Mechanisms of Cell–Cell Fusion in SARS-CoV-2: An Evolving Strategy for Transmission and Immune Evasion
et al., Viruses, doi:10.3390/v17111405, Oct 2025
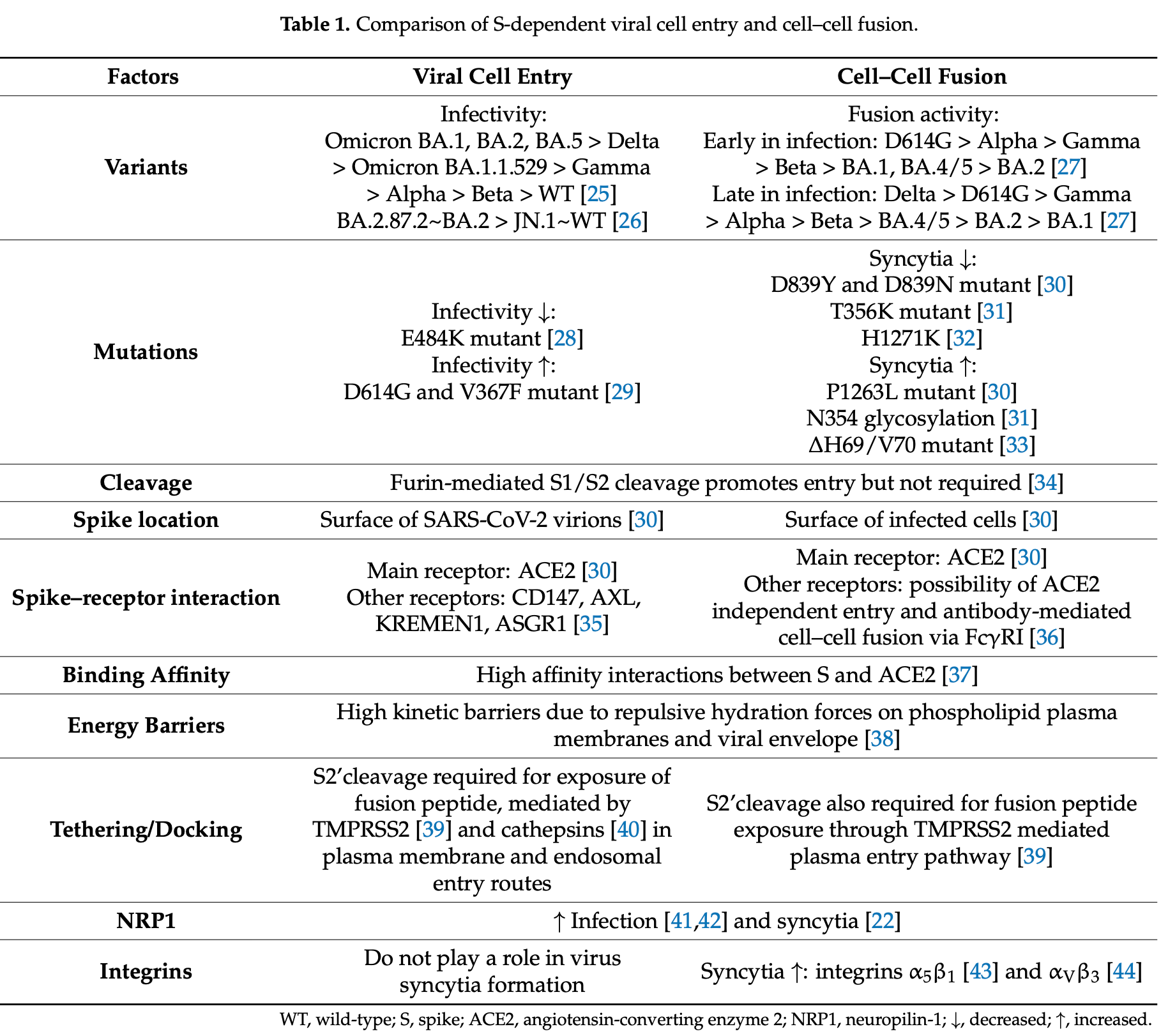
Review showing that SARS-CoV-2 cell-cell fusion represents an evolving strategy for immune evasion and viral persistence through syncytia formation. Authors note that spike protein-mediated fusion of infected cells with neighboring cells bypasses neutralizing antibodies while contributing to tissue damage and inflammatory responses. Fusion efficiency varies across SARS-CoV-2 variants, with Delta variants exhibiting enhanced fusogenicity compared to early Omicron variants, though recent Omicron subvariants have regained fusion capacity. The study implicates syncytia formation in lymphocyte depletion, pyroptotic cell death, and ongoing inflammation, suggesting that targeting fusion mechanisms could reduce COVID-19 pathogenicity.
Chiang et al., 22 Oct 2025, peer-reviewed, 5 authors.
Contact: rkg20@cam.ac.uk (corresponding author), kate.chiang@ucdconnect.ie.
Mechanisms of Cell–Cell Fusion in SARS-CoV-2: An Evolving Strategy for Transmission and Immune Evasion
Viruses, doi:10.3390/v17111405
Early studies on the evolution of SARS-CoV-2 revealed mutations that favored host transmission of the virus and more efficient viral entry. However, cell-free virus spread is vulnerable to host-neutralizing antibodies. As population immunity developed, mutations that confer escape from neutralization were selected. Notably, cell syncytia formation wherein an infected cell fuses with a noninfected cell is a more efficient route of transmission that bypasses humoral immunity. Cell syncytia formation has been implicated in the pathogenicity of SARS-CoV-2 infection whilst compromising host transmission due to impaired whole virion release. Therefore, understanding the mechanisms of virusmediated cell-cell fusion will aid in identifying and targeting more pathogenic strains of SARS-CoV-2. Whilst the general kinetics of cell-cell fusion have been known for decades, the specific mechanisms by which SARS-CoV-2 induces fusion are beginning to be elucidated. This is partially due to emergence of more reliable, high throughput methods of quantifying and comparing fusion efficiency in experimental models. Moreover, the ongoing inflammatory response and emerging health burden of long COVID may point to cell-cell fusion in the pathogenesis. In this review, we synthesize current understanding of SARS-CoV-2-mediated cell-cell fusion and its consequences on immune escape, viral persistence, and the innate immune response.
Abbreviations
References
Ackermann, Anders, Bilyy, Bowlin, Daniel et al., Patients with COVID-19: In the dark-NETs of neutrophils, Cell Death Differ, doi:10.1038/s41418-021-00805-z
Ackermann, Verleden, Kuehnel, Haverich, Welte et al., Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid-19, N. Engl. J. Med, doi:10.1056/NEJMoa2015432
Allolio, Harries, Calcium Ions Promote Membrane Fusion by Forming Negative-Curvature Inducing Clusters on Specific Anionic Lipids, ACS Nano, doi:10.1021/acsnano.0c08614
Amidei, Dobrovolny, Virus-mediated cell fusion of SARS-CoV-2 variants, Math. Biosci, doi:10.1016/j.mbs.2024.109144
Arora, Nehlmeier, Kempf, Cossmann, Schulz et al., Lung cell entry, cell-cell fusion capacity, and neutralisation sensitivity of omicron sublineage BA.2.75, Lancet Infect. Dis, doi:10.1016/S1473-3099(22)00591-6
Barrett, Neal, Edmonds, Moncman, Thompson et al., Effect of clinical isolate or cleavage site mutations in the SARS-CoV-2 spike protein on protein stability, cleavage, and cell-cell fusion, J. Biol. Chem, doi:10.1016/j.jbc.2021.100902
Barthe, Hertereau, Lamghari, Osman-Ponchet, Braud, Receptors and Cofactors That Contribute to SARS-CoV-2 Entry: Can Skin Be an Alternative Route of Entry?, Int. J. Mol. Sci, doi:10.3390/ijms24076253
Bertram, Dijkman, Habjan, Heurich, Gierer et al., TMPRSS2 Activates the Human Coronavirus 229E for Cathepsin-Independent Host Cell Entry and Is Expressed in Viral Target Cells in the Respiratory Epithelium, J. Virol, doi:10.1128/JVI.03372-12
Bhardwaj, Archana; Noumani, Himanshu, Chakravorty, Solanki, Recent advancement in the detection of potential cancer biomarkers using the nanomaterial integrated electrochemical sensing technique: A detailed review, Mater. Adv, doi:10.1039/D3MA00621B
Bignon, Gruet, Longhi, Split-GFP Reassembly Assay: Strengths and Caveats from a Multiparametric Analysis, Int. J. Mol. Sci, doi:10.3390/ijms232113167
Biswas, Yin, Blank, Zimmerberg, Cholesterol Promotes Hemifusion and Pore Widening in Membrane Fusion Induced by Influenza Hemagglutinin, J. Gen. Physiol, doi:10.1085/jgp.200709932
Blank, Burgoon, Baldridge, Mc, Urbach et al., None, J. Am. Med. Assoc, doi:10.1001/jama.1951.63670150005012b
Bolland, Marechal, Petiot, Porrot, Guivel-Benhassine et al., SARS-CoV-2 entry and fusion are independent of ACE2 localization to lipid rafts, bioRxiv, doi:10.1128/jvi.01823-24
Bosch, Van Der Zee, De Haan, Rottier, The Coronavirus Spike Protein Is a Class I Virus Fusion Protein: Structural and Functional Characterization of the Fusion Core Complex, J. Virol, doi:10.1128/JVI.77.16.8801-8811.2003
Boson, Legros, Zhou, Siret, Mathieu et al., The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles, J. Biol. Chem, doi:10.1074/jbc.RA120.016175
Bracq, Xie, Lambelé, Vu, Matz et al., T Cell-Macrophage Fusion Triggers Multinucleated Giant Cell Formation for HIV-1 Spreading, J. Virol, doi:10.1128/JVI.01237-17
Bratt, Gallaher, Comparison of Fusion from within and Fusion from without by Newcastle Disease Virus, Vitr. Cell. Dev. Biol. Plant, doi:10.1007/BF02616129
Bricogne, Fine, Pereira, Sung, Tijani et al., TMEM16F activation by Ca 2+ triggers plasma membrane expansion and directs PD-1 trafficking, Sci. Rep, doi:10.1038/s41598-018-37056-x
Buchrieser, Dufloo, Hubert, Monel, Planas et al., Syncytia formation by SARS--CoV--2--infected cells, EMBO J, doi:10.15252/embj.2020106267
Buonsenso, Tantisira, Long COVID and SARS-CoV-2 persistence: New answers, more questions, Lancet Infect. Dis, doi:10.1016/S1473-3099(24)00216-0
Burkova, Bakhno, Sequences in the Cytoplasmic Tail Contribute to the Intracellular Trafficking and the Cell Surface Localization of SARS-CoV-2 Spike Protein, Biomolecules, doi:10.3390/biom15020280
Bussani, Schneider, Zentilin, Collesi, Ali et al., Persistence of viral RNA, pneumocyte syncytia and thrombosis are hallmarks of advanced COVID-19 pathology, eBioMedicine, doi:10.1016/j.ebiom.2020.103104
Bussani, Zentilin, Correa, Colliva, Silvestri et al., Persistent SARS-CoV-2 infection in patients seemingly recovered from COVID-19, J. Pathol, doi:10.1002/path.6035
Cabantous, Nguyen, Pedelacq, Koraïchi, Chaudhary et al., A New Protein-Protein Interaction Sensor Based on Tripartite Split-GFP Association, Sci. Rep, doi:10.1038/srep02854
Cafaro, Schietroma, Sernicola, Belli, Campagna et al., Role of HIV-1 Tat Protein Interactions with Host Receptors in HIV Infection and Pathogenesis, Int. J. Mol. Sci, doi:10.3390/ijms25031704
Cantuti-Castelvetri, Ojha, Pedro, Djannatian, Franz et al., Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity, Science, doi:10.1126/science.abd2985
Carabelli, Peacock, Thorne, Harvey, Hughes et al., SARS-CoV-2 variant biology: Immune escape, transmission and fitness, Nat. Rev. Microbiol, doi:10.1038/s41579-022-00841-7
Carrascosa-Sàez, Marqués, Geller, Elena, Rahmeh et al., Cell type-specific adaptation of the SARS-CoV-2 spike, Virus Evol, doi:10.1093/ve/veae032
Carten, Khelashvili, Bidon, Straus, Tang et al., A Mechanistic Understanding of the Modes of Ca 2+ Ion Binding to the SARS-CoV-1 Fusion Peptide and Their Role in the Dynamics of Host Membrane Penetration, ACS Infect. Dis, doi:10.1021/acsinfecdis.3c00260
Cattin-Ortolá, Welch, Maslen, Papa, James et al., Sequences in the cytoplasmic tail of SARS-CoV-2 Spike facilitate expression at the cell surface and syncytia formation, Nat. Commun, doi:10.1038/s41467-021-25589-1
Chaudhary, Yadav, Kumar, Yadav, Ultrastructural study confirms the formation of single and heterotypic syncytial cells in bronchoalveolar fluids of COVID-19 patients, Virol. J, doi:10.1186/s12985-023-02062-7
Chen, Appelman, Willemen, Bos, Prado et al., Transfer of IgG from Long COVID patients induces symptomology in mice, bioRxiv, doi:10.1101/2024.05.30.596590
Cheng, Altaf, Castin, Reuschl, Sievers et al., Signatures of omicron-like adaptation in early SARS-CoV-2 variants and chronic infection, Cell Rep, doi:10.1016/j.celrep.2025.116135
Chippa, Aleem, Anjum, Postacute Coronavirus (COVID-19) Syndrome
Clemens, Ye, Zhou, Kim, Pease et al., SARS-CoV-2 spike protein-mediated cardiomyocyte fusion may contribute to increased arrhythmic risk in COVID-19, PLoS ONE, doi:10.1371/journal.pone.0282151
Coish, Macneil, Macneil, The SARS-CoV-2 antibody-dependent enhancement façade, Microbes Infect, doi:10.1016/j.micinf.2024.105464
Collier, De Marco, Ferreira, Meng, Datir et al., Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies, Nature, doi:10.1038/s41586-021-03412-7
Daly, Simonetti, Klein, Chen, Williamson et al., Neuropilin-1 is a host factor for SARS-CoV-2 infection, Science, doi:10.1126/science.abd3072
Dey, Qing, He, Chen, Jennings et al., A single C-terminal residue controls SARS-CoV-2 spike trafficking and incorporation into VLPs, Nat. Commun, doi:10.1038/s41467-023-44076-3
Dimitrov, Broder, Berger, Blumenthal, Calcium ions are required for cell fusion mediated by the CD4-human immunodeficiency virus type 1 envelope glycoprotein interaction, J. Virol, doi:10.1128/jvi.67.3.1647-1652.1993
Doherty, Mcmahon, Mechanisms of endocytosis, Annu. Rev. Biochem, doi:10.1146/annurev.biochem.78.081307.110540
Duelli, Lazebnik, Cell-to-cell fusion as a link between viruses and cancer, Nat. Rev. Cancer, doi:10.1038/nrc2272
Dufloo, Sanjuán, Temperature impacts SARS-CoV-2 spike fusogenicity and evolution, mBio, doi:10.1128/mbio.03360-23
Duncan, Fusogenic Reoviruses and Their Fusion-Associated Small Transmembrane (FAST) Proteins, Annu. Rev. Virol, doi:10.1146/annurev-virology-092818-015523
Duprex, Dutch, Paramyxoviruses, Pathogenesis, Vaccines, Antivirals, and Prototypes for Pandemic Preparedness, J. Infect. Dis, doi:10.1093/infdis/jiad123
El Najjar, Schmitt, Dutch, Paramyxovirus Glycoprotein Incorporation, Assembly and Budding: A Three Way Dance for Infectious Particle Production, Viruses, doi:10.3390/v6083019
Furnon, Cowton, De Lorenzo, Orton, Herder et al., Phenotypic evolution of SARS-CoV-2 spike during the COVID-19 pandemic, Nat. Microbiol, doi:10.1038/s41564-024-01878-5
García-Murria, Expósito-Domínguez, Duart, Mingarro, Martinez-Gil, A Bimolecular Multicellular Complementation System for the Detection of Syncytium Formation: A New Methodology for the Identification of Nipah Virus Entry Inhibitors, Viruses, doi:10.3390/v11030229
García-Murria, Gadea-Salom, Moreno, Rius-Salvador, Zaragoza et al., Identification of small molecules capable of enhancing viral membrane fusion, Virol. J, doi:10.1186/s12985-023-02068-1
Gatignol, Duarte, Daviet, Chang, Jeang, Sequential steps in Tat trans-activation of HIV-1 mediated through cellular DNA, RNA, and protein binding factors, Gene Expr
Graham, Seow, Huettner, Khan, Kouphou et al., Neutralization potency of monoclonal antibodies recognizing dominant and subdominant epitopes on SARS-CoV-2 Spike is impacted by the B.1.1.7 variant, Immunity, doi:10.1016/j.immuni.2021.03.023
Green, Sambrook, Screening Bacterial Colonies Using X-Gal and IPTG: α-Complementation, Cold Spring Harb. Protoc, doi:10.1101/pdb.prot101329
Hastie, Lowe, Mcauley, Mills, Winter et al., True prevalence of long-COVID in a nationwide, population cohort study, Nat. Commun, doi:10.1038/s41467-023-43661-w
Hernández, Podbilewicz, The hallmarks of cell-cell fusion, Development, doi:10.1242/dev.155523
Holland, Munk, Lucero, Nguyen, Landau, α-Complementation assay for HIV envelope glycoproteinmediated fusion, Virology, doi:10.1016/j.virol.2003.11.012
Hörnich, Großkopf, Schlagowski, Tenbusch, Kleine-Weber et al., SARS-CoV-2 and SARS-CoV Spike-Mediated Cell-Cell Fusion Differ in Their Requirements for Receptor Expression and Proteolytic Activation, J. Virol, doi:10.1128/JVI.00002-21
Jackson, Farzan, Chen, Choe, Mechanisms of SARS-CoV-2 entry into cells, Nat. Rev. Mol. Cell Biol, doi:10.1038/s41580-021-00418-x
Kelly, Yang, Application of Fluorescence-and Bioluminescence-Based Biosensors in Cancer Drug Discovery, Biosensors, doi:10.3390/bios14120570
Kemp, Collier, Datir, Ferreira, Gayed et al., SARS-CoV-2 evolution during treatment of chronic infection, Nature, doi:10.1038/s41586-021-03291-y
Kibria, Lavine, Tang, Wang, Gao et al., Antibody-mediated SARS-CoV-2 entry in cultured cells, EMBO Rep, doi:10.15252/embr.202357724
Kim, None
Labzin, Chew, Eschke, Wang, Esposito et al., Macrophage ACE2 is necessary for SARS-CoV-2 replication and subsequent cytokine responses that restrict continued virion release, Sci. Signal, doi:10.1126/scisignal.abq1366
Lai, Freed, SARS-CoV-2 Fusion Peptide has a Greater Membrane Perturbating Effect than SARS-CoV with Highly Specific Dependence on Ca 2+, J. Mol. Biol, doi:10.1016/j.jmb.2021.166946
Lamers, Haagmans, SARS-CoV-2 pathogenesis, Nat. Rev. Microbiol, doi:10.1038/s41579-022-00713-0
Lee, Menasche, Mavrikaki, Uyemura, Hong et al., Differences in syncytia formation by SARS-CoV-2 variants modify host chromatin accessibility and cellular senescence via TP53, Cell Rep, doi:10.1016/j.celrep.2023.113478
Lee, Schick, Calculation of free energy barriers to the fusion of small vesicles, Biophys J, doi:10.1529/biophysj.107.119511
Leroy, Han, Woottum, Bracq, Bouchet et al., Virus-Mediated Cell-Cell Fusion, Int. J. Mol. Sci, doi:10.3390/ijms21249644
Li, Liu, Faraone, Hsu, Chamblee et al., Distinct patterns of SARS-CoV-2 BA.2.87.1 and JN.1 variants in immune evasion, antigenicity, and cell-cell fusion, mBio, doi:10.1128/mbio.00751-24
Li, Liu, Zhang, Cytoplasmic tail determines the membrane trafficking and localization of SARS-CoV-2 spike protein Review, Front. Mol. Biosci, doi:10.3389/fmolb.2022.1004036
Li, Yang, Nan, Wang, Wang et al., SARS-CoV-2 spike host cell surface exposure promoted by a COPI sorting inhibitor, Acta Pharm. Sin. B, doi:10.1016/j.apsb.2023.04.007
Li, Yuan, Li, Wang, Spike protein mediated membrane fusion during SARS-CoV-2 infection, J. Med. Virol, doi:10.1002/jmv.28212
Lim, Zhang, Chang, ACE2-Independent Alternative Receptors for SARS-CoV-2, Viruses, doi:10.3390/v14112535
Lin, Li, Wang, Shi, Syncytia formation during SARS-CoV-2 lung infection: A disastrous unity to eliminate lymphocytes, Cell Death Differ, doi:10.1038/s41418-021-00795-y
Lin, Sha, Zhang, Adler, Vidal et al., A single mutation may contribute to accelerated evolution of SARS-CoV-2 toward Omicron, Nat. Commun, doi:10.1038/s41467-025-62300-0
Little, Rorick, Su, Baldock, Malhotra et al., Missense mutations that cause Van der Woude syndrome and popliteal pterygium syndrome affect the DNA-binding and transcriptional activation functions of IRF6, Hum. Mol. Genet, doi:10.1093/hmg/ddn381
Liu, Lu, Chen, Plow, Qin, Integrin mediates cell entry of the SARS-CoV-2 virus independent of cellular receptor ACE2, J. Biol. Chem, doi:10.1016/j.jbc.2022.101710
Liu, Wei, Xu, Zhao, Huang et al., SARS-CoV-2 spike protein-induced cell fusion activates the cGAS-STING pathway and the interferon response, Sci. Signal, doi:10.1126/scisignal.abg8744
Liu, Yue, Meng, Xiao, Yang et al., Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity, Natl. Sci. Rev, doi:10.1093/nsr/nwae206
Lustig, Ganga, Rodel, Tegally, Jackson et al., SARS-CoV-2 evolves increased infection elicited cell death and fusion in an immunosuppressed individual, medRxiv, doi:10.1101/2022.11.23.22282673
Ma, Ng, Zappia, Gearing, Olshansky et al., Unique Transcriptional Architecture in Airway Epithelial Cells and Macrophages Shapes Distinct Responses following Influenza Virus Infection Ex Vivo, J. Virol, doi:10.1128/JVI.01986-18
Ma, Zhu, Lin, Wang, Zhang et al., Pyroptosis of syncytia formed by fusion of SARS-CoV-2 spike and ACE2-expressing cells, Cell Discov, doi:10.1038/s41421-021-00310-0
Mangeney, Renard, Schlecht-Louf, Bouallaga, Heidmann et al., Placental syncytins: Genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins, Proc. Natl. Acad. Sci, doi:10.1073/pnas.0707873105
Martens, Mcmahon, Mechanisms of membrane fusion: Disparate players and common principles, Nat. Rev. Mol. Cell Biol, doi:10.1038/nrm2417
Martínez-Mármol, Giordano-Santini, Kaulich, Cho, Przybyla et al., SARS-CoV-2 infection and viral fusogens cause neuronal and glial fusion that compromises neuronal activity, Sci. Adv, doi:10.1126/sciadv.adg2248
Massoud, Paulmurugan, Chapter 47-Molecular Imaging of Protein-Protein Interactions and Protein Folding, Molecular Imaging
Mcbride, Li, Machamer, The Cytoplasmic Tail of the Severe Acute Respiratory Syndrome Coronavirus Spike Protein Contains a Novel Endoplasmic Reticulum Retrieval Signal That Binds COPI and Promotes Interaction with Membrane Protein, J. Virol, doi:10.1128/JVI.02146-06
Meng, Abdullahi, Ferreira, Goonawardane, Saito et al., Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity, Nature, doi:10.1038/s41586-022-04474-x
Meng, Kemp, Papa, Datir, Ferreira et al., Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1, Cell Rep, doi:10.1016/j.celrep.2021.109292
Mlcochova, Kemp, Dhar, Papa, Meng et al., SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion, Nature, doi:10.1038/s41586-021-03944-y
Müller, Igwe, Wiechert, Oldiges, Scaling production of GFP1-10 detector protein in E. coli for secretion screening by split GFP assay, Microb. Cell Factories, doi:10.1186/s12934-021-01672-6
Ou, Zhou, Dai, Zhang, Zhao et al., V367F Mutation in SARS-CoV-2 Spike RBD Emerging during the Early Transmission Phase Enhances Viral Infectivity through Increased Human ACE2 Receptor Binding Affinity, J Virol, doi:10.1128/JVI.00617-21
Papa, Mallery, Albecka, Welch, Cattin-Ortolá et al., Furin cleavage of SARS-CoV-2 Spike promotes but is not essential for infection and cell-cell fusion, PLoS Pathog, doi:10.1371/journal.ppat.1009246
Park, Acosta, Hoq, Khurana, Golding et al., Pyrogenic and inflammatory mediators are produced by polarized M1 and M2 macrophages activated with D-dimer and SARS-CoV-2 spike immune complexes, Cytokine, doi:10.1016/j.cyto.2023.156447
Peacock, Goldhill, Zhou, Baillon, Frise et al., The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets, Nat. Microbiol, doi:10.1038/s41564-021-00908-w
Pickering, Wilson, Bravo, Perera, Seow et al., Antibodies to the RBD of SARS-CoV-2 spike mediate productive infection of primary human macrophages, Nat. Commun, doi:10.1038/s41467-024-54458-w
Pipchuk, Yang, Monitoring Hippo Signaling Pathway Activity Using a Luciferase-Based Large Tumor Suppressor (LATS) Biosensor, Bioluminescence: Methods and Protocols
Prévost, Richard, Gasser, Ding, Fage et al., Impact of temperature on the affinity of SARS-CoV-2 Spike glycoprotein for host ACE2, J. Biol. Chem, doi:10.1016/j.jbc.2021.101151
Queiroz, Brito, Pereira, Pereira, Amoras et al., Severe COVID-19 and long COVID are associated with high expression of STING, cGAS and IFN-α, Sci. Rep, doi:10.1038/s41598-024-55696-0
Quinchia, Echeverri, Cruz-Pacheco, Maldonado, Orozco, Electrochemical Biosensors for Determination of Colorectal Tumor Biomarkers, Micromachines, doi:10.3390/mi11040411
Raman, Mccracken, Cassar, Moss, Finnigan et al., Multiorgan MRI findings after hospitalisation with COVID-19 in the UK (C-MORE): A prospective, multicentre, observational cohort study, Lancet Respir. Med, doi:10.1016/S2213-2600(23)00262-X
Rand, Parsegian, Physical force considerations in model and biological membranes, Can. J. Biochem. Cell Biol, doi:10.1139/o84-097
Reinholm, Maljanen, Jalkanen, Altan, Tauriainen et al., Neutralizing antibodies after the third COVID-19 vaccination in healthcare workers with or without breakthrough infection, Commun. Med, doi:10.1038/s43856-024-00457-3
Reuter, Chen, Kropff, Peter, Britt et al., SARS-CoV-2 Spike Protein Is Capable of Inducing Cell-Cell Fusions Independent from Its Receptor ACE2 and This Activity Can Be Impaired by Furin Inhibitors or a Subset of Monoclonal Antibodies, Viruses, doi:10.3390/v15071500
Reyes-Alcaraz, Garcia-Rojas, Merlinsky, Seong, Bond et al., A NanoBiT assay to monitor membrane proteins trafficking for drug discovery and drug development, Commun. Biol, doi:10.1038/s42003-022-03163-9
Rocheleau, Laroche, Fu, Stewart, Mohamud et al., Identification of a High-Frequency Intrahost SARS-CoV-2 Spike Variant with Enhanced Cytopathic and Fusogenic Effects, mBio, doi:10.1128/mBio.00788-21
Ross, Gambhir, None
Sanders, Jumper, Ackerman, Bracha, Donlic et al., SARS-CoV-2 requires cholesterol for viral entry and pathological syncytia formation, eLife, doi:10.7554/eLife.65962
Sapir, Avinoam, Podbilewicz, Chernomordik, Viral and Developmental Cell Fusion Mechanisms: Conservation and Divergence, Dev. Cell, doi:10.1016/j.devcel.2007.12.008
Schaefer, Jung, Hummer, Binding of SARS-CoV-2 Fusion Peptide to Host Endosome and Plasma Membrane, J. Phys. Chem. B, doi:10.1021/acs.jpcb.1c04176
Sefik, Qu, Junqueira, Kaffe, Mirza et al., Inflammasome activation in infected macrophages drives COVID-19 pathology, Nature, doi:10.1038/s41586-022-04802-1
Shi, Cai, Zhu, Peng, Voyer et al., Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane, Nature, doi:10.1038/s41586-023-06273-4
Shum, Lee, Tam, Xia, Chung et al., Binding affinity between coronavirus spike protein and human ACE2 receptor, Comput. Struct. Biotechnol. J, doi:10.1016/j.csbj.2024.01.009
Sievers, Cheng, Csiba, Meng, Gupta, SARS-CoV-2 and innate immunity: The good, the bad, and the "goldilocks, Cell. Mol. Immunol, doi:10.1038/s41423-023-01104-y
Sigrist, Bridge, Le Mercier, A potential role for integrins in host cell entry by SARS-CoV-2, Antivir. Res, doi:10.1016/j.antiviral.2020.104759
Sim, Shin, Park, Park, Bae et al., Amelioration of SARS-CoV-2 infection by ANO6 phospholipid scramblase inhibition, Cell Rep, doi:10.1016/j.celrep.2022.111117
Sultana, Nagesha, Reddy, Ramesh, Shyamalamma et al., Computational analysis of affinity dynamics between the variants of SARS-CoV-2 spike protein (RBD) and human ACE-2 receptor, Virol. J, doi:10.1186/s12985-024-02365-3
Syed, Taha, Tabata, Chen, Ciling et al., Rapid assessment of SARS-CoV-2-evolved variants using virus-like particles, Science, doi:10.1126/science.abl6184
Teo, Veryard, Barnes, An, Jones et al., Circular forms of unintegrated human immunodeficiency virus type 1 DNA and high levels of viral protein expression: Association with dementia and multinucleated giant cells in the brains of patients with AIDS, J. Virol, doi:10.1128/jvi.71.4.2928-2933.1997
Theuerkauf, Michels, Riechert, Maier, Flory et al., Quantitative assays reveal cell fusion at minimal levels of SARS-CoV-2 spike protein and fusion from without, iScience, doi:10.1016/j.isci.2021.102170
Tieu, Espey, Narayanan, Heise, Alem et al., SARS-CoV-2 S-protein expression drives syncytia formation in endothelial cells, Sci. Rep, doi:10.1038/s41598-025-86242-1
Tsai, Čiháková, Tucker, Cell-Specific Mechanisms in the Heart of COVID-19 Patients, Circ. Res, doi:10.1161/CIRCRESAHA.123.321876
Vanhulle, Doijen, Stroobants, Provinciael, Noppen et al., Cellular electrical impedance to profile SARS-CoV-2 fusion inhibitors and to assess the fusogenic potential of spike mutants, Antivir. Res, doi:10.1016/j.antiviral.2023.105587
Wang, Li, Hui, Tiwari, Zhang et al., Cholesterol 25-Hydroxylase inhibits SARS-CoV-2 and other coronaviruses by depleting membrane cholesterol, EMBO J, doi:10.15252/embj.2020106057
Wang, Yeh, Guo, Mohri, Yu et al., Impaired potency of neutralizing antibodies against cell-cell fusion mediated by SARS-CoV-2, Emerg. Microbes Infect, doi:10.1080/22221751.2023.2210237
Watterson, Robinson, Chappell, Butler, Edwards et al., A generic screening platform for inhibitors of virus induced cell fusion using cellular electrical impedance, Sci. Rep, doi:10.1038/srep22791
Whitlock, Chernomordik, Flagging fusion: Phosphatidylserine signaling in cell-cell fusion, J. Biol. Chem, doi:10.1016/j.jbc.2021.100411
Xia, Wang, Jiao, Yu, Xu et al., SARS-CoV-2 Omicron subvariants exhibit distinct fusogenicity, but similar sensitivity, to pan-CoV fusion inhibitors, Emerg. Microbes Infect, doi:10.1080/22221751.2023.2178241
Xie, Zou, Shan, Yang, Kum et al., Virus Replicons for Drug Discovery, EBioMedicine, doi:10.1016/j.ebiom.2016.09.013
Yu, Zheng, Zhou, Gao, Zhou et al., Antibody-mediated spike activation promotes cell-cell transmission of SARS-CoV-2, PLoS Pathog, doi:10.1371/journal.ppat.1011789
Zang, Case, Yutuc, Ma, Shen et al., Cholesterol 25-hydroxylase suppresses SARS-CoV-2 replication by blocking membrane fusion, Proc. Natl. Acad. Sci, doi:10.1073/pnas.2012197117
Zang, Castro, Mccune, Zeng, Rothlauf et al., TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes, Sci. Immunol, doi:10.1126/sciimmunol.abc3582
Zhang, Gong, Jiao, Neutralization heterogeneity of circulating SARS-CoV-2 variants to sera elicited by a vaccinee or convalescent, Future Virol, doi:10.2217/fvl-2021-0100
Zhang, Ma, Yang, Fan, Tian et al., Cell Fusion-Related Proteins and Signaling Pathways, and Their Roles in the Development and Progression of Cancer, Front. Cell Dev. Biol, doi:10.3389/fcell.2021.809668
Zhang, Wang, Nguyen, Watson, Lao et al., Integrin α5β1 contributes to cell fusion and inflammation mediated by SARS-CoV-2 spike via RGD-independent interaction, Proc. Natl. Acad. Sci, doi:10.1073/pnas.2311913120
Zhang, Zhang, Regulation of cGAS-STING signalling and its diversity of cellular outcomes, Nat. Rev. Immunol, doi:10.1038/s41577-024-01112-7
Zhang, Zheng, Niu, Zhang, Wang et al., SARS-CoV-2 spike protein dictates syncytium-mediated lymphocyte elimination, Cell Death Differ, doi:10.1038/s41418-021-00782-3
Zhao, Su, Castro, Tripler, Hu et al., Rapid, reliable, and reproducible cell fusion assay to quantify SARS-Cov-2 spike interaction with hACE2, PLoS Pathog, doi:10.1371/journal.ppat.1009683
Zhao, Yang, Yang, Zhang, Huang et al., Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development, Signal Transduct. Target. Ther, doi:10.1038/s41392-021-00558-8
Zheng, Zhao, Zheng, Chen, Du et al., Bat SARS-Like WIV1 coronavirus uses the ACE2 of multiple animal species as receptor and evades IFITM3 restriction via TMPRSS2 activation of membrane fusion, Emerg. Microbes Infect, doi:10.1080/22221751.2020.1787797
Zhuang, Tsukuda, Wrensch, Wing, Schilling et al., The circadian clock component BMAL1 regulates SARS-CoV-2 entry and replication in lung epithelial cells, iScience, doi:10.1016/j.isci.2021.103144
DOI record:
{
"DOI": "10.3390/v17111405",
"ISSN": [
"1999-4915"
],
"URL": "http://dx.doi.org/10.3390/v17111405",
"abstract": "<jats:p>Early studies on the evolution of SARS-CoV-2 revealed mutations that favored host transmission of the virus and more efficient viral entry. However, cell-free virus spread is vulnerable to host-neutralizing antibodies. As population immunity developed, mutations that confer escape from neutralization were selected. Notably, cell syncytia formation wherein an infected cell fuses with a noninfected cell is a more efficient route of transmission that bypasses humoral immunity. Cell syncytia formation has been implicated in the pathogenicity of SARS-CoV-2 infection whilst compromising host transmission due to impaired whole virion release. Therefore, understanding the mechanisms of virus-mediated cell–cell fusion will aid in identifying and targeting more pathogenic strains of SARS-CoV-2. Whilst the general kinetics of cell–cell fusion have been known for decades, the specific mechanisms by which SARS-CoV-2 induces fusion are beginning to be elucidated. This is partially due to emergence of more reliable, high throughput methods of quantifying and comparing fusion efficiency in experimental models. Moreover, the ongoing inflammatory response and emerging health burden of long COVID may point to cell–cell fusion in the pathogenesis. In this review, we synthesize current understanding of SARS-CoV-2-mediated cell–cell fusion and its consequences on immune escape, viral persistence, and the innate immune response.</jats:p>",
"alternative-id": [
"v17111405"
],
"author": [
{
"ORCID": "https://orcid.org/0000-0003-0999-7516",
"affiliation": [
{
"name": "University College Dublin School of Medicine, Belfield, 4 Dublin, Ireland"
},
{
"name": "Department of Medicine, University of Cambridge, Cambridge CB2 0QQ, UK"
},
{
"name": "Cambridge Institute of Therapeutic Immunology & Infectious Disease (CITIID), Cambridge CB2 0AW, UK"
}
],
"authenticated-orcid": false,
"family": "Chiang",
"given": "Kate Chander",
"sequence": "first"
},
{
"ORCID": "https://orcid.org/0009-0002-2430-2495",
"affiliation": [
{
"name": "Department of Medicine, University of Cambridge, Cambridge CB2 0QQ, UK"
},
{
"name": "Cambridge Institute of Therapeutic Immunology & Infectious Disease (CITIID), Cambridge CB2 0AW, UK"
}
],
"authenticated-orcid": false,
"family": "Chiu",
"given": "Cheng En Nicole",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0009-0006-0557-5489",
"affiliation": [
{
"name": "Department of Medicine, University of Cambridge, Cambridge CB2 0QQ, UK"
},
{
"name": "Cambridge Institute of Therapeutic Immunology & Infectious Disease (CITIID), Cambridge CB2 0AW, UK"
}
],
"authenticated-orcid": false,
"family": "Altaf",
"given": "Mazharul",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-4323-5574",
"affiliation": [
{
"name": "Department of Medicine, University of Cambridge, Cambridge CB2 0QQ, UK"
},
{
"name": "Cambridge Institute of Therapeutic Immunology & Infectious Disease (CITIID), Cambridge CB2 0AW, UK"
},
{
"name": "Cambridge University Hospitals NHS Foundation Trust, Cambridge CB2 0QQ, UK"
}
],
"authenticated-orcid": false,
"family": "Cheng",
"given": "Mark Tsz Kin",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-9751-1808",
"affiliation": [
{
"name": "Department of Medicine, University of Cambridge, Cambridge CB2 0QQ, UK"
},
{
"name": "Cambridge Institute of Therapeutic Immunology & Infectious Disease (CITIID), Cambridge CB2 0AW, UK"
},
{
"name": "Cambridge University Hospitals NHS Foundation Trust, Cambridge CB2 0QQ, UK"
},
{
"name": "Africa Health Research Institute, Durban KZN 031, South Africa"
},
{
"name": "Hong Kong Jockey Club Global Health Institute, Hong Kong, China"
}
],
"authenticated-orcid": false,
"family": "Gupta",
"given": "Ravindra K.",
"sequence": "additional"
}
],
"container-title": "Viruses",
"container-title-short": "Viruses",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2025,
10,
23
]
],
"date-time": "2025-10-23T01:14:02Z",
"timestamp": 1761182042000
},
"deposited": {
"date-parts": [
[
2025,
10,
23
]
],
"date-time": "2025-10-23T01:27:44Z",
"timestamp": 1761182864000
},
"indexed": {
"date-parts": [
[
2025,
10,
23
]
],
"date-time": "2025-10-23T02:30:52Z",
"timestamp": 1761186652052,
"version": "build-2065373602"
},
"is-referenced-by-count": 0,
"issue": "11",
"issued": {
"date-parts": [
[
2025,
10,
22
]
]
},
"journal-issue": {
"issue": "11",
"published-online": {
"date-parts": [
[
2025,
11
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
10,
22
]
],
"date-time": "2025-10-22T00:00:00Z",
"timestamp": 1761091200000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/1999-4915/17/11/1405/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "1405",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2025,
10,
22
]
]
},
"published-online": {
"date-parts": [
[
2025,
10,
22
]
]
},
"publisher": "MDPI AG",
"reference": [
{
"DOI": "10.1038/s43856-024-00457-3",
"article-title": "Neutralizing antibodies after the third COVID-19 vaccination in healthcare workers with or without breakthrough infection",
"author": "Reinholm",
"doi-asserted-by": "crossref",
"first-page": "28",
"journal-title": "Commun. Med.",
"key": "ref_1",
"volume": "4",
"year": "2024"
},
{
"DOI": "10.2217/fvl-2021-0100",
"article-title": "Neutralization heterogeneity of circulating SARS-CoV-2 variants to sera elicited by a vaccinee or convalescent",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "403",
"journal-title": "Future Virol.",
"key": "ref_2",
"volume": "17",
"year": "2022"
},
{
"DOI": "10.1038/s41579-022-00713-0",
"article-title": "SARS-CoV-2 pathogenesis",
"author": "Lamers",
"doi-asserted-by": "crossref",
"first-page": "270",
"journal-title": "Nat. Rev. Microbiol.",
"key": "ref_3",
"volume": "20",
"year": "2022"
},
{
"DOI": "10.1038/s41580-021-00418-x",
"article-title": "Mechanisms of SARS-CoV-2 entry into cells",
"author": "Jackson",
"doi-asserted-by": "crossref",
"first-page": "3",
"journal-title": "Nat. Rev. Mol. Cell Biol.",
"key": "ref_4",
"volume": "23",
"year": "2022"
},
{
"DOI": "10.1021/acs.jpcb.1c04176",
"article-title": "Binding of SARS-CoV-2 Fusion Peptide to Host Endosome and Plasma Membrane",
"author": "Schaefer",
"doi-asserted-by": "crossref",
"first-page": "7732",
"journal-title": "J. Phys. Chem. B",
"key": "ref_5",
"volume": "125",
"year": "2021"
},
{
"DOI": "10.1002/jmv.28212",
"article-title": "Spike protein mediated membrane fusion during SARS-CoV-2 infection",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "e28212",
"journal-title": "J. Med. Virol.",
"key": "ref_6",
"volume": "95",
"year": "2023"
},
{
"DOI": "10.1073/pnas.0707873105",
"article-title": "Placental syncytins: Genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins",
"author": "Mangeney",
"doi-asserted-by": "crossref",
"first-page": "20534",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_7",
"volume": "104",
"year": "2007"
},
{
"DOI": "10.3389/fcell.2021.809668",
"doi-asserted-by": "crossref",
"key": "ref_8",
"unstructured": "Zhang, H., Ma, H., Yang, X., Fan, L., Tian, S., Niu, R., Yan, M., Zheng, M., and Zhang, S. (2022). Cell Fusion-Related Proteins and Signaling Pathways, and Their Roles in the Development and Progression of Cancer. Front. Cell Dev. Biol., 9."
},
{
"DOI": "10.1038/nrc2272",
"article-title": "Cell-to-cell fusion as a link between viruses and cancer",
"author": "Duelli",
"doi-asserted-by": "crossref",
"first-page": "968",
"journal-title": "Nat. Rev. Cancer",
"key": "ref_9",
"volume": "7",
"year": "2007"
},
{
"DOI": "10.1007/BF02616129",
"article-title": "Comparison of Fusion from within and Fusion from without by Newcastle Disease Virus",
"author": "Bratt",
"doi-asserted-by": "crossref",
"first-page": "3",
"journal-title": "Vitr. Cell. Dev. Biol. Plant",
"key": "ref_10",
"volume": "6",
"year": "1970"
},
{
"DOI": "10.3390/ijms21249644",
"doi-asserted-by": "crossref",
"key": "ref_11",
"unstructured": "Leroy, H., Han, M., Woottum, M., Bracq, L., Bouchet, J., Xie, M., and Benichou, S. (2020). Virus-Mediated Cell-Cell Fusion. Int. J. Mol. Sci., 21."
},
{
"DOI": "10.1080/22221751.2023.2210237",
"article-title": "Impaired potency of neutralizing antibodies against cell–cell fusion mediated by SARS-CoV-2",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "2210237",
"journal-title": "Emerg. Microbes Infect.",
"key": "ref_12",
"volume": "12",
"year": "2023"
},
{
"DOI": "10.1001/jama.1951.63670150005012b",
"article-title": "CYTOLOGIC SMEARS IN DIAGNOSIS OF HERPES SIMPLEX, HERPES ZOSTER, AND VARICELLA",
"author": "Blank",
"doi-asserted-by": "crossref",
"first-page": "1410",
"journal-title": "J. Am. Med. Assoc.",
"key": "ref_13",
"volume": "146",
"year": "1951"
},
{
"DOI": "10.1128/JVI.01237-17",
"article-title": "T Cell-Macrophage Fusion Triggers Multinucleated Giant Cell Formation for HIV-1 Spreading",
"author": "Bracq",
"doi-asserted-by": "crossref",
"first-page": "e01237-17",
"journal-title": "J. Virol.",
"key": "ref_14",
"volume": "91",
"year": "2017"
},
{
"DOI": "10.1128/jvi.71.4.2928-2933.1997",
"article-title": "Circular forms of unintegrated human immunodeficiency virus type 1 DNA and high levels of viral protein expression: Association with dementia and multinucleated giant cells in the brains of patients with AIDS",
"author": "Teo",
"doi-asserted-by": "crossref",
"first-page": "2928",
"journal-title": "J. Virol.",
"key": "ref_15",
"volume": "71",
"year": "1997"
},
{
"DOI": "10.1016/j.ebiom.2020.103104",
"doi-asserted-by": "crossref",
"key": "ref_16",
"unstructured": "Bussani, R., Schneider, E., Zentilin, L., Collesi, C., Ali, H., Braga, L., Volpe, M.C., Colliva, A., Zanconati, F., and Berlot, G. (2020). Persistence of viral RNA, pneumocyte syncytia and thrombosis are hallmarks of advanced COVID-19 pathology. eBioMedicine, 61."
},
{
"DOI": "10.1186/s12985-023-02062-7",
"article-title": "Ultrastructural study confirms the formation of single and heterotypic syncytial cells in bronchoalveolar fluids of COVID-19 patients",
"author": "Chaudhary",
"doi-asserted-by": "crossref",
"first-page": "97",
"journal-title": "Virol. J.",
"key": "ref_17",
"volume": "20",
"year": "2023"
},
{
"DOI": "10.1016/j.mbs.2024.109144",
"doi-asserted-by": "crossref",
"key": "ref_18",
"unstructured": "Amidei, A., and Dobrovolny, H.M. (2024). Virus-mediated cell fusion of SARS-CoV-2 variants. Math. Biosci., 369."
},
{
"DOI": "10.1101/2022.11.23.22282673",
"doi-asserted-by": "crossref",
"key": "ref_19",
"unstructured": "Lustig, G., Ganga, Y., Rodel, H., Tegally, H., Jackson, L., Cele, S., Khan, K., Jule, Z., Reedoy, K., and Karim, F. (2022). SARS-CoV-2 evolves increased infection elicited cell death and fusion in an immunosuppressed individual. medRxiv."
},
{
"key": "ref_20",
"unstructured": "Chippa, V., Aleem, A., and Anjum, F. (2024). Postacute Coronavirus (COVID-19) Syndrome, StatPearls Publishing LLC."
},
{
"DOI": "10.1161/CIRCRESAHA.123.321876",
"article-title": "Cell-Specific Mechanisms in the Heart of COVID-19 Patients",
"author": "Tsai",
"doi-asserted-by": "crossref",
"first-page": "1290",
"journal-title": "Circ. Res.",
"key": "ref_21",
"volume": "132",
"year": "2023"
},
{
"DOI": "10.1126/sciadv.adg2248",
"article-title": "SARS-CoV-2 infection and viral fusogens cause neuronal and glial fusion that compromises neuronal activity",
"author": "Kaulich",
"doi-asserted-by": "crossref",
"first-page": "eadg2248",
"journal-title": "Sci. Adv.",
"key": "ref_22",
"volume": "9",
"year": "2023"
},
{
"DOI": "10.1038/s41598-024-55696-0",
"doi-asserted-by": "crossref",
"key": "ref_23",
"unstructured": "Queiroz, M.A.F., Brito, W.R.d.S., Pereira, K.A.S., Pereira, L.M.S., Amoras, E.d.S.G., Lima, S.S., Santos, E.F.d., Costa, F.P.d., Sarges, K.M.L.d., and Cantanhede, M.H.D. (2024). Severe COVID-19 and long COVID are associated with high expression of STING, cGAS and IFN-α. Sci. Rep., 14."
},
{
"DOI": "10.1126/scisignal.abg8744",
"article-title": "SARS-CoV-2 spike protein–induced cell fusion activates the cGAS-STING pathway and the interferon response",
"author": "Liu",
"doi-asserted-by": "crossref",
"first-page": "eabg8744",
"journal-title": "Sci. Signal.",
"key": "ref_24",
"volume": "15",
"year": "2022"
},
{
"DOI": "10.1186/s12985-024-02365-3",
"article-title": "Computational analysis of affinity dynamics between the variants of SARS-CoV-2 spike protein (RBD) and human ACE-2 receptor",
"author": "Sultana",
"doi-asserted-by": "crossref",
"first-page": "88",
"journal-title": "Virol. J.",
"key": "ref_25",
"volume": "21",
"year": "2024"
},
{
"DOI": "10.1128/mbio.00751-24",
"article-title": "Distinct patterns of SARS-CoV-2 BA.2.87.1 and JN.1 variants in immune evasion, antigenicity, and cell-cell fusion",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "e00751-24",
"journal-title": "mBio",
"key": "ref_26",
"volume": "15",
"year": "2024"
},
{
"DOI": "10.1038/s41564-024-01878-5",
"article-title": "Phenotypic evolution of SARS-CoV-2 spike during the COVID-19 pandemic",
"author": "Furnon",
"doi-asserted-by": "crossref",
"first-page": "77",
"journal-title": "Nat. Microbiol.",
"key": "ref_27",
"volume": "10",
"year": "2025"
},
{
"DOI": "10.1038/s41586-021-03412-7",
"article-title": "Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies",
"author": "Collier",
"doi-asserted-by": "crossref",
"first-page": "136",
"journal-title": "Nature",
"key": "ref_28",
"volume": "593",
"year": "2021"
},
{
"DOI": "10.1128/JVI.00617-21",
"article-title": "V367F Mutation in SARS-CoV-2 Spike RBD Emerging during the Early Transmission Phase Enhances Viral Infectivity through Increased Human ACE2 Receptor Binding Affinity",
"author": "Ou",
"doi-asserted-by": "crossref",
"first-page": "e0061721",
"journal-title": "J Virol.",
"key": "ref_29",
"volume": "95",
"year": "2021"
},
{
"DOI": "10.1101/2021.01.24.428007",
"doi-asserted-by": "crossref",
"key": "ref_30",
"unstructured": "Barrett, C.T., Neal, H.E., Edmonds, K., Moncman, C.L., Thompson, R., Branttie, J.M., Boggs, K.B., Wu, C.Y., Leung, D.W., and Dutch, R.E. (2021). Effect of clinical isolate or cleavage site mutations in the SARS-CoV-2 spike protein on protein stability, cleavage, and cell-cell fusion. J. Biol. Chem., 297."
},
{
"DOI": "10.1093/nsr/nwae206",
"article-title": "Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity",
"author": "Liu",
"doi-asserted-by": "crossref",
"first-page": "nwae206",
"journal-title": "Natl. Sci. Rev.",
"key": "ref_31",
"volume": "11",
"year": "2024"
},
{
"DOI": "10.1038/s41467-021-25589-1",
"article-title": "Sequences in the cytoplasmic tail of SARS-CoV-2 Spike facilitate expression at the cell surface and syncytia formation",
"author": "Welch",
"doi-asserted-by": "crossref",
"first-page": "5333",
"journal-title": "Nat. Commun.",
"key": "ref_32",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1016/j.celrep.2021.109292",
"article-title": "Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7",
"author": "Meng",
"doi-asserted-by": "crossref",
"first-page": "109292",
"journal-title": "Cell Rep.",
"key": "ref_33",
"volume": "35",
"year": "2021"
},
{
"DOI": "10.1371/journal.ppat.1009246",
"doi-asserted-by": "crossref",
"key": "ref_34",
"unstructured": "Papa, G., Mallery, D.L., Albecka, A., Welch, L.G., Cattin-Ortolá, J., Luptak, J., Paul, D., McMahon, H.T., Goodfellow, I.G., and Carter, A. (2021). Furin cleavage of SARS-CoV-2 Spike promotes but is not essential for infection and cell-cell fusion. PLoS Pathog., 17."
},
{
"DOI": "10.3390/ijms24076253",
"doi-asserted-by": "crossref",
"key": "ref_35",
"unstructured": "Barthe, M., Hertereau, L., Lamghari, N., Osman-Ponchet, H., and Braud, V.M. (2023). Receptors and Cofactors That Contribute to SARS-CoV-2 Entry: Can Skin Be an Alternative Route of Entry?. Int. J. Mol. Sci., 24."
},
{
"DOI": "10.15252/embr.202357724",
"article-title": "Antibody-mediated SARS-CoV-2 entry in cultured cells",
"author": "Kibria",
"doi-asserted-by": "crossref",
"first-page": "e57724",
"journal-title": "EMBO Rep.",
"key": "ref_36",
"volume": "24",
"year": "2023"
},
{
"DOI": "10.1016/j.csbj.2024.01.009",
"article-title": "Binding affinity between coronavirus spike protein and human ACE2 receptor",
"author": "Shum",
"doi-asserted-by": "crossref",
"first-page": "759",
"journal-title": "Comput. Struct. Biotechnol. J.",
"key": "ref_37",
"volume": "23",
"year": "2024"
},
{
"DOI": "10.1139/o84-097",
"article-title": "Physical force considerations in model and biological membranes",
"author": "Rand",
"doi-asserted-by": "crossref",
"first-page": "752",
"journal-title": "Can. J. Biochem. Cell Biol.",
"key": "ref_38",
"volume": "62",
"year": "1984"
},
{
"DOI": "10.1128/JVI.00002-21",
"article-title": "SARS-CoV-2 and SARS-CoV Spike-Mediated Cell-Cell Fusion Differ in Their Requirements for Receptor Expression and Proteolytic Activation",
"author": "Schlagowski",
"doi-asserted-by": "crossref",
"first-page": "e00002-21",
"journal-title": "J. Virol.",
"key": "ref_39",
"volume": "95",
"year": "2021"
},
{
"DOI": "10.1038/s41392-021-00558-8",
"article-title": "Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development",
"author": "Zhao",
"doi-asserted-by": "crossref",
"first-page": "134",
"journal-title": "Signal Transduct. Target. Ther.",
"key": "ref_40",
"volume": "6",
"year": "2021"
},
{
"DOI": "10.1126/science.abd3072",
"article-title": "Neuropilin-1 is a host factor for SARS-CoV-2 infection",
"author": "Daly",
"doi-asserted-by": "crossref",
"first-page": "861",
"journal-title": "Science",
"key": "ref_41",
"volume": "370",
"year": "2020"
},
{
"DOI": "10.1126/science.abd2985",
"article-title": "Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity",
"author": "Ojha",
"doi-asserted-by": "crossref",
"first-page": "856",
"journal-title": "Science",
"key": "ref_42",
"volume": "370",
"year": "2020"
},
{
"DOI": "10.1073/pnas.2311913120",
"article-title": "Integrin α5β1 contributes to cell fusion and inflammation mediated by SARS-CoV-2 spike via RGD-independent interaction",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "e2311913120",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_43",
"volume": "120",
"year": "2023"
},
{
"DOI": "10.1016/j.antiviral.2020.104759",
"article-title": "A potential role for integrins in host cell entry by SARS-CoV-2",
"author": "Sigrist",
"doi-asserted-by": "crossref",
"first-page": "104759",
"journal-title": "Antivir. Res.",
"key": "ref_44",
"volume": "177",
"year": "2020"
},
{
"DOI": "10.1056/NEJMoa2015432",
"article-title": "Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid-19",
"author": "Ackermann",
"doi-asserted-by": "crossref",
"first-page": "120",
"journal-title": "N. Engl. J. Med.",
"key": "ref_45",
"volume": "383",
"year": "2020"
},
{
"DOI": "10.7554/eLife.65962",
"article-title": "SARS-CoV-2 requires cholesterol for viral entry and pathological syncytia formation",
"author": "Sanders",
"doi-asserted-by": "crossref",
"first-page": "e65962",
"journal-title": "eLife",
"key": "ref_46",
"volume": "10",
"year": "2021"
},
{
"DOI": "10.1371/journal.pone.0282151",
"doi-asserted-by": "crossref",
"key": "ref_47",
"unstructured": "Clemens, D.J., Ye, D., Zhou, W., Kim, C.S.J., Pease, D.R., Navaratnarajah, C.K., Barkhymer, A., Tester, D.J., Nelson, T.J., and Cattaneo, R. (2023). SARS-CoV-2 spike protein-mediated cardiomyocyte fusion may contribute to increased arrhythmic risk in COVID-19. PLoS ONE, 18."
},
{
"DOI": "10.1016/S2213-2600(23)00262-X",
"article-title": "Multiorgan MRI findings after hospitalisation with COVID-19 in the UK (C-MORE): A prospective, multicentre, observational cohort study",
"author": "Raman",
"doi-asserted-by": "crossref",
"first-page": "1003",
"journal-title": "Lancet Respir. Med.",
"key": "ref_48",
"volume": "11",
"year": "2023"
},
{
"DOI": "10.1038/s41564-021-00908-w",
"article-title": "The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets",
"author": "Peacock",
"doi-asserted-by": "crossref",
"first-page": "899",
"journal-title": "Nat. Microbiol.",
"key": "ref_49",
"volume": "6",
"year": "2021"
},
{
"DOI": "10.1038/s41586-021-03944-y",
"article-title": "SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion",
"author": "Mlcochova",
"doi-asserted-by": "crossref",
"first-page": "114",
"journal-title": "Nature",
"key": "ref_50",
"volume": "599",
"year": "2021"
},
{
"DOI": "10.1038/s41586-021-03291-y",
"article-title": "SARS-CoV-2 evolution during treatment of chronic infection",
"author": "Kemp",
"doi-asserted-by": "crossref",
"first-page": "277",
"journal-title": "Nature",
"key": "ref_51",
"volume": "592",
"year": "2021"
},
{
"DOI": "10.1038/s41586-022-04474-x",
"article-title": "Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity",
"author": "Meng",
"doi-asserted-by": "crossref",
"first-page": "706",
"journal-title": "Nature",
"key": "ref_52",
"volume": "603",
"year": "2022"
},
{
"DOI": "10.1080/22221751.2023.2178241",
"article-title": "SARS-CoV-2 Omicron subvariants exhibit distinct fusogenicity, but similar sensitivity, to pan-CoV fusion inhibitors",
"author": "Xia",
"doi-asserted-by": "crossref",
"first-page": "2178241",
"journal-title": "Emerg. Microbes Infect.",
"key": "ref_53",
"volume": "12",
"year": "2023"
},
{
"DOI": "10.1016/j.celrep.2025.116135",
"article-title": "Signatures of omicron-like adaptation in early SARS-CoV-2 variants and chronic infection",
"author": "Cheng",
"doi-asserted-by": "crossref",
"first-page": "116135",
"journal-title": "Cell Rep.",
"key": "ref_54",
"volume": "44",
"year": "2025"
},
{
"DOI": "10.1038/s41467-025-62300-0",
"article-title": "A single mutation may contribute to accelerated evolution of SARS-CoV-2 toward Omicron",
"author": "Lin",
"doi-asserted-by": "crossref",
"first-page": "6951",
"journal-title": "Nat. Commun.",
"key": "ref_55",
"volume": "16",
"year": "2025"
},
{
"DOI": "10.1038/nrm2417",
"article-title": "Mechanisms of membrane fusion: Disparate players and common principles",
"author": "Martens",
"doi-asserted-by": "crossref",
"first-page": "543",
"journal-title": "Nat. Rev. Mol. Cell Biol.",
"key": "ref_56",
"volume": "9",
"year": "2008"
},
{
"DOI": "10.1146/annurev-virology-092818-015523",
"article-title": "Fusogenic Reoviruses and Their Fusion-Associated Small Transmembrane (FAST) Proteins",
"author": "Duncan",
"doi-asserted-by": "crossref",
"first-page": "341",
"journal-title": "Annu. Rev. Virol.",
"key": "ref_57",
"volume": "6",
"year": "2019"
},
{
"DOI": "10.1128/JVI.77.16.8801-8811.2003",
"article-title": "The Coronavirus Spike Protein Is a Class I Virus Fusion Protein: Structural and Functional Characterization of the Fusion Core Complex",
"author": "Bosch",
"doi-asserted-by": "crossref",
"first-page": "8801",
"journal-title": "J. Virol.",
"key": "ref_58",
"volume": "77",
"year": "2003"
},
{
"DOI": "10.1016/j.devcel.2007.12.008",
"article-title": "Viral and Developmental Cell Fusion Mechanisms: Conservation and Divergence",
"author": "Sapir",
"doi-asserted-by": "crossref",
"first-page": "11",
"journal-title": "Dev. Cell",
"key": "ref_59",
"volume": "14",
"year": "2008"
},
{
"DOI": "10.1093/infdis/jiad123",
"article-title": "Paramyxoviruses: Pathogenesis, Vaccines, Antivirals, and Prototypes for Pandemic Preparedness",
"author": "Duprex",
"doi-asserted-by": "crossref",
"first-page": "S390",
"journal-title": "J. Infect. Dis.",
"key": "ref_60",
"volume": "228",
"year": "2023"
},
{
"DOI": "10.3390/v6083019",
"article-title": "Paramyxovirus Glycoprotein Incorporation, Assembly and Budding: A Three Way Dance for Infectious Particle Production",
"author": "Schmitt",
"doi-asserted-by": "crossref",
"first-page": "3019",
"journal-title": "Viruses",
"key": "ref_61",
"volume": "6",
"year": "2014"
},
{
"DOI": "10.1128/JVI.02146-06",
"article-title": "The Cytoplasmic Tail of the Severe Acute Respiratory Syndrome Coronavirus Spike Protein Contains a Novel Endoplasmic Reticulum Retrieval Signal That Binds COPI and Promotes Interaction with Membrane Protein",
"author": "McBride",
"doi-asserted-by": "crossref",
"first-page": "2418",
"journal-title": "J. Virol.",
"key": "ref_62",
"volume": "81",
"year": "2007"
},
{
"DOI": "10.3389/fmolb.2022.1004036",
"doi-asserted-by": "crossref",
"key": "ref_63",
"unstructured": "Li, Q., Liu, Y., and Zhang, L. (2022). Cytoplasmic tail determines the membrane trafficking and localization of SARS-CoV-2 spike protein Review. Front. Mol. Biosci., 9."
},
{
"DOI": "10.3390/biom15020280",
"doi-asserted-by": "crossref",
"key": "ref_64",
"unstructured": "Burkova, E.E., and Bakhno, I.A. (2025). Sequences in the Cytoplasmic Tail Contribute to the Intracellular Trafficking and the Cell Surface Localization of SARS-CoV-2 Spike Protein. Biomolecules, 15."
},
{
"DOI": "10.1038/s41467-023-44076-3",
"article-title": "A single C-terminal residue controls SARS-CoV-2 spike trafficking and incorporation into VLPs",
"author": "Dey",
"doi-asserted-by": "crossref",
"first-page": "8358",
"journal-title": "Nat. Commun.",
"key": "ref_65",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.1016/j.apsb.2023.04.007",
"article-title": "SARS-CoV-2 spike host cell surface exposure promoted by a COPI sorting inhibitor",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "3043",
"journal-title": "Acta Pharm. Sin. B",
"key": "ref_66",
"volume": "13",
"year": "2023"
},
{
"DOI": "10.1074/jbc.RA120.016175",
"doi-asserted-by": "crossref",
"key": "ref_67",
"unstructured": "Boson, B., Legros, V., Zhou, B., Siret, E., Mathieu, C., Cosset, F.-L., Lavillette, D., and Denolly, S. (2021). The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles. J. Biol. Chem., 296."
},
{
"DOI": "10.1128/mBio.00788-21",
"doi-asserted-by": "crossref",
"key": "ref_68",
"unstructured": "Rocheleau, L., Laroche, G., Fu, K., Stewart, C.M., Mohamud, A.O., Côté, M., Giguère, P.M., Langlois, M.-A., and Pelchat, M. (2021). Identification of a High-Frequency Intrahost SARS-CoV-2 Spike Variant with Enhanced Cytopathic and Fusogenic Effects. mBio, 12."
},
{
"DOI": "10.1126/science.abl6184",
"article-title": "Rapid assessment of SARS-CoV-2–evolved variants using virus-like particles",
"author": "Syed",
"doi-asserted-by": "crossref",
"first-page": "1626",
"journal-title": "Science",
"key": "ref_69",
"volume": "374",
"year": "2021"
},
{
"article-title": "SARS-CoV-2 variant biology: Immune escape, transmission and fitness",
"author": "Carabelli",
"first-page": "162",
"journal-title": "Nat. Rev. Microbiol.",
"key": "ref_70",
"volume": "21",
"year": "2023"
},
{
"DOI": "10.1038/s41586-023-06273-4",
"article-title": "Cryo-EM structure of SARS-CoV-2 postfusion spike in membrane",
"author": "Shi",
"doi-asserted-by": "crossref",
"first-page": "403",
"journal-title": "Nature",
"key": "ref_71",
"volume": "619",
"year": "2023"
},
{
"DOI": "10.1002/path.6035",
"article-title": "Persistent SARS-CoV-2 infection in patients seemingly recovered from COVID-19",
"author": "Bussani",
"doi-asserted-by": "crossref",
"first-page": "254",
"journal-title": "J. Pathol.",
"key": "ref_72",
"volume": "259",
"year": "2023"
},
{
"DOI": "10.1016/j.celrep.2022.111117",
"article-title": "Amelioration of SARS-CoV-2 infection by ANO6 phospholipid scramblase inhibition",
"author": "Sim",
"doi-asserted-by": "crossref",
"first-page": "111117",
"journal-title": "Cell Rep.",
"key": "ref_73",
"volume": "40",
"year": "2022"
},
{
"DOI": "10.1016/j.isci.2021.102170",
"article-title": "Quantitative assays reveal cell fusion at minimal levels of SARS-CoV-2 spike protein and fusion from without",
"author": "Theuerkauf",
"doi-asserted-by": "crossref",
"first-page": "102170",
"journal-title": "iScience",
"key": "ref_74",
"volume": "24",
"year": "2021"
},
{
"DOI": "10.1371/journal.ppat.1009683",
"doi-asserted-by": "crossref",
"key": "ref_75",
"unstructured": "Zhao, M., Su, P.Y., Castro, D.A., Tripler, T.N., Hu, Y., Cook, M., Ko, A.I., Farhadian, S.F., Israelow, B., and Dela Cruz, C.S. (2021). Rapid, reliable, and reproducible cell fusion assay to quantify SARS-Cov-2 spike interaction with hACE2. PLoS Pathog., 17."
},
{
"DOI": "10.1186/s12985-023-02068-1",
"article-title": "Identification of small molecules capable of enhancing viral membrane fusion",
"author": "Moreno",
"doi-asserted-by": "crossref",
"first-page": "99",
"journal-title": "Virol. J.",
"key": "ref_76",
"volume": "20",
"year": "2023"
},
{
"DOI": "10.15252/embj.2020106267",
"article-title": "Syncytia formation by SARS--CoV--2--infected cells",
"author": "Buchrieser",
"doi-asserted-by": "crossref",
"first-page": "e106267",
"journal-title": "EMBO J.",
"key": "ref_77",
"volume": "39",
"year": "2020"
},
{
"DOI": "10.1038/s41418-021-00782-3",
"article-title": "SARS-CoV-2 spike protein dictates syncytium-mediated lymphocyte elimination",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "2765",
"journal-title": "Cell Death Differ.",
"key": "ref_78",
"volume": "28",
"year": "2021"
},
{
"DOI": "10.1016/j.celrep.2023.113478",
"article-title": "Differences in syncytia formation by SARS-CoV-2 variants modify host chromatin accessibility and cellular senescence via TP53",
"author": "Lee",
"doi-asserted-by": "crossref",
"first-page": "113478",
"journal-title": "Cell Rep.",
"key": "ref_79",
"volume": "42",
"year": "2023"
},
{
"DOI": "10.1016/j.antiviral.2023.105587",
"article-title": "Cellular electrical impedance to profile SARS-CoV-2 fusion inhibitors and to assess the fusogenic potential of spike mutants",
"author": "Vanhulle",
"doi-asserted-by": "crossref",
"first-page": "105587",
"journal-title": "Antivir. Res.",
"key": "ref_80",
"volume": "213",
"year": "2023"
},
{
"DOI": "10.1093/ve/veae032",
"article-title": "Cell type-specific adaptation of the SARS-CoV-2 spike",
"author": "Geller",
"doi-asserted-by": "crossref",
"first-page": "veae032",
"journal-title": "Virus Evol.",
"key": "ref_81",
"volume": "10",
"year": "2024"
},
{
"DOI": "10.1038/s41598-025-86242-1",
"doi-asserted-by": "crossref",
"key": "ref_82",
"unstructured": "Tieu, K.V., Espey, M., Narayanan, A., Heise, R.L., Alem, F., and Conway, D.E. (2025). SARS-CoV-2 S-protein expression drives syncytia formation in endothelial cells. Sci. Rep., 15."
},
{
"DOI": "10.1529/biophysj.107.119511",
"article-title": "Calculation of free energy barriers to the fusion of small vesicles",
"author": "Lee",
"doi-asserted-by": "crossref",
"first-page": "1699",
"journal-title": "Biophys J.",
"key": "ref_83",
"volume": "94",
"year": "2008"
},
{
"DOI": "10.1242/dev.155523",
"article-title": "The hallmarks of cell-cell fusion",
"author": "Podbilewicz",
"doi-asserted-by": "crossref",
"first-page": "4481",
"journal-title": "Development",
"key": "ref_84",
"volume": "144",
"year": "2017"
},
{
"DOI": "10.1101/2021.01.04.425297",
"doi-asserted-by": "crossref",
"key": "ref_85",
"unstructured": "Lai, A.L., and Freed, J.H. (2021). SARS-CoV-2 Fusion Peptide has a Greater Membrane Perturbating Effect than SARS-CoV with Highly Specific Dependence on Ca2+. J. Mol. Biol., 433."
},
{
"DOI": "10.1021/acsinfecdis.3c00260",
"article-title": "A Mechanistic Understanding of the Modes of Ca2+ Ion Binding to the SARS-CoV-1 Fusion Peptide and Their Role in the Dynamics of Host Membrane Penetration",
"author": "Carten",
"doi-asserted-by": "crossref",
"first-page": "398",
"journal-title": "ACS Infect. Dis.",
"key": "ref_86",
"volume": "10",
"year": "2024"
},
{
"DOI": "10.1021/acsnano.0c08614",
"article-title": "Calcium Ions Promote Membrane Fusion by Forming Negative-Curvature Inducing Clusters on Specific Anionic Lipids",
"author": "Allolio",
"doi-asserted-by": "crossref",
"first-page": "12880",
"journal-title": "ACS Nano",
"key": "ref_87",
"volume": "15",
"year": "2021"
},
{
"DOI": "10.1128/jvi.67.3.1647-1652.1993",
"article-title": "Calcium ions are required for cell fusion mediated by the CD4-human immunodeficiency virus type 1 envelope glycoprotein interaction",
"author": "Dimitrov",
"doi-asserted-by": "crossref",
"first-page": "1647",
"journal-title": "J. Virol.",
"key": "ref_88",
"volume": "67",
"year": "1993"
},
{
"DOI": "10.1085/jgp.200709932",
"article-title": "Cholesterol Promotes Hemifusion and Pore Widening in Membrane Fusion Induced by Influenza Hemagglutinin",
"author": "Biswas",
"doi-asserted-by": "crossref",
"first-page": "503",
"journal-title": "J. Gen. Physiol.",
"key": "ref_89",
"volume": "131",
"year": "2008"
},
{
"DOI": "10.1073/pnas.2012197117",
"article-title": "Cholesterol 25-hydroxylase suppresses SARS-CoV-2 replication by blocking membrane fusion",
"author": "Zang",
"doi-asserted-by": "crossref",
"first-page": "32105",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_90",
"volume": "117",
"year": "2020"
},
{
"DOI": "10.15252/embj.2020106057",
"article-title": "Cholesterol 25-Hydroxylase inhibits SARS-CoV-2 and other coronaviruses by depleting membrane cholesterol",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "e106057",
"journal-title": "EMBO J.",
"key": "ref_91",
"volume": "39",
"year": "2020"
},
{
"DOI": "10.1038/s41598-018-37056-x",
"doi-asserted-by": "crossref",
"key": "ref_92",
"unstructured": "Bricogne, C., Fine, M., Pereira, P.M., Sung, J., Tijani, M., Wang, Y., Henriques, R., Collins, M.K., and Hilgemann, D.W. (2019). TMEM16F activation by Ca2+ triggers plasma membrane expansion and directs PD-1 trafficking. Sci. Rep., 9."
},
{
"DOI": "10.1016/j.jbc.2021.100411",
"doi-asserted-by": "crossref",
"key": "ref_93",
"unstructured": "Whitlock, J.M., and Chernomordik, L.V. (2021). Flagging fusion: Phosphatidylserine signaling in cell–cell fusion. J. Biol. Chem., 296."
},
{
"DOI": "10.1101/2024.07.13.603361",
"doi-asserted-by": "crossref",
"key": "ref_94",
"unstructured": "Bolland, W., Marechal, I., Petiot, C., Porrot, F., Guivel-Benhassine, F., Casartelli, N., Schwartz, O., and Buchrieser, J. (2024). SARS-CoV-2 entry and fusion are independent of ACE2 localization to lipid rafts. bioRxiv."
},
{
"DOI": "10.1016/j.jbc.2022.101710",
"doi-asserted-by": "crossref",
"key": "ref_95",
"unstructured": "Liu, J., Lu, F., Chen, Y., Plow, E., and Qin, J. (2022). Integrin mediates cell entry of the SARS-CoV-2 virus independent of cellular receptor ACE2. J. Biol. Chem., 298."
},
{
"DOI": "10.1038/s41418-021-00805-z",
"article-title": "Patients with COVID-19: In the dark-NETs of neutrophils",
"author": "Ackermann",
"doi-asserted-by": "crossref",
"first-page": "3125",
"journal-title": "Cell Death Differ.",
"key": "ref_96",
"volume": "28",
"year": "2021"
},
{
"DOI": "10.1038/s41586-022-04802-1",
"article-title": "Inflammasome activation in infected macrophages drives COVID-19 pathology",
"author": "Sefik",
"doi-asserted-by": "crossref",
"first-page": "585",
"journal-title": "Nature",
"key": "ref_97",
"volume": "606",
"year": "2022"
},
{
"DOI": "10.1126/scisignal.abq1366",
"article-title": "Macrophage ACE2 is necessary for SARS-CoV-2 replication and subsequent cytokine responses that restrict continued virion release",
"author": "Labzin",
"doi-asserted-by": "crossref",
"first-page": "eabq1366",
"journal-title": "Sci. Signal.",
"key": "ref_98",
"volume": "16",
"year": "2023"
},
{
"DOI": "10.1371/journal.ppat.1011789",
"doi-asserted-by": "crossref",
"key": "ref_99",
"unstructured": "Yu, S., Zheng, X., Zhou, Y., Gao, Y., Zhou, B., Zhao, Y., Li, T., Li, Y., Mou, J., and Cui, X. (2023). Antibody-mediated spike activation promotes cell-cell transmission of SARS-CoV-2. PLoS Pathog., 19."
},
{
"DOI": "10.1016/j.micinf.2024.105464",
"article-title": "The SARS-CoV-2 antibody-dependent enhancement façade",
"author": "Coish",
"doi-asserted-by": "crossref",
"first-page": "105464",
"journal-title": "Microbes Infect",
"key": "ref_100",
"volume": "27",
"year": "2025"
},
{
"DOI": "10.1038/s41467-024-54458-w",
"article-title": "Antibodies to the RBD of SARS-CoV-2 spike mediate productive infection of primary human macrophages",
"author": "Pickering",
"doi-asserted-by": "crossref",
"first-page": "10764",
"journal-title": "Nat. Commun.",
"key": "ref_101",
"volume": "15",
"year": "2024"
},
{
"DOI": "10.1016/j.immuni.2021.03.023",
"article-title": "Neutralization potency of monoclonal antibodies recognizing dominant and subdominant epitopes on SARS-CoV-2 Spike is impacted by the B.1.1.7 variant",
"author": "Graham",
"doi-asserted-by": "crossref",
"first-page": "1276",
"journal-title": "Immunity",
"key": "ref_102",
"volume": "54",
"year": "2021"
},
{
"DOI": "10.3390/v15071500",
"doi-asserted-by": "crossref",
"key": "ref_103",
"unstructured": "Reuter, N., Chen, X., Kropff, B., Peter, A.S., Britt, W.J., Mach, M., Überla, K., and Thomas, M. (2023). SARS-CoV-2 Spike Protein Is Capable of Inducing Cell–Cell Fusions Independent from Its Receptor ACE2 and This Activity Can Be Impaired by Furin Inhibitors or a Subset of Monoclonal Antibodies. Viruses, 15."
},
{
"DOI": "10.3390/v14112535",
"doi-asserted-by": "crossref",
"key": "ref_104",
"unstructured": "Lim, S., Zhang, M., and Chang, T.L. (2022). ACE2-Independent Alternative Receptors for SARS-CoV-2. Viruses, 14."
},
{
"DOI": "10.1038/s41418-021-00795-y",
"article-title": "Syncytia formation during SARS-CoV-2 lung infection: A disastrous unity to eliminate lymphocytes",
"author": "Lin",
"doi-asserted-by": "crossref",
"first-page": "2019",
"journal-title": "Cell Death Differ.",
"key": "ref_105",
"volume": "28",
"year": "2021"
},
{
"DOI": "10.1126/sciimmunol.abc3582",
"article-title": "TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes",
"author": "Zang",
"doi-asserted-by": "crossref",
"first-page": "eabc3582",
"journal-title": "Sci. Immunol.",
"key": "ref_106",
"volume": "5",
"year": "2020"
},
{
"DOI": "10.1080/22221751.2020.1787797",
"article-title": "Bat SARS-Like WIV1 coronavirus uses the ACE2 of multiple animal species as receptor and evades IFITM3 restriction via TMPRSS2 activation of membrane fusion",
"author": "Zheng",
"doi-asserted-by": "crossref",
"first-page": "1567",
"journal-title": "Emerg. Microbes Infect.",
"key": "ref_107",
"volume": "9",
"year": "2020"
},
{
"DOI": "10.1128/JVI.03372-12",
"article-title": "TMPRSS2 Activates the Human Coronavirus 229E for Cathepsin-Independent Host Cell Entry and Is Expressed in Viral Target Cells in the Respiratory Epithelium",
"author": "Bertram",
"doi-asserted-by": "crossref",
"first-page": "6150",
"journal-title": "J. Virol.",
"key": "ref_108",
"volume": "87",
"year": "2013"
},
{
"DOI": "10.1016/j.cyto.2023.156447",
"article-title": "Pyrogenic and inflammatory mediators are produced by polarized M1 and M2 macrophages activated with D-dimer and SARS-CoV-2 spike immune complexes",
"author": "Park",
"doi-asserted-by": "crossref",
"first-page": "156447",
"journal-title": "Cytokine",
"key": "ref_109",
"volume": "173",
"year": "2024"
},
{
"DOI": "10.1101/2021.07.09.451812",
"doi-asserted-by": "crossref",
"key": "ref_110",
"unstructured": "Prévost, J., Richard, J., Gasser, R., Ding, S., Fage, C., Anand, S.P., Adam, D., Gupta Vergara, N., Tauzin, A., and Benlarbi, M. (2021). Impact of temperature on the affinity of SARS-CoV-2 Spike glycoprotein for host ACE2. J. Biol. Chem., 297."
},
{
"DOI": "10.1128/mbio.03360-23",
"article-title": "Temperature impacts SARS-CoV-2 spike fusogenicity and evolution",
"author": "Dufloo",
"doi-asserted-by": "crossref",
"first-page": "e03360-23",
"journal-title": "mBio",
"key": "ref_111",
"volume": "15",
"year": "2024"
},
{
"DOI": "10.1038/s41421-021-00310-0",
"article-title": "Pyroptosis of syncytia formed by fusion of SARS-CoV-2 spike and ACE2-expressing cells",
"author": "Ma",
"doi-asserted-by": "crossref",
"first-page": "73",
"journal-title": "Cell Discov.",
"key": "ref_112",
"volume": "7",
"year": "2021"
},
{
"DOI": "10.1016/j.isci.2021.103144",
"article-title": "The circadian clock component BMAL1 regulates SARS-CoV-2 entry and replication in lung epithelial cells",
"author": "Zhuang",
"doi-asserted-by": "crossref",
"first-page": "103144",
"journal-title": "iScience",
"key": "ref_113",
"volume": "24",
"year": "2021"
},
{
"DOI": "10.1038/srep02854",
"doi-asserted-by": "crossref",
"key": "ref_114",
"unstructured": "Cabantous, S., Nguyen, H.B., Pedelacq, J.-D., Koraïchi, F., Chaudhary, A., Ganguly, K., Lockard, M.A., Favre, G., Terwilliger, T.C., and Waldo, G.S. (2013). A New Protein-Protein Interaction Sensor Based on Tripartite Split-GFP Association. Sci. Rep., 3."
},
{
"DOI": "10.1186/s12934-021-01672-6",
"article-title": "Scaling production of GFP1-10 detector protein in E. coli for secretion screening by split GFP assay",
"author": "Igwe",
"doi-asserted-by": "crossref",
"first-page": "191",
"journal-title": "Microb. Cell Factories",
"key": "ref_115",
"volume": "20",
"year": "2021"
},
{
"DOI": "10.3390/ijms232113167",
"doi-asserted-by": "crossref",
"key": "ref_116",
"unstructured": "Bignon, C., Gruet, A., and Longhi, S. (2022). Split-GFP Reassembly Assay: Strengths and Caveats from a Multiparametric Analysis. Int. J. Mol. Sci., 23."
},
{
"DOI": "10.3390/ijms25031704",
"doi-asserted-by": "crossref",
"key": "ref_117",
"unstructured": "Cafaro, A., Schietroma, I., Sernicola, L., Belli, R., Campagna, M., Mancini, F., Farcomeni, S., Pavone-Cossut, M.R., Borsetti, A., and Monini, P. (2024). Role of HIV-1 Tat Protein Interactions with Host Receptors in HIV Infection and Pathogenesis. Int. J. Mol. Sci., 25."
},
{
"article-title": "Sequential steps in Tat trans-activation of HIV-1 mediated through cellular DNA, RNA, and protein binding factors",
"author": "Gatignol",
"first-page": "217",
"journal-title": "Gene Expr.",
"key": "ref_118",
"volume": "5",
"year": "1996"
},
{
"DOI": "10.1016/j.virol.2003.11.012",
"article-title": "α-Complementation assay for HIV envelope glycoprotein-mediated fusion",
"author": "Holland",
"doi-asserted-by": "crossref",
"first-page": "343",
"journal-title": "Virology",
"key": "ref_119",
"volume": "319",
"year": "2004"
},
{
"DOI": "10.1101/pdb.prot101329",
"article-title": "Screening Bacterial Colonies Using X-Gal and IPTG: α-Complementation",
"author": "Green",
"doi-asserted-by": "crossref",
"first-page": "pdb-prot101329",
"journal-title": "Cold Spring Harb. Protoc.",
"key": "ref_120",
"volume": "2019",
"year": "2019"
},
{
"DOI": "10.1101/522540",
"doi-asserted-by": "crossref",
"key": "ref_121",
"unstructured": "García-Murria, M.J., Expósito-Domínguez, N., Duart, G., Mingarro, I., and Martinez-Gil, L. (2019). A Bimolecular Multicellular Complementation System for the Detection of Syncytium Formation: A New Methodology for the Identification of Nipah Virus Entry Inhibitors. Viruses, 11."
},
{
"key": "ref_122",
"unstructured": "Kim, S.-B. (2022). Monitoring Hippo Signaling Pathway Activity Using a Luciferase-Based Large Tumor Suppressor (LATS) Biosensor. Bioluminescence: Methods and Protocols, Volume 2, Springer."
},
{
"DOI": "10.3390/bios14120570",
"doi-asserted-by": "crossref",
"key": "ref_123",
"unstructured": "Kelly, T., and Yang, X. (2024). Application of Fluorescence- and Bioluminescence-Based Biosensors in Cancer Drug Discovery. Biosensors, 14."
},
{
"DOI": "10.1038/s42003-022-03163-9",
"doi-asserted-by": "crossref",
"key": "ref_124",
"unstructured": "Reyes-Alcaraz, A., Garcia-Rojas, E.Y.L., Merlinsky, E.A., Seong, J.Y., Bond, R.A., and McConnell, B.K. (2022). A NanoBiT assay to monitor membrane proteins trafficking for drug discovery and drug development. Commun. Biol., 5."
},
{
"DOI": "10.1093/hmg/ddn381",
"article-title": "Missense mutations that cause Van der Woude syndrome and popliteal pterygium syndrome affect the DNA-binding and transcriptional activation functions of IRF6",
"author": "Little",
"doi-asserted-by": "crossref",
"first-page": "535",
"journal-title": "Hum. Mol. Genet.",
"key": "ref_125",
"volume": "18",
"year": "2008"
},
{
"key": "ref_126",
"unstructured": "Ross, B.D., and Gambhir, S.S. (2021). Chapter 47—Molecular Imaging of Protein–Protein Interactions and Protein Folding. Molecular Imaging, Academic Press. [2nd ed.]."
},
{
"DOI": "10.1128/JVI.01986-18",
"article-title": "Unique Transcriptional Architecture in Airway Epithelial Cells and Macrophages Shapes Distinct Responses following Influenza Virus Infection Ex Vivo",
"author": "Ma",
"doi-asserted-by": "crossref",
"first-page": "10-1128",
"journal-title": "J. Virol.",
"key": "ref_127",
"volume": "93",
"year": "2019"
},
{
"DOI": "10.3390/mi11040411",
"doi-asserted-by": "crossref",
"key": "ref_128",
"unstructured": "Quinchia, J., Echeverri, D., Cruz-Pacheco, A.F., Maldonado, M.E., and Orozco, J. (2020). Electrochemical Biosensors for Determination of Colorectal Tumor Biomarkers. Micromachines, 11."
},
{
"DOI": "10.1039/D3MA00621B",
"article-title": "Recent advancement in the detection of potential cancer biomarkers using the nanomaterial integrated electrochemical sensing technique: A detailed review",
"author": "Bhardwaj",
"doi-asserted-by": "crossref",
"first-page": "475",
"journal-title": "Mater. Adv.",
"key": "ref_129",
"volume": "5",
"year": "2024"
},
{
"DOI": "10.1016/j.ebiom.2016.09.013",
"article-title": "Zika Virus Replicons for Drug Discovery",
"author": "Xie",
"doi-asserted-by": "crossref",
"first-page": "156",
"journal-title": "EBioMedicine",
"key": "ref_130",
"volume": "12",
"year": "2016"
},
{
"DOI": "10.1038/srep22791",
"doi-asserted-by": "crossref",
"key": "ref_131",
"unstructured": "Watterson, D., Robinson, J., Chappell, K.J., Butler, M.S., Edwards, D.J., Fry, S.R., Bermingham, I.M., Cooper, M.A., and Young, P.R. (2016). A generic screening platform for inhibitors of virus induced cell fusion using cellular electrical impedance. Sci. Rep., 6."
},
{
"DOI": "10.1016/S1473-3099(22)00591-6",
"article-title": "Lung cell entry, cell–cell fusion capacity, and neutralisation sensitivity of omicron sublineage BA.2.75",
"author": "Arora",
"doi-asserted-by": "crossref",
"first-page": "1537",
"journal-title": "Lancet Infect. Dis.",
"key": "ref_132",
"volume": "22",
"year": "2022"
},
{
"DOI": "10.1146/annurev.biochem.78.081307.110540",
"article-title": "Mechanisms of endocytosis",
"author": "Doherty",
"doi-asserted-by": "crossref",
"first-page": "857",
"journal-title": "Annu. Rev. Biochem.",
"key": "ref_133",
"volume": "78",
"year": "2009"
},
{
"DOI": "10.1038/s41423-023-01104-y",
"article-title": "SARS-CoV-2 and innate immunity: The good, the bad, and the “goldilocks”",
"author": "Sievers",
"doi-asserted-by": "crossref",
"first-page": "171",
"journal-title": "Cell. Mol. Immunol.",
"key": "ref_134",
"volume": "21",
"year": "2024"
},
{
"DOI": "10.1038/s41577-024-01112-7",
"article-title": "Regulation of cGAS–STING signalling and its diversity of cellular outcomes",
"author": "Zhang",
"doi-asserted-by": "crossref",
"first-page": "425",
"journal-title": "Nat. Rev. Immunol.",
"key": "ref_135",
"volume": "25",
"year": "2025"
},
{
"DOI": "10.1038/s41467-023-43661-w",
"article-title": "True prevalence of long-COVID in a nationwide, population cohort study",
"author": "Hastie",
"doi-asserted-by": "crossref",
"first-page": "7892",
"journal-title": "Nat. Commun.",
"key": "ref_136",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.1016/S1473-3099(24)00216-0",
"article-title": "Long COVID and SARS-CoV-2 persistence: New answers, more questions",
"author": "Buonsenso",
"doi-asserted-by": "crossref",
"first-page": "796",
"journal-title": "Lancet Infect. Dis.",
"key": "ref_137",
"volume": "24",
"year": "2024"
},
{
"DOI": "10.1101/2024.05.30.596590",
"doi-asserted-by": "crossref",
"key": "ref_138",
"unstructured": "Chen, H.-J., Appelman, B., Willemen, H., Bos, A., Prado, J., Geyer, C.E., Santos Ribeiro, P.S., Versteeg, S., Larsen, M., and Schüchner, E. (2024). Transfer of IgG from Long COVID patients induces symptomology in mice. bioRxiv."
}
],
"reference-count": 138,
"references-count": 138,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.mdpi.com/1999-4915/17/11/1405"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Mechanisms of Cell–Cell Fusion in SARS-CoV-2: An Evolving Strategy for Transmission and Immune Evasion",
"type": "journal-article",
"update-policy": "https://doi.org/10.3390/mdpi_crossmark_policy",
"volume": "17"
}