In silico Evaluation of H1-Antihistamine as Potential Inhibitors of SARS-CoV-2 RNA-dependent RNA Polymerase: Repurposing Study of COVID-19 Therapy
et al., Turkish Journal of Pharmaceutical Sciences, doi:10.4274/tjps.galenos.2024.49768, Jan 2025
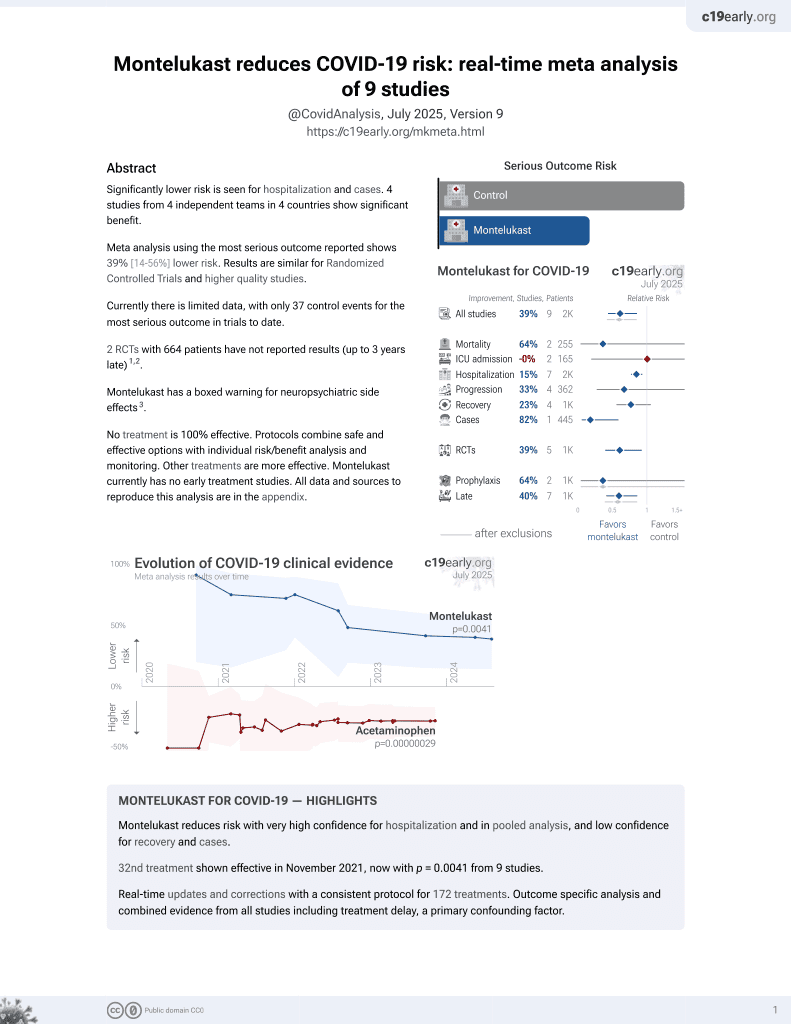
32nd treatment shown to reduce risk in
November 2021, now with p = 0.0041 from 9 studies.
Lower risk for hospitalization and cases.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
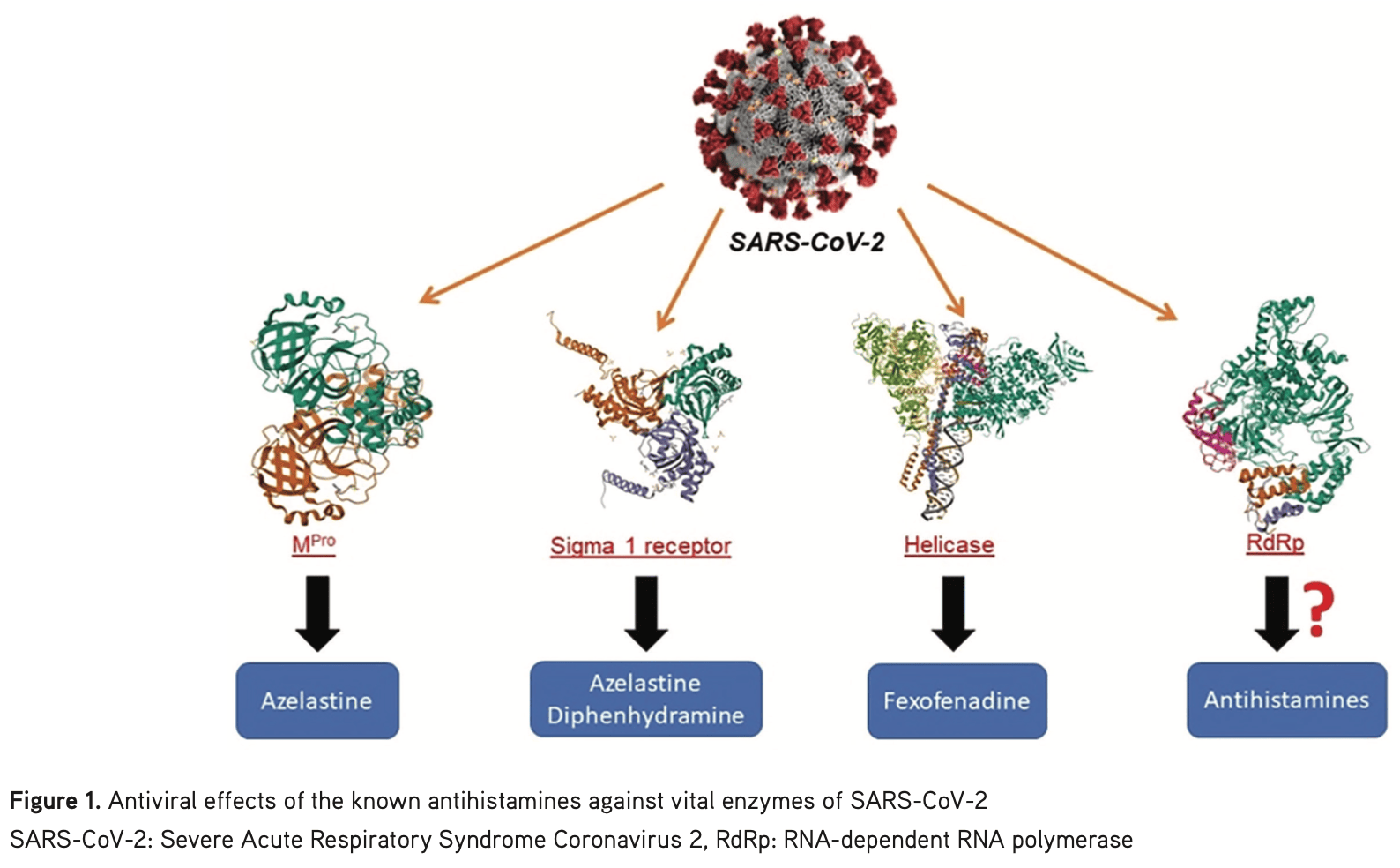
In silico study showing that H1RA antihistamines, including bilastine, fexofenadine, mizolastine, rupatadine, terfenadine, and the leukotriene receptor antagonists montelukast and zafirlukast, may inhibit SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) and potentially treat COVID-19. Docking analysis revealed that these drugs bind to RdRp with higher affinity than molnupiravir and its derivatives. The drugs interacted with key RdRp residues through hydrogen bonds, electrostatic interactions, and hydrophobic contacts.
5 preclinical studies support the efficacy of montelukast for COVID-19:
In silico studies predict inhibition of SARS-CoV-2 with montelukast or metabolites via binding to the spikeA,2 (and specifically the receptor binding domainB,3), MproC,2, RNA-dependent RNA polymeraseD,1,2 , PLproE,2, nucleocapsidF,2, and helicaseG,2 proteins.
Montelukast inhibits SARS-CoV-2 omicron infection in Vero cells
at 1μM3 and inhibits platelet activation induced by plasma from
COVID-19 patients4.
1.
Hamdan et al., In silico Evaluation of H1-Antihistamine as Potential Inhibitors of SARS-CoV-2 RNA-dependent RNA Polymerase: Repurposing Study of COVID-19 Therapy, Turkish Journal of Pharmaceutical Sciences, doi:10.4274/tjps.galenos.2024.49768.
2.
Haque et al., Exploring potential therapeutic candidates against COVID-19: a molecular docking study, Discover Molecules, doi:10.1007/s44345-024-00005-5.
a.
The trimeric spike (S) protein is a glycoprotein that mediates viral entry by binding to the host ACE2 receptor, is critical for SARS-CoV-2's ability to infect host cells, and is a target of neutralizing antibodies. Inhibition of the spike protein prevents viral attachment, halting infection at the earliest stage.
b.
The receptor binding domain is a specific region of the spike protein that binds ACE2 and is a major target of neutralizing antibodies. Focusing on the precise binding site allows highly specific disruption of viral attachment with reduced potential for off-target effects.
c.
The main protease or Mpro, also known as 3CLpro or nsp5, is a cysteine protease that cleaves viral polyproteins into functional units needed for replication. Inhibiting Mpro disrupts the SARS-CoV-2 lifecycle within the host cell, preventing the creation of new copies.
d.
RNA-dependent RNA polymerase (RdRp), also called nsp12, is the core enzyme of the viral replicase-transcriptase complex that copies the positive-sense viral RNA genome into negative-sense templates for progeny RNA synthesis. Inhibiting RdRp blocks viral genome replication and transcription.
e.
The papain-like protease (PLpro) has multiple functions including cleaving viral polyproteins and suppressing the host immune response by deubiquitination and deISGylation of host proteins. Inhibiting PLpro may block viral replication and help restore normal immune responses.
f.
The nucleocapsid (N) protein binds and encapsulates the viral genome by coating the viral RNA. N enables formation and release of infectious virions and plays additional roles in viral replication and pathogenesis. N is also an immunodominant antigen used in diagnostic assays.
g.
The helicase, or nsp13, protein unwinds the double-stranded viral RNA, a crucial step in replication and transcription. Inhibition may prevent viral genome replication and the creation of new virus components.
Hamdan et al., 10 Jan 2025, peer-reviewed, 3 authors.
Contact: ilkay.kucukguzel@fbu.edu.tr (corresponding author).
In silico studies are an important part of preclinical research, however results may be very different in vivo.
In silico Evaluation of H1-Antihistamine as Potential Inhibitors of SARS-CoV-2 RNA-dependent RNA Polymerase: Repurposing Study of COVID-19 Therapy
Turkish Journal of Pharmaceutical Sciences, doi:10.4274/tjps.galenos.2024.49768
Introduction: Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), from the family Coronaviridae, is the seventh known coronavirus to infect humans and cause acute respiratory syndrome. Although vaccination efforts have been conducted against this virus, which emerged in Wuhan, China, in December 2019 and has spread rapidly around the world, the lack of an Food and Drug Administration-approved antiviral agent has made drug repurposing an important approach for emergency response during the COVID-19 pandemic. The aim of this study was to investigate the potential of H1-antihistamines as antiviral agents against SARS-CoV-2 RNA-dependent RNA polymerase enzyme. Materials and Methods: Using molecular docking techniques, we explored the interactions between H1-antihistamines and RNA-dependent RNA polymerase (RdRp), a key enzyme involved in viral replication. The three-dimensional structure of 37 H1-antihistamine molecules was drawn and their energies were minimized using Spartan 0.4. Subsequently, we conducted a docking study with Autodock Vina to assess the binding affinity of these molecules to the target site. The docking scores and conformations were then visualized using Discovery Studio.
Results: The results examined showed that the docking scores of the H1-antihistamines were between 5.0 and 8.3 kcal/mol. These findings suggested that among all the analyzed drugs, bilastine, fexofenadine, montelukast, zafirlukast, mizolastine, and rupatadine might bind with the best binding energy (< -7.0 kcal/mol) and inhibit RdRp, potentially halting the replication of the virus.
Conclusion: This study highlights the potential of H1-antihistamines in combating COVID-19 and underscores the value of computational approaches in rapid drug discovery and repurposing efforts. Finally, experimental studies are required to measure the potency of H1-antihistamines before their clinical use against COVID-19 as RdRp inhibitors.
Ethics Ethics Committee Approval: Not required. Informed Consent: Not required.
Authorship Contributions Concept: İ.K., Design: M.H., N.K., İ.K., Data Collection or Processing: M.H., N.K., Analysis or Interpretation: M.H., N.K., İ.K., Literature Search: M.H., N.K., İ.K., Writing: M.H., N.K., İ.K.
Conflict of Interest: The authors have no conflicts of interest to declare.
Financial Disclosure: The authors declared that this study received no financial support.
References
Choi, Azmat, Leukotriene Receptor Antagonists
Church, Allergy, histamine and antihistamines, Handb Exp Pharmacol
De Farias, Santos Junior, Rêgo, José, Origin and evolution of RNA-dependent RNA polymerase, Front Genet
Drayman, Jones, Azizi, Froggatt, Tan et al., Drug repurposing screen identifies masitinib as a 3CLpro inhibitor that blocks replication of SARS-CoV-2 in vitro, bioRxiv
Dyall, Gross, Kindrachuk, Johnson, Olinger et al., Middle East respiratory syndrome and severe acute respiratory syndrome: current therapeutic options and potential targets for novel therapies, Drugs
Elfiky, Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study, Life Sci
Gao, Yan, Huang, Liu, Zhao et al., Structure of the RNA-dependent RNA polymerase from COVID-19 virus, Science
Ghahremanpour, Tirado-Rives, Deshmukh, Ippolito, Zhang et al., Identification of 14 known drugs as inhibitors of the main protease of SARS-CoV-2, bioRxiv
Ghobain, Rebh, Saad, Alhuraiji, Zahrani et al., The efficacy of Zafirlukast as a SARS-CoV-2 helicase inhibitor in adult patients with moderate COVID-19 Pneumonia (pilot randomized clinical trial), J Infect Public Health
Hillen, Kokic, Farnung, Dienemann, Tegunov et al., Structure of replicating SARS-CoV-2 polymerase, Nature
Kirchdoerfer, Ward, Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors, Nat Commun
Kitchen, Decornez, Furr, Bajorath, Docking and scoring in virtual screening for drug discovery: methods and applications, Nat Rev Drug Discov
Krause, Smolle, Covid-19 mortality and local burden of infectious diseases: a worldwide country-by-country analysis, J Infect Public Health
Kulabaş, Yeşil, Küçükgüzel, Evaluation of molnupiravir analogues as novel coronavirus (SARS-CoV-2) RNA-dependent RNA polymerase (RdRp) inhibitors-an in silico docking and ADMET simulation study, J Res Pharm
Lee, Kim, Jang, DOVE: An infectious disease outbreak statistics visualization system, IEEE Access
Liu, Cao, Xu, Wang, Zhang et al., Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro, Cell Discov
Lung, Lin, Yang, Chou, Shu et al., The potential chemical structure of anti-SARS-CoV-2 RNA-dependent RNA polymerase, J Med Virol
Mccarthy, Morrison, Persistent RNA virus infections: do PAMPS drive chronic disease?, Curr Opin Virol
Mingorance, Friesland, Coto-Llerena, Pérez-Del-Pulgar, Boix et al., Selective inhibition of hepatitis C virus infection by hydroxyzine and benztropine, Antimicrob Agents Chemother
Morris, Huey, Lindstrom, Sanner, Belew et al., Software news and updates gabedit-a graphical user interface for computational chemistry softwares, J Comput Chem
Oh, Adnan, Cho, Network pharmacology study to elucidate the key targets of underlying antihistamines against COVID-19, Curr Issues Mol Biol
Piplani, Singh, Winkler, Petrovsky, Potential COVID-19 therapies from computational repurposing of drugs and natural products against the SARS-CoV-2 helicase, Int J Mol Sci
Reznikov, Norris, Vashisht, Gabizon, Lee et al., Identification of antiviral antihistamines for COVID-19 repurposing, Biochem Biophys Res Commun
Seliem, Panda, Girgis, Hassaneen, Shaldam et al., New quinoline-triazole conjugates: Synthesis, and antiviral properties against SARS-CoV-2, Bioorg Chem
Simons, Advances in H1-antihistamines, N Engl J Med
Stewart, Optimization of parameters for semiempirical methods V: modification of NDDO approximations and application to 70 elements, J Mol Model
Tian, Qiang, Liang, Ren, Jia et al., RNA-dependent RNA polymerase (RdRp) inhibitors: The current landscape and repurposing for the COVID-19 pandemic, Eur J Med Chem
Trott, Olson, AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J Comput Chem
Wang, Cao, Zhang, Yang, Liu et al., Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro, Cell Res
Zheng, Zhang, Huang, Nandakumar, Liu et al., Potential treatment methods targeting 2019-nCoV infection, Eur J Med Chem
Zhou, Yang, Wang, Hu, Zhang et al., A pneumonia outbreak associated with a new coronavirus of probable bat origin, Nature
Zongyi, Adam, Xin, Xiaolin, Dalia et al., Chlorcyclizine inhibits viral fusion of hepatitis C virus entry by directly targeting HCV envelope glycoprotein 1, Cell Chem Biol
DOI record:
{
"DOI": "10.4274/tjps.galenos.2024.49768",
"ISSN": [
"1304-530X",
"2148-6247"
],
"URL": "http://dx.doi.org/10.4274/tjps.galenos.2024.49768",
"author": [
{
"affiliation": [],
"family": "HAMDAN",
"given": "Mazin",
"sequence": "first"
},
{
"affiliation": [],
"family": "KULABAŞ",
"given": "Necla",
"sequence": "additional"
},
{
"affiliation": [],
"family": "KÜÇÜKGÜZEL",
"given": "İlkay",
"sequence": "additional"
}
],
"container-title": "Turkish Journal of Pharmaceutical Sciences",
"container-title-short": "tjps",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2025,
1,
10
]
],
"date-time": "2025-01-10T12:41:13Z",
"timestamp": 1736512873000
},
"deposited": {
"date-parts": [
[
2025,
1,
10
]
],
"date-time": "2025-01-10T12:41:19Z",
"timestamp": 1736512879000
},
"indexed": {
"date-parts": [
[
2025,
1,
11
]
],
"date-time": "2025-01-11T05:26:30Z",
"timestamp": 1736573190860,
"version": "3.32.0"
},
"is-referenced-by-count": 0,
"issue": "6",
"issued": {
"date-parts": [
[
2025,
1,
10
]
]
},
"journal-issue": {
"issue": "6",
"published-online": {
"date-parts": [
[
2025,
1,
10
]
]
}
},
"member": "2811",
"original-title": [],
"page": "566-576",
"prefix": "10.4274",
"published": {
"date-parts": [
[
2025,
1,
10
]
]
},
"published-online": {
"date-parts": [
[
2025,
1,
10
]
]
},
"publisher": "Galenos Yayinevi",
"reference-count": 0,
"references-count": 0,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.turkjps.org/articles/lessemgreaterin-silico-lessemgreaterevaluation-of-h1-antihistamine-as-potential-inhibitors-of-sars-cov-2-rna-dependent-rna-polymerase-repurposing-study-of-covid-19-therapy/doi/tjps.galenos.2024.49768"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "In silico Evaluation of H1-Antihistamine as Potential Inhibitors of SARS-CoV-2 RNA-dependent RNA Polymerase: Repurposing Study of COVID-19 Therapy",
"type": "journal-article",
"volume": "21"
}
hamdan2