Enisamium Inhibits SARS-CoV-2 RNA Synthesis
et al., Biomedicines, doi:10.3390/biomedicines9091254, Sep 2021
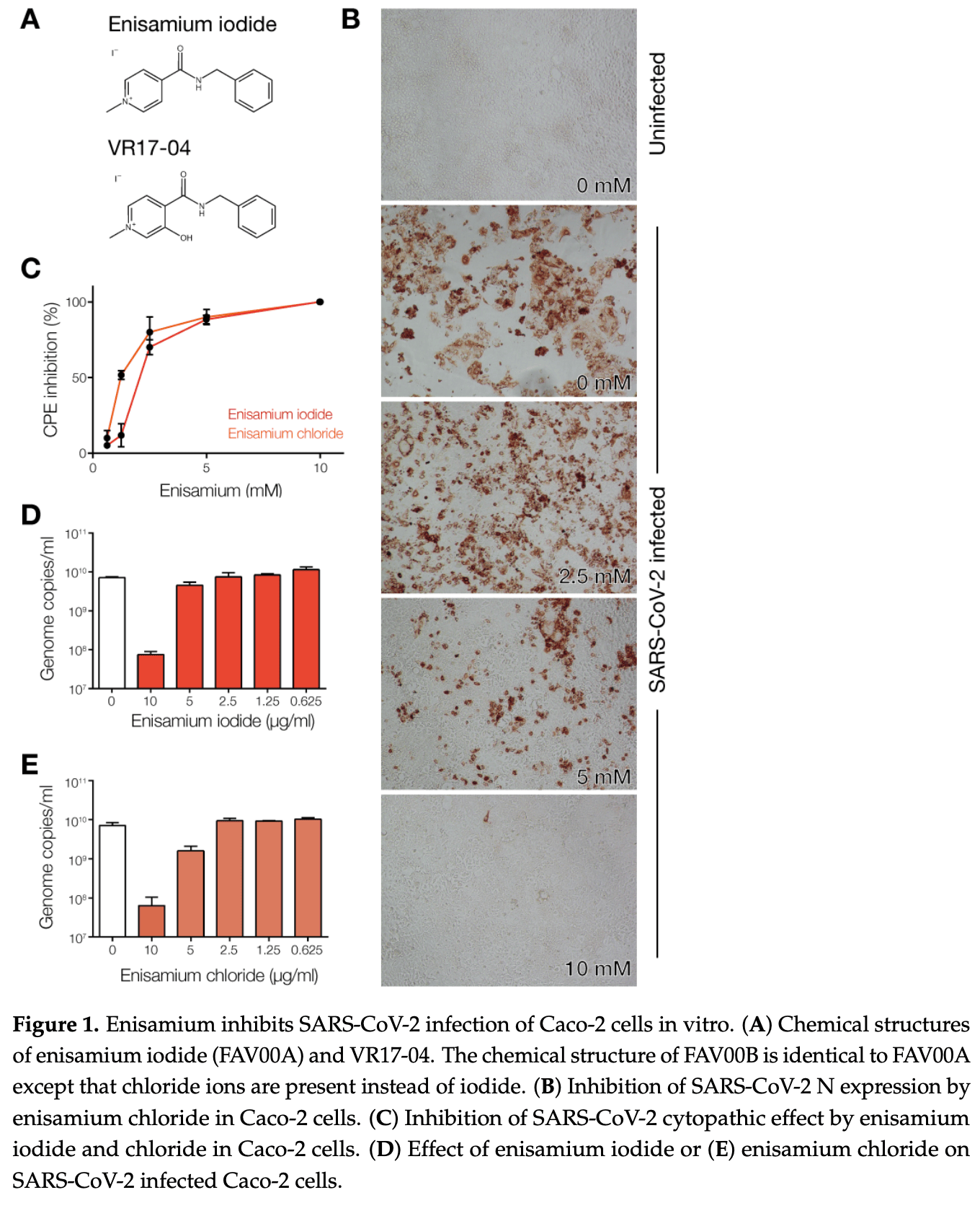
In vitro study showing inhibition of SARS-CoV-2 RNA synthesis by enisamium and its metabolite VR17-04. Authors showed that enisamium inhibited SARS-CoV-2 infection in Caco-2 cells with an IC50 of 1.2 mM and coronavirus HCoV-NL63 infection in normal human bronchial epithelial (NHBE) cells with an IC50 of ~60 μg/mL. Molecular dynamics simulations suggest VR17-04 works by forming hydrogen bonds with cytosine or adenine bases in the viral RNA template, preventing GTP and UTP incorporation during RNA synthesis. The active metabolite VR17-04 showed stronger binding to the viral RNA polymerase complex than unmetabolized enisamium.
Elli et al., 17 Sep 2021, USA, peer-reviewed, 15 authors.
Contact: lutz.mueller@regenold.com (corresponding author), elli@ronzoni.it, sala@ronzoni.it, cosentino@ronzoni.it, guerrini@ronzoni.it, denisa.bojkova@kgu.de, marco.bechtel94@gmx.de, tv293@cam.ac.uk, dboltz@iitri.org, mmuzzio@iitri.org, xpeng@iitri.org, a.goy@farmak.ua, v.margitich@farmak.ua, cinatl@em.uni-frankfurt.de, ajwt6@cam.ac.uk.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Enisamium Inhibits SARS-CoV-2 RNA Synthesis
Biomedicines, doi:10.3390/biomedicines9091254
Pandemic SARS-CoV-2 causes a mild to severe respiratory disease called coronavirus disease 2019 . While control of the SARS-CoV-2 spread partly depends on vaccineinduced or naturally acquired protective herd immunity, antiviral strategies are still needed to manage COVID-19. Enisamium is an inhibitor of influenza A and B viruses in cell culture and clinically approved in countries of the Commonwealth of Independent States. In vitro, enisamium acts through metabolite VR17-04 and inhibits the activity of the influenza A virus RNA polymerase. Here we show that enisamium can inhibit coronavirus infections in NHBE and Caco-2 cells, and the activity of the SARS-CoV-2 RNA polymerase in vitro. Docking and molecular dynamics simulations provide insight into the mechanism of action and indicate that enisamium metabolite VR17-04 prevents GTP and UTP incorporation. Overall, these results suggest that enisamium is an inhibitor of SARS-CoV-2 RNA synthesis in vitro.
References
Beigel, Tomashek, Dodd, Mehta, Zingman et al., Remdesivir for the Treatment of Covid-19-Final Report, N. Engl. J. Med, doi:10.1056/NEJMoa2007764
Bojkova, Bechtel, Mclaughlin, Mcgreig, Klann et al., Aprotinin Inhibits SARS-CoV-2 Replication, Cells, doi:10.3390/cells9112377
Bojkova, Klann, Koch, Widera, Krause et al., Proteomics of SARS-CoV-2-infected host cells reveals therapy targets, Nature, doi:10.1038/s41586-020-2332-7
Boltz, Peng, Muzzio, Dash, Thomas et al., Activity of enisamium, an isonicotinic acid derivative, against influenza viruses in differentiated normal human bronchial epithelial cells, Antivir Chem. Chemother, doi:10.1177/2040206618811416
Case, Cheatham, Darden, Gohlke, Luo et al., The Amber biomolecular simulation programs, J. Comput. Chem, doi:10.1002/jcc.20290
Chen, Malone, Llewellyn, Grasso, Shelton et al., Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex, Cell, doi:10.1016/j.cell.2020.07.033
Cox, Wolf, Plemper, Therapeutically administered ribonucleoside analogue MK-4482/EIDD-2801 blocks SARS-CoV-2 transmission in ferrets, Nat. Microbiol, doi:10.1038/s41564-020-00835-2
Frisch, Trucks, Schlegel, Scuseria, Robb et al., Revision C.01
Gesteiger, Marsili, Iterative partial equalization of orbital electronegativity a rapid access to atomic charges, Tetrahedron, doi:10.1016/0040-4020(80)80168-2
Goldhill, .; Te, Velthuis, Fletcher, Langat et al., The mechanism of resistance to favipiravir in influenza, Proc. Natl. Acad. Sci, doi:10.1073/pnas.1811345115
Hartenian, Nandakumar, Lari, Ly, Tucker et al., The molecular virology of coronaviruses, J. Biol. Chem, doi:10.1074/jbc.REV120.013930
Hillen, Kokic, Farnung, Dienemann, Tegunov et al., Structure of replicating SARS-CoV-2 polymerase, Nature, doi:10.1038/s41586-020-2368-8
Humphrey, Dalke, Schulten, Vmd, Visual molecular dynamics, J. Mol. Graph, doi:10.1016/0263-7855(96)00018-5
Jorgensen, Chandrasekhar, Madura, Impey, Klein, Comparison of simple potential functions for simulating liquid water, J. Chem. Phys, doi:10.1063/1.445869
Morris, Huey, Lindstrom, Sanner, Belew et al., AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility, J. Comput. Chem, doi:10.1002/jcc.21256
Phillips, Braun, Wang, Gumbart, Tajkhorshid et al., Scalable molecular dynamics with NAMD, J. Comput. Chem, doi:10.1002/jcc.20289
Shannon, Selisko, Le, Huchting, Touret et al., Rapid incorporation of Favipiravir by the fast and permissive viral RNA polymerase complex results in SARS-CoV-2 lethal mutagenesis, Nat. Commun, doi:10.1038/s41467-020-18463-z
Sheahan, Sims, Zhou, Graham, Pruijssers et al., An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice, Sci. Transl. Med, doi:10.1126/scitranslmed.abb5883
Srinivasan, Cheatham, Iii; Cieplak, Kollman, Case, Continuum solvent studies of the stability of DNA, RNA, and phosphoramidate-DNA helices, J. Am. Chem. Soc, doi:10.1021/ja981844+
Subissi, Imbert, Ferron, Collet, Coutard et al., SARS-CoV ORF1b-encoded nonstructural proteins 12-16: Replicative enzymes as antiviral targets, Antivir. Res, doi:10.1016/j.antiviral.2013.11.006
Subissi, Posthuma, Collet, Zevenhoven-Dobbe, Gorbalenya et al., One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities, Proc. Natl. Acad. Sci, doi:10.1073/pnas.1323705111
Toptan, Hoehl, Westhaus, Bojkova, Berger et al., Optimized qRT-PCR Approach for the Detection of Intra-and Extra-Cellular SARS-CoV-2 RNAs, Int. J. Mol. Sci, doi:10.3390/ijms21124396
Velthuis, Arnold, Cameron, Van Den Worm, Snijder, The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent, Nucleic Acids Res, doi:10.1093/nar/gkp904
Velthuis, Zubkova, Shaw, Mehle, Boltz et al., Enisamium Reduces Influenza Virus Shedding and Improves Patient Recovery by Inhibiting Viral RNA Polymerase Activity, Antimicrob Agents Chemother, doi:10.1128/AAC.02605-20
Vial, Oade, Russell, Eggink, Velthuis, A SARS-CoV-2 mini-genome assay based on negative-sense RNA to study replication inhibitors and emerging mutations, BioRxiv, doi:10.1101/2021.06.28.450211
Walker, Fan, Keown, Margitich, Grimes et al., Enisamium is a small molecule inhibitor of the influenza A virus and SARS-CoV-2 RNA polymerases, bioRxiv, doi:10.1101/2020.04.21.053017
Weis, Katebzadeh, Söderhjelm, Nilsson, Ryde, Ligand affinities predicted with the MM/PBSA method: Dependence on the simulation method and the force field, J. Med. Chem, doi:10.1021/jm0608210
Yan, Zhang, Ge, Zheng, Gao et al., Architecture of a SARS-CoV-2 mini replication and transcription complex, Nat. Commun, doi:10.1038/s41467-020-19770-1
Yin, Mao, Luan, Shen, Shen et al., Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir, Science, doi:10.1126/science.abc1560
Zurbaev, Slita, Sinegubova, Muryleva, Lavrentieva, Anti-viral activity of enisamium iodide against viruses of influenza and ARVI's on different cell lines, Ther. Arch, doi:10.26442/00403660.2020.11.000872
DOI record:
{
"DOI": "10.3390/biomedicines9091254",
"ISSN": [
"2227-9059"
],
"URL": "http://dx.doi.org/10.3390/biomedicines9091254",
"abstract": "<jats:p>Pandemic SARS-CoV-2 causes a mild to severe respiratory disease called coronavirus disease 2019 (COVID-19). While control of the SARS-CoV-2 spread partly depends on vaccine-induced or naturally acquired protective herd immunity, antiviral strategies are still needed to manage COVID-19. Enisamium is an inhibitor of influenza A and B viruses in cell culture and clinically approved in countries of the Commonwealth of Independent States. In vitro, enisamium acts through metabolite VR17-04 and inhibits the activity of the influenza A virus RNA polymerase. Here we show that enisamium can inhibit coronavirus infections in NHBE and Caco-2 cells, and the activity of the SARS-CoV-2 RNA polymerase in vitro. Docking and molecular dynamics simulations provide insight into the mechanism of action and indicate that enisamium metabolite VR17-04 prevents GTP and UTP incorporation. Overall, these results suggest that enisamium is an inhibitor of SARS-CoV-2 RNA synthesis in vitro.</jats:p>",
"alternative-id": [
"biomedicines9091254"
],
"author": [
{
"ORCID": "https://orcid.org/0000-0003-0686-2480",
"affiliation": [
{
"name": "Istituto di Ricerche Chimiche e Biochimiche “G. Ronzoni”, Via Giuseppe Colombo 81, 20133 Milano, Italy"
}
],
"authenticated-orcid": false,
"family": "Elli",
"given": "Stefano",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Institute of Medical Virology, University Hospital Frankfurt am Main, Goethe University, Paul-Ehrlich-Straße 40, 60596 Frankfurt am Main, Germany"
}
],
"family": "Bojkova",
"given": "Denisa",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Medical Virology, University Hospital Frankfurt am Main, Goethe University, Paul-Ehrlich-Straße 40, 60596 Frankfurt am Main, Germany"
}
],
"family": "Bechtel",
"given": "Marco",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Division of Virology, Department of Pathology, Addenbrooke’s Hospital, University of Cambridge, Hills Road, Cambridge CB2 2QQ, UK"
}
],
"family": "Vial",
"given": "Thomas",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "IIT Research Institute, 10 W 35th St, Chicago, IL 60616, USA"
}
],
"family": "Boltz",
"given": "David",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "IIT Research Institute, 10 W 35th St, Chicago, IL 60616, USA"
}
],
"family": "Muzzio",
"given": "Miguel",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "IIT Research Institute, 10 W 35th St, Chicago, IL 60616, USA"
}
],
"family": "Peng",
"given": "Xinjian",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Istituto di Ricerche Chimiche e Biochimiche “G. Ronzoni”, Via Giuseppe Colombo 81, 20133 Milano, Italy"
}
],
"family": "Sala",
"given": "Federico",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Istituto di Ricerche Chimiche e Biochimiche “G. Ronzoni”, Via Giuseppe Colombo 81, 20133 Milano, Italy"
}
],
"family": "Cosentino",
"given": "Cesare",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Farmak Joint Stock Company, Kyrylivska Street, 04080 Kyiv, Ukraine"
}
],
"family": "Goy",
"given": "Andrew",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Istituto di Ricerche Chimiche e Biochimiche “G. Ronzoni”, Via Giuseppe Colombo 81, 20133 Milano, Italy"
}
],
"family": "Guerrini",
"given": "Marco",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-3633-1309",
"affiliation": [
{
"name": "Regenold GmbH, Zöllinplatz 4, 79410 Badenweiler, Germany"
}
],
"authenticated-orcid": false,
"family": "Müller",
"given": "Lutz",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Institute of Medical Virology, University Hospital Frankfurt am Main, Goethe University, Paul-Ehrlich-Straße 40, 60596 Frankfurt am Main, Germany"
}
],
"family": "Cinatl",
"given": "Jindrich",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Farmak Joint Stock Company, Kyrylivska Street, 04080 Kyiv, Ukraine"
}
],
"family": "Margitich",
"given": "Victor",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-5129-3953",
"affiliation": [
{
"name": "Division of Virology, Department of Pathology, Addenbrooke’s Hospital, University of Cambridge, Hills Road, Cambridge CB2 2QQ, UK"
}
],
"authenticated-orcid": false,
"family": "te Velthuis",
"given": "Aartjan J. W.",
"sequence": "additional"
}
],
"container-title": "Biomedicines",
"container-title-short": "Biomedicines",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2021,
9,
18
]
],
"date-time": "2021-09-18T01:23:29Z",
"timestamp": 1631928209000
},
"deposited": {
"date-parts": [
[
2024,
7,
18
]
],
"date-time": "2024-07-18T23:31:43Z",
"timestamp": 1721345503000
},
"funder": [
{
"DOI": "10.13039/100004440",
"award": [
"206579/Z/17/Z"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/100004440",
"id-type": "DOI"
}
],
"name": "Wellcome"
},
{
"DOI": "10.13039/501100001826",
"award": [
"10430 01 201 0018"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100001826",
"id-type": "DOI"
}
],
"name": "ZonMw"
}
],
"indexed": {
"date-parts": [
[
2025,
2,
21
]
],
"date-time": "2025-02-21T20:46:35Z",
"timestamp": 1740170795183,
"version": "3.37.3"
},
"is-referenced-by-count": 5,
"issue": "9",
"issued": {
"date-parts": [
[
2021,
9,
17
]
]
},
"journal-issue": {
"issue": "9",
"published-online": {
"date-parts": [
[
2021,
9
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2021,
9,
17
]
],
"date-time": "2021-09-17T00:00:00Z",
"timestamp": 1631836800000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/2227-9059/9/9/1254/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "1254",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2021,
9,
17
]
]
},
"published-online": {
"date-parts": [
[
2021,
9,
17
]
]
},
"publisher": "MDPI AG",
"reference": [
{
"DOI": "10.1126/scitranslmed.abb5883",
"article-title": "An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice",
"author": "Sheahan",
"doi-asserted-by": "crossref",
"first-page": "541",
"journal-title": "Sci. Transl. Med.",
"key": "ref_1",
"volume": "12",
"year": "2020"
},
{
"DOI": "10.1038/s41564-020-00835-2",
"article-title": "Therapeutically administered ribonucleoside analogue MK-4482/EIDD-2801 blocks SARS-CoV-2 transmission in ferrets",
"author": "Cox",
"doi-asserted-by": "crossref",
"first-page": "11",
"journal-title": "Nat. Microbiol.",
"key": "ref_2",
"volume": "6",
"year": "2021"
},
{
"DOI": "10.1038/s41467-020-18463-z",
"article-title": "Rapid incorporation of Favipiravir by the fast and permissive viral RNA polymerase complex results in SARS-CoV-2 lethal mutagenesis",
"author": "Shannon",
"doi-asserted-by": "crossref",
"first-page": "1",
"journal-title": "Nat. Commun.",
"key": "ref_3",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1073/pnas.1811345115",
"article-title": "The mechanism of resistance to favipiravir in influenza",
"author": "Goldhill",
"doi-asserted-by": "crossref",
"first-page": "11613",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_4",
"volume": "115",
"year": "2018"
},
{
"DOI": "10.1074/jbc.REV120.013930",
"article-title": "The molecular virology of coronaviruses",
"author": "Hartenian",
"doi-asserted-by": "crossref",
"first-page": "12910",
"journal-title": "J. Biol. Chem.",
"key": "ref_5",
"volume": "295",
"year": "2020"
},
{
"DOI": "10.1038/s41564-020-0695-z",
"doi-asserted-by": "crossref",
"key": "ref_6",
"unstructured": "Coronaviridae Study Group of the International Committee on Taxonomy of V (2020). The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol., 5, 536–544."
},
{
"DOI": "10.1016/j.antiviral.2013.11.006",
"article-title": "SARS-CoV ORF1b-encoded nonstructural proteins 12-16: Replicative enzymes as antiviral targets",
"author": "Subissi",
"doi-asserted-by": "crossref",
"first-page": "122",
"journal-title": "Antivir. Res.",
"key": "ref_7",
"volume": "101",
"year": "2014"
},
{
"DOI": "10.1093/nar/gkp904",
"article-title": "The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent",
"author": "Arnold",
"doi-asserted-by": "crossref",
"first-page": "203",
"journal-title": "Nucleic Acids Res.",
"key": "ref_8",
"volume": "38",
"year": "2010"
},
{
"DOI": "10.1073/pnas.1323705111",
"article-title": "One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities",
"author": "Subissi",
"doi-asserted-by": "crossref",
"first-page": "E3900",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_9",
"volume": "111",
"year": "2014"
},
{
"DOI": "10.1038/s41467-020-19770-1",
"article-title": "Architecture of a SARS-CoV-2 mini replication and transcription complex",
"author": "Yan",
"doi-asserted-by": "crossref",
"first-page": "5874",
"journal-title": "Nat. Commun.",
"key": "ref_10",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1038/s41586-020-2368-8",
"article-title": "Structure of replicating SARS-CoV-2 polymerase",
"author": "Hillen",
"doi-asserted-by": "crossref",
"first-page": "154",
"journal-title": "Nature",
"key": "ref_11",
"volume": "584",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.07.033",
"article-title": "Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex",
"author": "Chen",
"doi-asserted-by": "crossref",
"first-page": "1560",
"journal-title": "Cell",
"key": "ref_12",
"volume": "182",
"year": "2020"
},
{
"DOI": "10.1128/AAC.02605-20",
"doi-asserted-by": "crossref",
"key": "ref_13",
"unstructured": "Te Velthuis, A.J.W., Zubkova, T.G., Shaw, M., Mehle, A., Boltz, D., Gmeinwieser, N., Stammer, H., Milde, J., Muller, L., and Margitich, V. (2021). Enisamium Reduces Influenza Virus Shedding and Improves Patient Recovery by Inhibiting Viral RNA Polymerase Activity. Antimicrob Agents Chemother, 65."
},
{
"DOI": "10.1101/2021.06.28.450211",
"doi-asserted-by": "crossref",
"key": "ref_14",
"unstructured": "Vial, T., Oade, M.S., Russell, C.A., Eggink, D., and te Velthuis, A.J.W. (2021). A SARS-CoV-2 mini-genome assay based on negative-sense RNA to study replication inhibitors and emerging mutations. BioRxiv."
},
{
"DOI": "10.1101/2020.04.21.053017",
"doi-asserted-by": "crossref",
"key": "ref_15",
"unstructured": "Walker, A.P., Fan, H., Keown, J.R., Margitich, V., Grimes, J.M., Fodor, E., and Te Velthuis, A.J.W. (2021). Enisamium is a small molecule inhibitor of the influenza A virus and SARS-CoV-2 RNA polymerases. bioRxiv."
},
{
"DOI": "10.1056/NEJMoa2007764",
"article-title": "Remdesivir for the Treatment of Covid-19—Final Report",
"author": "Beigel",
"doi-asserted-by": "crossref",
"first-page": "1813",
"journal-title": "N. Engl. J. Med.",
"key": "ref_16",
"volume": "383",
"year": "2020"
},
{
"DOI": "10.1177/2040206618811416",
"article-title": "Activity of enisamium, an isonicotinic acid derivative, against influenza viruses in differentiated normal human bronchial epithelial cells",
"author": "Boltz",
"doi-asserted-by": "crossref",
"first-page": "2040206618811416",
"journal-title": "Antivir Chem. Chemother.",
"key": "ref_17",
"volume": "26",
"year": "2018"
},
{
"DOI": "10.26442/00403660.2020.11.000872",
"article-title": "Anti-viral activity of enisamium iodide against viruses of influenza and ARVI’s on different cell lines",
"author": "Zurbaev",
"doi-asserted-by": "crossref",
"first-page": "45",
"journal-title": "Ther. Arch.",
"key": "ref_18",
"volume": "92",
"year": "2020"
},
{
"DOI": "10.1038/s41586-020-2332-7",
"article-title": "Proteomics of SARS-CoV-2-infected host cells reveals therapy targets",
"author": "Bojkova",
"doi-asserted-by": "crossref",
"first-page": "469",
"journal-title": "Nature",
"key": "ref_19",
"volume": "583",
"year": "2020"
},
{
"DOI": "10.3390/cells9112377",
"doi-asserted-by": "crossref",
"key": "ref_20",
"unstructured": "Bojkova, D., Bechtel, M., McLaughlin, K.M., McGreig, J.E., Klann, K., Bellinghausen, C., Rohde, G., Jonigk, D., Braubach, P., and Ciesek, S. (2020). Aprotinin Inhibits SARS-CoV-2 Replication. Cells, 9."
},
{
"DOI": "10.1101/2020.04.20.052258",
"doi-asserted-by": "crossref",
"key": "ref_21",
"unstructured": "Toptan, T., Hoehl, S., Westhaus, S., Bojkova, D., Berger, A., Rotter, B., Hoffmeier, K., Cinatl, J., Ciesek, S., and Widera, M. (2020). Optimized qRT-PCR Approach for the Detection of Intra- and Extra-Cellular SARS-CoV-2 RNAs. Int. J. Mol. Sci., 21."
},
{
"key": "ref_22",
"unstructured": "Frisch, M.J., Trucks, G.W., Schlegel, H.B., Scuseria, G.E., Robb, M.A., Cheeseman, J.R., Scalmani, G., Barone, V., Petersson, G.A., and Nakatsuji, H. (2016). Gaussian 16, Revision C.01, Gaussian Inc."
},
{
"DOI": "10.1002/jcc.21256",
"article-title": "AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility",
"author": "Morris",
"doi-asserted-by": "crossref",
"first-page": "2785",
"journal-title": "J. Comput. Chem.",
"key": "ref_23",
"volume": "30",
"year": "2009"
},
{
"DOI": "10.1126/science.abc1560",
"article-title": "Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir",
"author": "Yin",
"doi-asserted-by": "crossref",
"first-page": "1499",
"journal-title": "Science",
"key": "ref_24",
"volume": "368",
"year": "2020"
},
{
"DOI": "10.1016/0040-4020(80)80168-2",
"article-title": "Iterative partial equalization of orbital electronegativity a rapid access to atomic charges",
"author": "Gesteiger",
"doi-asserted-by": "crossref",
"first-page": "3219",
"journal-title": "Tetrahedron",
"key": "ref_25",
"volume": "36",
"year": "1980"
},
{
"DOI": "10.1002/jcc.20289",
"article-title": "Scalable molecular dynamics with NAMD",
"author": "Phillips",
"doi-asserted-by": "crossref",
"first-page": "1781",
"journal-title": "J. Comput. Chem.",
"key": "ref_26",
"volume": "26",
"year": "2005"
},
{
"DOI": "10.1002/jcc.20290",
"article-title": "The Amber biomolecular simulation programs",
"author": "Case",
"doi-asserted-by": "crossref",
"first-page": "1668",
"journal-title": "J. Comput. Chem.",
"key": "ref_27",
"volume": "26",
"year": "2005"
},
{
"DOI": "10.1063/1.445869",
"article-title": "Comparison of simple potential functions for simulating liquid water",
"author": "Jorgensen",
"doi-asserted-by": "crossref",
"first-page": "926",
"journal-title": "J. Chem. Phys.",
"key": "ref_28",
"volume": "79",
"year": "1983"
},
{
"DOI": "10.1016/0263-7855(96)00018-5",
"article-title": "VMD: Visual molecular dynamics",
"author": "Humphrey",
"doi-asserted-by": "crossref",
"first-page": "27",
"journal-title": "J. Mol. Graph.",
"key": "ref_29",
"volume": "14",
"year": "1996"
},
{
"DOI": "10.1021/jm0608210",
"article-title": "Ligand affinities predicted with the MM/PBSA method: Dependence on the simulation method and the force field",
"author": "Weis",
"doi-asserted-by": "crossref",
"first-page": "6596",
"journal-title": "J. Med. Chem.",
"key": "ref_30",
"volume": "49",
"year": "2006"
},
{
"DOI": "10.1021/ja981844+",
"article-title": "Continuum solvent studies of the stability of DNA, RNA, and phosphoramidate–DNA helices",
"author": "Srinivasan",
"doi-asserted-by": "crossref",
"first-page": "9401",
"journal-title": "J. Am. Chem. Soc.",
"key": "ref_31",
"volume": "120",
"year": "1998"
}
],
"reference-count": 31,
"references-count": 31,
"relation": {
"has-preprint": [
{
"asserted-by": "object",
"id": "10.1101/2021.01.05.21249237",
"id-type": "doi"
}
]
},
"resource": {
"primary": {
"URL": "https://www.mdpi.com/2227-9059/9/9/1254"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Enisamium Inhibits SARS-CoV-2 RNA Synthesis",
"type": "journal-article",
"volume": "9"
}