.1038/s41467-024-48198-0
Lu, Van Laethem, Bhattacharya, Craveiro, Saba et al., Molecular constraints on CDR3 for thymic selection of MHC-restricted TCRs from a random preselection repertoire, Nat Commun, doi:10.1038/s41467-019-08906-7
Magazine, Zhang, Bungwon, Mcgee, Wu et al., Immune epitopes of SARS-CoV-2 spike protein and considerations for universal vaccine development, Immunohorizons, doi:10.4049/immunohorizons.2400003
Mauri, Elli, Caviglia, Uboldi, Azzi, Rawgraphs: A visualisation platform to create open outputs, doi:10.1145/3125571.3125585
Mazzotti, Gaimari, Bravaccini, Maltoni, Cerchione et al., Tcell receptor repertoire sequencing and its applications: focus on infectious diseases and cancer, Int J Mol Sci, doi:10.3390/ijms23158590
Netea, Rovina, Akinosoglou, Antoniadou, Antonakos, Complex immune dysregulation in COVID-19 patients with severe respiratory failure, Cell Host Microbe, doi:10.1016/j.chom.2020.04.009
Nolan, Vignali, Klinger, Dines, Kaplan et al., A large-scale database of T-cell receptor beta (TCRb) sequences and binding associations from natural and synthetic exposure to SARS-CoV-2, Res Sq, doi:10.21203/rs.3.rs-51964/v1
Petrova, Ferrante, Gorski, Cross-reactivity of T cells and its role in the immune system, Crit Rev Immunol, doi:10.1615/CritRevImmunol.v32.i4.50
Pilkinton, Mcdonnell, Barnett, Chopra, Gangula et al., In chronic infection, HIV gag-specific CD4+ T cell receptor diversity is higher than CD8+ T cell receptor diversity and is associated with less HIV quasispecies diversity, J Virol, doi:10.1128/JVI.02380-20
Qi, Liu, Cheng, Glanville, Zhang et al., Diversity and clonal selection in the human T-cell repertoire, Proc Natl Acad Sci U S A, doi:10.1073/pnas.1409155111
Ray, Paul, Bandopadhyay, 'rozario, Sarif et al., A phase 2 single center open label randomised control trial for convalescent plasma therapy in patients with severe COVID-19, Nat Commun, doi:10.1038/s41467-022-28064-7
Raychaudhuri, Bandopadhyay, 'rozario, Sarif, Ray et al., Clinical trial subgroup analyses to investigate clinical and immunological outcomes of convalescent plasma therapy in severe COVID-19, Mayo Clin Proc Innov Qual Outcomes, doi:10.1016/j.mayocpiqo.2022.09.001
Ren, Li, He, Hu, Teng, Longitudinal change trend of the TCR repertoire reveals the immune response intensity of the inactivated COVID-19 vaccine, Mol Immunol, doi:10.1016/j.molimm.2023.09.006
Richman, Aggen, Dossett, Donermeyer, Allen et al., Structural features of T cell receptor variable regions that enhance domain stability and enable expression as single-chain ValphaVbeta fragments, Mol Immunol, doi:10.1016/j.molimm.2008.09.021
Schultheiß, Paschold, Simnica, Mohme, Willscher et al., Next-generation sequencing of T and B cell receptor repertoires from COVID-19 patients showed signatures associated with severity of disease, Immunity, doi:10.1016/j.immuni.2020.06.024
Shi, Strasser, Green, Latz, Mantovani et al., Legacy of the discovery of the T-cell receptor: 40 years of shaping basic immunology and translational work to develop novel therapies, Cell Mol Immunol, doi:10.1038/s41423-024-01168-4
Shomuradova, Vagida, Sheetikov, Zornikova, Kiryukhin et al., SARS-CoV-2 epitopes are recognized by a public and diverse repertoire of human T cell receptors, Immunity, doi:10.1016/j.immuni.2020.11.004
Shugay, Bagaev, Turchaninova, Bolotin, Britanova et al., VDJtools: unifying post-analysis of T cell receptor repertoires, PLoS Comput Biol, doi:10.1371/journal.pcbi.1004503
Shugay, Bagaev, Zvyagin, Vroomans, Crawford et al., VDJdb: a curated database of T-cell receptor sequences with known antigen specificity, Nucleic Acids Res, doi:10.1093/nar/gkx760
Tang, Chen, Huang, Zhang, Zeng et al., SRplot: A free online platform for data visualization and graphing, PloS One, doi:10.1371/journal.pone.0294236
Tarke, Ramezani-Rad, Pereira Neto, Lee, Silva-Moraes et al., SARS-CoV-2 breakthrough infections enhance T cell response magnitude, breadth, and epitope repertoire, Cell Rep Med, doi:10.1016/j.xcrm.2024.101583
Tickotsky, Sagiv, Prilusky, Shifrut, Friedman, McPAS-TCR: a manually curated catalogue of pathology-associated T cell receptor sequences, Bioinformatics, doi:10.1093/bioinformatics/btx286
Tippalagama, Chihab, Kearns, Lewis, Panda et al., Antigen-specificity measurements are the key to understanding T cell responses, Front Immunol, doi:10.3389/fimmu.2023.1127470
Turner, Gruta, Kedzierska, Thomas, Doherty, Functional implications of T cell receptor diversity, Curr Opin Immunol, doi:10.1016/j.coi.2009.05.004
Wang, Dash, Mccullers, Doherty, Thomas, T cell receptor ab diversity inversely correlates with pathogen-specific antibody levels in human cytomegalovirus infection, Sci Transl Med, doi:10.1126/scitranslmed.3003647
Wang, Zhou, Luo, Xu, Xu, Comprehensive analysis of TCR repertoire in COVID-19 using single cell sequencing, Genomics, doi:10.1016/j.ygeno.2020.12.036
Wo, Zhang, Li, Sun, Zhang et al., The role of gamma-delta T cells in diseases of the central nervous system, Front Immunol, doi:10.3389/fimmu.2020.580304
Xu, Li, Yuan, Li, Yang et al., T cell receptor b repertoires in patients with COVID-19 reveal disease severity signatures, Front Immunol, doi:10.3389/fimmu.2023.1190844
Yang, Wang, Sun, Qin, Yang et al., SARS-CoV-2 epitopespecific T cells: Immunity response feature, TCR repertoire characteristics and crossreactivity, Front Immunol, doi:10.3389/fimmu.2023.1146196
DOI record:
{
"DOI": "10.3389/fimmu.2025.1582949",
"ISSN": [
"1664-3224"
],
"URL": "http://dx.doi.org/10.3389/fimmu.2025.1582949",
"abstract": "<jats:sec><jats:title>Introduction</jats:title><jats:p>Severe COVID-19 is characterized by immune dysregulation, with T cells playing a central role in disease progression and recovery. However, the longitudinal dynamics of the T cell receptor (TCR) repertoire during the course of severe illness remain unclear.</jats:p></jats:sec><jats:sec><jats:title>Methods</jats:title><jats:p>To investigate temporal changes in adaptive immunity, we analyzed peripheral blood samples from the ICU-admitted severe COVID-19 patients (n = 36) collected at three time points: Day 1 (T1), Day 4 (T2), and Day 7 (T3). Bulk RNA-sequencing was performed to extract TCR repertoires, and cytokine profiles were assessed in parallel. TCR clonotypes were annotated using VDJdb and TCRex to infer potential epitope specificities.</jats:p></jats:sec><jats:sec><jats:title>Results</jats:title><jats:p>By T3, we observed a 2.3-fold expansion in TCR clonotypes along with increased TCR-β (TRB) chain usage, indicating the emergence of a broad polyclonal T cell response. In contrast, TCR-γ (TRG) chain prevalence declined. Pro-inflammatory cytokines, including IL-1β and IL-6, were reduced over time, marking a shift toward immune resolution. Changes in CDR3 motifs and preferential TRBV gene segment usage were detected, suggesting repertoire adaptation. Additionally, annotated TCR clonotypes at T3 mapped to SARS-CoV-2 and other pathogen-associated epitopes (e.g., CMV, Plasmodium), reflecting possible cross-reactivity or memory T cell recruitment.</jats:p></jats:sec><jats:sec><jats:title>Discussion</jats:title><jats:p>These findings suggest a coordinated transition from immune dysfunction to recovery in severe COVID-19, marked by expanding TCR diversity, reduced inflammation, and predicted broadening of antigen recognition. The integrated analysis of TCR repertoire dynamics and cytokine profiles provides insights into the adaptive immune mechanisms underlying viral clearance and immune stabilization.</jats:p></jats:sec>",
"alternative-id": [
"10.3389/fimmu.2025.1582949"
],
"article-number": "1582949",
"author": [
{
"affiliation": [],
"family": "Khare",
"given": "Kriti",
"sequence": "first"
},
{
"affiliation": [],
"family": "Yadav",
"given": "Sunita",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Halder",
"given": "Sayanti",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Ray",
"given": "Yogiraj",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Ganguly",
"given": "Dipyaman",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Pandey",
"given": "Rajesh",
"sequence": "additional"
}
],
"container-title": "Frontiers in Immunology",
"container-title-short": "Front. Immunol.",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"frontiersin.org"
]
},
"created": {
"date-parts": [
[
2025,
7,
10
]
],
"date-time": "2025-07-10T04:12:13Z",
"timestamp": 1752120733000
},
"deposited": {
"date-parts": [
[
2025,
7,
10
]
],
"date-time": "2025-07-10T04:12:16Z",
"timestamp": 1752120736000
},
"indexed": {
"date-parts": [
[
2025,
7,
10
]
],
"date-time": "2025-07-10T04:40:02Z",
"timestamp": 1752122402709,
"version": "3.41.2"
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2025,
7,
10
]
]
},
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
7,
10
]
],
"date-time": "2025-07-10T00:00:00Z",
"timestamp": 1752105600000
}
}
],
"link": [
{
"URL": "https://www.frontiersin.org/articles/10.3389/fimmu.2025.1582949/full",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1965",
"original-title": [],
"prefix": "10.3389",
"published": {
"date-parts": [
[
2025,
7,
10
]
]
},
"published-online": {
"date-parts": [
[
2025,
7,
10
]
]
},
"publisher": "Frontiers Media SA",
"reference": [
{
"DOI": "10.1016/j.intcar.2020.100003",
"article-title": "COVID-19 Disease: a critical care perspective",
"author": "Carter",
"doi-asserted-by": "publisher",
"first-page": "100003",
"journal-title": "Clinics Integrated Care",
"key": "B1",
"volume": "1",
"year": "2020"
},
{
"DOI": "10.1016/j.pulmoe.2021.03.008",
"article-title": "The immune response to SARS-CoV-2 and COVID-19 immunopathology - Current perspectives",
"author": "Boechat",
"doi-asserted-by": "publisher",
"journal-title": "Pulmonology",
"key": "B2",
"volume": "27",
"year": "2021"
},
{
"DOI": "10.1038/s41579-020-00459-7",
"article-title": "Characteristics of SARS-CoV-2 and COVID-19",
"author": "Hu",
"doi-asserted-by": "publisher",
"journal-title": "Nat Rev Microbiol",
"key": "B3",
"volume": "19",
"year": "2020"
},
{
"DOI": "10.1038/s41579-022-00713-0",
"article-title": "SARS-CoV-2 pathogenesis",
"author": "Lamers",
"doi-asserted-by": "publisher",
"journal-title": "Nat Rev Microbiol",
"key": "B4",
"volume": "20",
"year": "2022"
},
{
"DOI": "10.1111/cei.12622",
"article-title": "The T cell antigen receptor: the Swiss army knife of the immune system",
"author": "Attaf",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Clin Exp Immunol",
"key": "B5",
"volume": "181",
"year": "2015"
},
{
"DOI": "10.1038/s41577-018-0007-5",
"article-title": "Understanding the drivers of MHC restriction of T cell receptors",
"author": "La Gruta",
"doi-asserted-by": "publisher",
"journal-title": "Nat Rev Immunol",
"key": "B6",
"volume": "18",
"year": "2018"
},
{
"DOI": "10.3389/fimmu.2023.1127470",
"article-title": "Antigen-specificity measurements are the key to understanding T cell responses",
"author": "Tippalagama",
"doi-asserted-by": "publisher",
"journal-title": "Front Immunol",
"key": "B7",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.1016/j.coi.2009.05.004",
"article-title": "Functional implications of T cell receptor diversity",
"author": "Turner",
"doi-asserted-by": "publisher",
"journal-title": "Curr Opin Immunol",
"key": "B8",
"volume": "21",
"year": "2009"
},
{
"DOI": "10.1126/scitranslmed.3003647",
"article-title": "T cell receptor αβ diversity inversely correlates with pathogen-specific antibody levels in human cytomegalovirus infection",
"author": "Wang",
"doi-asserted-by": "publisher",
"first-page": "128ra42",
"journal-title": "Sci Transl Med",
"key": "B9",
"volume": "4",
"year": "2012"
},
{
"DOI": "10.1128/JVI.02380-20",
"article-title": "In chronic infection, HIV gag-specific CD4+ T cell receptor diversity is higher than CD8+ T cell receptor diversity and is associated with less HIV quasispecies diversity",
"author": "Pilkinton",
"doi-asserted-by": "publisher",
"journal-title": "J Virol",
"key": "B10",
"volume": "95",
"year": "2021"
},
{
"DOI": "10.1038/nm0597-533",
"article-title": "HIV infection induces changes in CD4+ T-cell phenotype and depletions within the CD4+ T-cell repertoire that are not immediately restored by antiviral or immune-based therapies",
"author": "Connors",
"doi-asserted-by": "publisher",
"journal-title": "Nat Med",
"key": "B11",
"volume": "3",
"year": "1997"
},
{
"DOI": "10.1016/j.immuni.2020.11.004",
"article-title": "SARS-CoV-2 epitopes are recognized by a public and diverse repertoire of human T cell receptors",
"author": "Shomuradova",
"doi-asserted-by": "publisher",
"first-page": "1245",
"journal-title": "Immunity",
"key": "B12",
"volume": "53",
"year": "2020"
},
{
"DOI": "10.1016/j.chom.2021.05.010",
"article-title": "SARS-CoV-2 human T cell epitopes: Adaptive immune response against COVID-19",
"author": "Grifoni",
"doi-asserted-by": "publisher",
"journal-title": "Cell Host Microbe",
"key": "B13",
"volume": "29",
"year": "2021"
},
{
"DOI": "10.1016/j.ijid.2021.10.033",
"article-title": "T-cell receptor repertoires as potential diagnostic markers for patients with COVID-19",
"author": "Hou",
"doi-asserted-by": "publisher",
"journal-title": "Int J Infect Dis",
"key": "B14",
"volume": "113",
"year": "2021"
},
{
"DOI": "10.1016/j.xcrm.2024.101583",
"article-title": "SARS-CoV-2 breakthrough infections enhance T cell response magnitude, breadth, and epitope repertoire",
"author": "Tarke",
"doi-asserted-by": "publisher",
"journal-title": "Cell Rep Med",
"key": "B15",
"volume": "5",
"year": "2024"
},
{
"DOI": "10.1016/j.tips.2020.06.001",
"article-title": "Deciphering the TCR repertoire to solve the COVID-19 mystery",
"author": "Gutierrez",
"doi-asserted-by": "publisher",
"journal-title": "Trends Pharmacol Sci",
"key": "B16",
"volume": "41",
"year": "2020"
},
{
"DOI": "10.1172/jci.insight.151849",
"article-title": "Longitudinal analysis of T cell receptor repertoires reveals shared patterns of antigen-specific response to SARS-CoV-2 infection",
"author": "Gittelman",
"doi-asserted-by": "publisher",
"first-page": "e151849",
"journal-title": "JCI Insight",
"key": "B17",
"volume": "7",
"year": "2022"
},
{
"DOI": "10.1016/j.molimm.2023.09.006",
"article-title": "Longitudinal change trend of the TCR repertoire reveals the immune response intensity of the inactivated COVID-19 vaccine",
"author": "Ren",
"doi-asserted-by": "publisher",
"first-page": "39",
"journal-title": "Mol Immunol",
"key": "B18",
"volume": "163",
"year": "2023"
},
{
"DOI": "10.1016/j.mayocpiqo.2022.09.001",
"article-title": "Clinical trial subgroup analyses to investigate clinical and immunological outcomes of convalescent plasma therapy in severe COVID-19",
"author": "Raychaudhuri",
"doi-asserted-by": "publisher",
"journal-title": "Mayo Clin Proc Innov Qual Outcomes",
"key": "B19",
"volume": "6",
"year": "2022"
},
{
"DOI": "10.1038/s41467-022-28064-7",
"article-title": "A phase 2 single center open label randomised control trial for convalescent plasma therapy in patients with severe COVID-19",
"author": "Ray",
"doi-asserted-by": "publisher",
"first-page": "383",
"journal-title": "Nat Commun",
"key": "B20",
"volume": "13",
"year": "2022"
},
{
"key": "B21",
"unstructured": "FastQC: a quality control tool for high throughput sequence data – ScienceOpen"
},
{
"DOI": "10.1093/bioinformatics/btu170",
"article-title": "Trimmomatic: A flexible trimmer for Illumina sequence data",
"author": "Bolger",
"doi-asserted-by": "publisher",
"journal-title": "Bioinformatics",
"key": "B22",
"volume": "30",
"year": "2014"
},
{
"DOI": "10.1038/nmeth.3364",
"article-title": "MiXCR: software for comprehensive adaptive immunity profiling",
"author": "Bolotin",
"doi-asserted-by": "publisher",
"journal-title": "Nat Methods",
"key": "B23",
"volume": "12",
"year": "2015"
},
{
"DOI": "10.1371/journal.pcbi.1004503",
"article-title": "VDJtools: unifying post-analysis of T cell receptor repertoires",
"author": "Shugay",
"doi-asserted-by": "publisher",
"journal-title": "PLoS Comput Biol",
"key": "B24",
"volume": "11",
"year": "2015"
},
{
"DOI": "10.1101/gr.849004",
"article-title": "WebLogo: a sequence logo generator",
"author": "Crooks",
"doi-asserted-by": "publisher",
"journal-title": "Genome Res",
"key": "B25",
"volume": "14",
"year": "2004"
},
{
"DOI": "10.1093/nar/gkx760",
"article-title": "VDJdb: a curated database of T-cell receptor sequences with known antigen specificity",
"author": "Shugay",
"doi-asserted-by": "publisher",
"journal-title": "Nucleic Acids Res",
"key": "B26",
"volume": "46",
"year": "2018"
},
{
"DOI": "10.3389/fimmu.2019.02820",
"article-title": "Detection of enriched T cell epitope specificity in full T cell receptor sequence repertoires",
"author": "Gielis",
"doi-asserted-by": "publisher",
"journal-title": "Front Immunol",
"key": "B27",
"volume": "10",
"year": "2019"
},
{
"DOI": "10.1093/bioinformatics/btx286",
"article-title": "McPAS-TCR: a manually curated catalogue of pathology-associated T cell receptor sequences",
"author": "Tickotsky",
"doi-asserted-by": "publisher",
"journal-title": "Bioinformatics",
"key": "B28",
"volume": "33",
"year": "2017"
},
{
"DOI": "10.21203/rs.3.rs-51964/v1",
"article-title": "A large-scale database of T-cell receptor beta (TCRβ) sequences and binding associations from natural and synthetic exposure to SARS-CoV-2",
"author": "Nolan",
"doi-asserted-by": "publisher",
"journal-title": "Res Sq",
"key": "B29",
"year": "2020"
},
{
"article-title": "Rawgraphs: A visualisation platform to create open outputs",
"author": "Mauri",
"first-page": "1",
"key": "B30",
"volume-title": "Proceedings of the 12th Biannual Conference on Italian SIGCHI Chapter - CHItaly ‘17",
"year": "2017"
},
{
"DOI": "10.1371/journal.pone.0294236",
"article-title": "SRplot: A free online platform for data visualization and graphing",
"author": "Tang",
"doi-asserted-by": "publisher",
"journal-title": "PloS One",
"key": "B31",
"volume": "18",
"year": "2023"
},
{
"DOI": "10.4061/2011/970968",
"article-title": "Control of V(D)J recombination through transcriptional elongation and changes in locus chromatin structure and nuclear organization",
"author": "Del Blanco",
"doi-asserted-by": "publisher",
"journal-title": "Genet Res Int",
"key": "B32",
"volume": "2011",
"year": "2011"
},
{
"DOI": "10.1038/s41467-024-48198-0",
"article-title": "Designing meaningful continuous representations of T cell receptor sequences with deep generative models",
"author": "Leary",
"doi-asserted-by": "publisher",
"first-page": "4271",
"journal-title": "Nat Commun",
"key": "B33",
"volume": "15",
"year": "2024"
},
{
"DOI": "10.3390/vaccines11010101",
"article-title": "Updated Insights into the T Cell-Mediated Immune Response against SARS-CoV-2: A Step towards Efficient and Reliable Vaccines",
"author": "Dhawan",
"doi-asserted-by": "publisher",
"first-page": "699",
"journal-title": "Vaccines (Basel)",
"key": "B34",
"volume": "11",
"year": "2023"
},
{
"DOI": "10.1084/jem.182.6.1703",
"article-title": "T cell receptor repertoire for a viral epitope in humans is diversified by tolerance to a background major histocompatibility complex antigen",
"author": "Burrows",
"doi-asserted-by": "publisher",
"journal-title": "J Exp Med",
"key": "B35",
"volume": "182",
"year": "1995"
},
{
"DOI": "10.1615/CritRevImmunol.v32.i4.50",
"article-title": "Cross-reactivity of T cells and its role in the immune system",
"author": "Petrova",
"doi-asserted-by": "publisher",
"journal-title": "Crit Rev Immunol",
"key": "B36",
"volume": "32",
"year": "2012"
},
{
"DOI": "10.1016/j.celrep.2017.03.072",
"article-title": "Sequence and structural analyses reveal distinct and highly diverse human CD8+ TCR repertoires to immunodominant viral antigens",
"author": "Chen",
"doi-asserted-by": "publisher",
"journal-title": "Cell Rep",
"key": "B37",
"volume": "19",
"year": "2017"
},
{
"DOI": "10.3389/fimmu.2023.1146196",
"article-title": "SARS-CoV-2 epitope-specific T cells: Immunity response feature, TCR repertoire characteristics and cross-reactivity",
"author": "Yang",
"doi-asserted-by": "publisher",
"journal-title": "Front Immunol",
"key": "B38",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.1084/jem.20100603",
"article-title": "Allelic polymorphism in the T cell receptor and its impact on immune responses",
"author": "Gras",
"doi-asserted-by": "publisher",
"journal-title": "J Exp Med",
"key": "B39",
"volume": "207",
"year": "2010"
},
{
"DOI": "10.1038/s41423-024-01168-4",
"article-title": "Legacy of the discovery of the T-cell receptor: 40 years of shaping basic immunology and translational work to develop novel therapies",
"author": "Shi",
"doi-asserted-by": "publisher",
"journal-title": "Cell Mol Immunol",
"key": "B40",
"volume": "21",
"year": "2024"
},
{
"DOI": "10.3390/ijms22020523",
"article-title": "Role of CD4+ T cells in the control of viral infections: recent advances and open questions",
"author": "Kervevan",
"doi-asserted-by": "publisher",
"first-page": "523",
"journal-title": "Int J Mol Sci",
"key": "B41",
"volume": "22",
"year": "2021"
},
{
"DOI": "10.3389/fimmu.2023.1190844",
"article-title": "T cell receptor β repertoires in patients with COVID-19 reveal disease severity signatures",
"author": "Xu",
"doi-asserted-by": "publisher",
"journal-title": "Front Immunol",
"key": "B42",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.1016/j.ygeno.2020.12.036",
"article-title": "Comprehensive analysis of TCR repertoire in COVID-19 using single cell sequencing",
"author": "Wang",
"doi-asserted-by": "publisher",
"journal-title": "Genomics",
"key": "B43",
"volume": "113",
"year": "2021"
},
{
"DOI": "10.1016/j.it.2021.06.005",
"article-title": "New insights into TCR β-selection",
"author": "Dutta",
"doi-asserted-by": "publisher",
"journal-title": "Trends Immunol",
"key": "B44",
"volume": "42",
"year": "2021"
},
{
"DOI": "10.1016/j.chom.2020.04.009",
"article-title": "Complex immune dysregulation in COVID-19 patients with severe respiratory failure",
"author": "Giamarellos-Bourboulis",
"doi-asserted-by": "publisher",
"first-page": "992",
"journal-title": "Cell Host Microbe",
"key": "B45",
"volume": "27",
"year": "2020"
},
{
"DOI": "10.3389/fimmu.2020.580304",
"article-title": "The role of gamma-delta T cells in diseases of the central nervous system",
"author": "Wo",
"doi-asserted-by": "publisher",
"journal-title": "Front Immunol",
"key": "B46",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1073/pnas.1409155111",
"article-title": "Diversity and clonal selection in the human T-cell repertoire",
"author": "Qi",
"doi-asserted-by": "publisher",
"journal-title": "Proc Natl Acad Sci U S A",
"key": "B47",
"volume": "111",
"year": "2014"
},
{
"DOI": "10.3390/ijms23158590",
"article-title": "T-cell receptor repertoire sequencing and its applications: focus on infectious diseases and cancer",
"author": "Mazzotti",
"doi-asserted-by": "publisher",
"first-page": "8590",
"journal-title": "Int J Mol Sci",
"key": "B48",
"volume": "23",
"year": "2022"
},
{
"DOI": "10.1016/j.isci.2024.110983",
"article-title": "TCR repertoire dynamics and their responses underscores dengue severity",
"author": "Khare",
"doi-asserted-by": "publisher",
"journal-title": "iScience",
"key": "B49",
"volume": "27",
"year": "2024"
},
{
"DOI": "10.1038/s41467-019-08906-7",
"article-title": "Molecular constraints on CDR3 for thymic selection of MHC-restricted TCRs from a random pre-selection repertoire",
"author": "Lu",
"doi-asserted-by": "publisher",
"first-page": "1019",
"journal-title": "Nat Commun",
"key": "B50",
"volume": "10",
"year": "2019"
},
{
"DOI": "10.1016/j.molimm.2008.09.021",
"article-title": "Structural features of T cell receptor variable regions that enhance domain stability and enable expression as single-chain ValphaVbeta fragments",
"author": "Richman",
"doi-asserted-by": "publisher",
"journal-title": "Mol Immunol",
"key": "B51",
"volume": "46",
"year": "2009"
},
{
"DOI": "10.23812/CONTI-E",
"article-title": "Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies",
"author": "Conti",
"doi-asserted-by": "publisher",
"journal-title": "J Biol Regul Homeost Agents",
"key": "B52",
"volume": "34",
"year": "2020"
},
{
"DOI": "10.3390/tropicalmed9010013",
"article-title": "Pro-inflammatory and anti-inflammatory interleukins in infectious diseases: A comprehensive review",
"author": "Al-Qahtani",
"doi-asserted-by": "publisher",
"first-page": "13",
"journal-title": "Trop Med Infect Dis",
"key": "B53",
"volume": "9",
"year": "2024"
},
{
"DOI": "10.1016/j.cell.2024.03.037",
"article-title": "Principles and therapeutic applications of adaptive immunity",
"author": "Chi",
"doi-asserted-by": "publisher",
"journal-title": "Cell",
"key": "B54",
"volume": "187",
"year": "2024"
},
{
"DOI": "10.4049/immunohorizons.2400003",
"article-title": "Immune epitopes of SARS-CoV-2 spike protein and considerations for universal vaccine development",
"author": "Magazine",
"doi-asserted-by": "publisher",
"journal-title": "Immunohorizons",
"key": "B55",
"volume": "8",
"year": "2024"
},
{
"DOI": "10.3389/fimmu.2022.1011935",
"article-title": "Cross-reactivity influences changes in human influenza A virus and Epstein Barr virus specific CD8 memory T cell receptor alpha and beta repertoires between young and old",
"author": "Clark",
"doi-asserted-by": "publisher",
"journal-title": "Front Immunol",
"key": "B56",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.1016/j.immuni.2020.06.024",
"article-title": "Next-generation sequencing of T and B cell receptor repertoires from COVID-19 patients showed signatures associated with severity of disease",
"author": "Schultheiß",
"doi-asserted-by": "publisher",
"first-page": "442",
"journal-title": "Immunity",
"key": "B57",
"volume": "53",
"year": "2020"
}
],
"reference-count": 57,
"references-count": 57,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.frontiersin.org/articles/10.3389/fimmu.2025.1582949/full"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Temporal TCR dynamics and epitope diversity mark recovery in severe COVID-19 patients",
"type": "journal-article",
"update-policy": "https://doi.org/10.3389/crossmark-policy",
"volume": "16"
}