SARS-CoV-2 genomic evolution during a severe and long-lasting omicron infection under antiviral therapy
et al., BMC Infectious Diseases, doi:10.1186/s12879-025-10740-w, Mar 2025
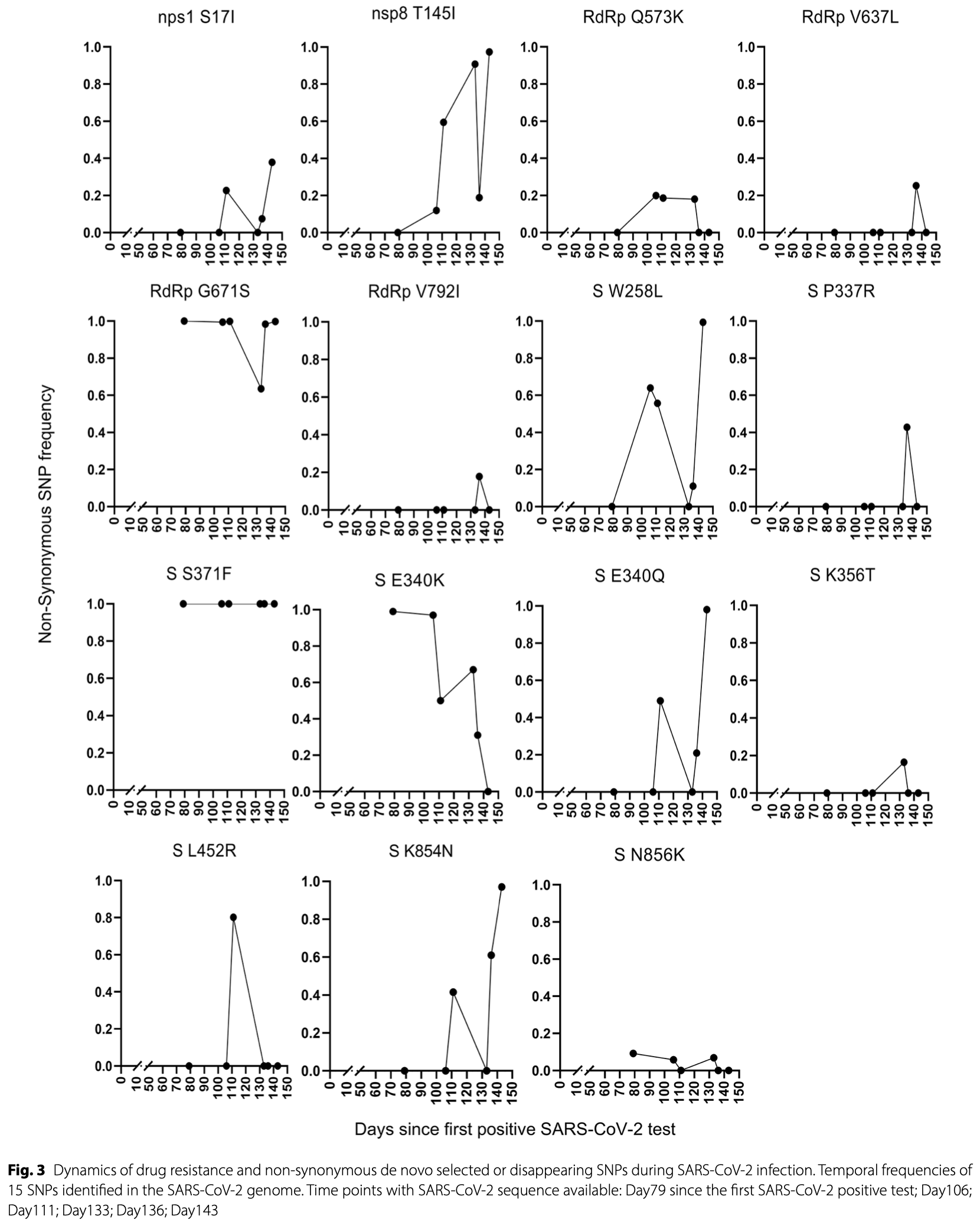
Case study of a 72-year-old immunocompromised male demonstrating SARS-CoV-2 genomic evolution during a 149-day persistent Omicron infection while undergoing antiviral therapy. The patient experienced three episodes of severe COVID-19 despite multiple courses of sotrovimab/remdesivir combined therapy. Genome sequencing at six time-points revealed significant viral evolution, including emergence of drug-resistance mutations after antiviral treatments and fixation of five non-synonymous mutations over time.
Bolis et al., 13 Mar 2025, Italy, peer-reviewed, 11 authors.
Contact: claudia.alteri@unimi.it.
SARS-CoV-2 genomic evolution during a severe and long-lasting omicron infection under antiviral therapy
BMC Infectious Diseases, doi:10.1186/s12879-025-10740-w
Background Prolonged SARS-CoV-2 infection observed in immunocompromised individuals even in the presence of antiviral treatment provides opportunities for viruses to evolve in immune escape and drug-resistant variants. Case presentation A 72-year-old male with IgG4-related disease was admitted to the Emergency Department of a city Hospital in Milan and then transferred to Fondazione IRCCS Ca' Granda Ospedale Maggiore Policlinico in December 2023, due to respiratory distress due to SARS-CoV-2 infection diagnosed in November 2023. After 117 days since the onset of the infection, and two cycles of sotrovimab/remdesivir combined therapy, the clinical improvement allowed the hospital discharge, notwithstanding the persistent SARS-CoV-2 positivity. Fifteen days later, the patient was re-admitted to the hospital due to worsening clinical conditions. After a third cycle of sotrovimab/remdesivir combined therapy prolonged with nirmatrelvir/ritonavir, nasopharyngeal load dropped and clinical conditions improved, ending with a successful discharge. SARS-CoV-2 whole genome sequences, obtained at six time-points of infection, showed an FL.1.5.1 recombinant form infection and a genetic distance of median (IQR) 0.00052 (0.00041-0.00066) similar to the genetic distance observed among the 43 contemporaneous FL.1.5.1 recombinant forms (p = 0.098). De novo SNPs were observed at all time points, with a peak (n = 70) at day 133 of infection, corresponding to the time of the second hospitalization. Six non-synonymous mutations (three in the RdRp and three in the spike protein, four of them known to be associated with drug resistance) appeared transiently, after the third and fourth course of sotrovimab 500 mg/remdesivir combination. Five de novo SNPs, three of them in the spike protein, were fixed over the long-lasting infection. The spike N856K, associated with reduced fusogenicity and infectivity in Omicron BA.1, was completely replaced by constitutive N at day 136. Conclusions This clinical case confirms the intra-host evolution dynamics of SARS-CoV-2 in an immunocompromised, prolonged-infected individual, involving positions associated with drug resistance and fusogenic traits of SARS-CoV-2. These results underscore the importance of the early detection of SARS-CoV-2
Supplementary Information The online version contains supplementary material available at h t t p s : / / d o i . o r g / 1 0 . 1 1 8 6 / s 1 2 8 7 9 -0 2 5 -1 0 7 4 0 -w.
Supplementary Material 1 Author contributions MB and SUR performed data analysis, data interpretation, and writing; LA and AL performed clinical evaluation and help in data interpretation and writing; BZP and AP helped in data processing; SUR and AP processed samples; AG and AB critically revised the manuscript; AM and CA conceived the clinical case. All authors revised and approved the manuscript.
Declarations Ethics approval and consent to participate The study protocol has been approved by the Territorial Ethical Committee Lombardy 3 (5164_Case.Report_11.09.2024_P). This study was conducted following the principles of the 1964 Declaration of Helsinki. The data used in this study were anonymized before use.
Consent for publication A written informed consent was obtained by the patient.
Competing interests The authors declare no competing interests.
Publisher's note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Aydillo, Gonzalez-Reiche, Aslam, Van De Guchte, Khan et al., Shedding of viable SARS-CoV-2 after immunosuppressive therapy for Cancer, N Engl J Med, doi:10.1056/NEJMc2031670
Baang, Smith, Mirabelli, Valesano, Manthei et al., Prolonged severe acute respiratory syndrome coronavirus 2 replication in an immunocompromised patient, J Infect Dis, doi:10.1093/infdis/jiaa666
Baj, Novazzi, Drago Ferrante, Genoni, Tettamanzi et al., Spike protein evolution in the SARS-CoV-2 Delta variant of concern: a case series from Northern Lombardy, Emerg Microbes Infect, doi:10.1080/22221751.2021
Chaguza, Hahn, Petrone, Accelerated SARS-CoV-2 intrahost evolution leading to distinct genotypes during chronic infection, Cell Rep Med
Chaguza, Hahn, Petrone, Zhou, Ferguson et al., Accelerated SARS-CoV-2 intrahost evolution leading to distinct genotypes during chronic infection, Cell Rep Med, doi:10.1016/j.xcrm.2023.100943
Choi, Choudhary, Regan, Sparks, Padera et al., Persistence and evolution of SARS-CoV-2 in an immunocompromised host, N Engl J Med, doi:10.1056/NEJMc2031364
Choi, Wan, Wong, Chan, Chu et al., Comparative effectiveness of combination therapy with nirmatrelvir-ritonavir and Remdesivir versus monotherapy with Remdesivir or nirmatrelvirritonavir in patients hospitalised with COVID-19: a target trial emulation study, Lancet Infect Dis, doi:10.1016/S1473-3099(24)00353-0
Ciccozzi, Pascarella, Two sides of the same coin: the N-terminal and the receptor binding domains of SARS-CoV-2 Spike, Future Virol, doi:10.2217/fvl-2022-0181
Consortium, Bonsall, Fraser, Golubchik, SARS-CoV-2 within-host diversity and transmission, Science, doi:10.1126/science.abg0821
Dhama, Nainu, Frediansyah, Yatoo, Mohapatra et al., Global emerging Omicron variant of SARS-CoV-2: impacts, challenges and strategies, J Infect Public Health, doi:10.1016/j.jiph.2022.11.024
Farjo, Koelle, Martin, Gibson, Walden et al., Within-host evolutionary dynamics and tissue compartmentalization during acute SARS-CoV-2 infection, J Virol, doi:10.1128/jvi.01618-23
Ghafari, Liu, Dhillon, Katzourakis, Weissman, Investigating the evolutionary origins of the first three SARS-CoV-2 variants of concern, Front Virol
Hogan, Duerr, Dimartino, Marier, Hochman et al., Remdesivir resistance in transplant recipients with persistent coronavirus disease 2019, Clin Infect Dis, doi:10.1093/cid/ciac769
Hou, Shi, Gong, Wen, Lan et al., Intra-vs. Interhost evolution of SARS-CoV-2 driven by uncorrelated Selection-The evolution thwarted, Mol Biol Evol, doi:10.1093/molbev/msad204
Igari, Sakao, Ishige, Dynamic diversity of SARS-CoV-2 genetic mutations in a lung transplantation patient with persistent COVID-19, Nat Commun, doi:10.1038/s41467-024-47941-x
Kang, Kim, Kim, Lim, Jang et al., Characteristics and risk factors of prolonged viable virus shedding in immunocompromised patients with COVID-19: a prospective cohort study, J Infect, doi:10.1016/j.jinf.2023.01.024
Kemp, Collier, Datir, Ferreira, Gayed et al., SARS-CoV-2 evolution during treatment of chronic infection, Nature
Li, Du, Yang, Zhang, Song et al., Two-step fitness selection for intra-host variations in SARS-CoV-2, Cell Rep, doi:10.1016/j.celrep.2021.110205
Lythgoe, Hall, Ferretti, De Cesare, Macintyre-Cockett et al., Oxford Virus Sequencing Analysis Group
Machkovech, Hahn, Wang, Grubaugh, Halfmann et al., Persistent SARS-CoV-2 infection: significance and implications, Lancet Infect Dis, doi:10.1016/S1473-3099(23)00815-0
Markov, Ghafari, Beer, The evolution of SARS-CoV-2, Nat Rev Microbiol
Marques, Graham-Wooten, Fitzgerald, Sobel Leonard, Cook et al., SARS-CoV-2 evolution during prolonged infection in immunocompromised patients, mBio, doi:10.1128/mbio.00110-24
Palomino-Cabrera, Tejerina, Molero-Salinas, Ferris, Veintimilla et al., Microbiology-ID COVID 19 study group. Frequent emergence of resistance mutations following complex Intra-Host genomic dynamics in SARS-CoV-2 patients receiving Sotrovimab, Antimicrob Agents Chemother, doi:10.1128/aac.00266-23
Park, Khan, Chiliveri, SARS-CoV-2 Omicron variants harbor Spike protein mutations responsible for their attenuated fusogenic phenotype, Commun Biol, doi:10.1038/s42003-023-04923-x
Paul, Pyne, Paul, Mutation profile of SARS-CoV-2 Spike protein and identification of potential multiple epitopes within Spike protein for vaccine development against SARS-CoV-2, Virus Disease, doi:10.1007/s13337-021-00747-7
Qiu, Wen, Wang, Sun, Li et al., Real-world effectiveness and safety of nirmatrelvir-ritonavir (Paxlovid)-treated for COVID-19 patients with onset of more than 5 days: a retrospective cohort study, Front Pharmacol, doi:10.3389/fphar.2024.1401658
Raglow, Surie, Chappell, Zhu, Martin et al., Investigating respiratory viruses in the acutely ill (IVY) network. SARS-CoV-2 shedding and evolution in patients who were immunocompromised during the Omicron period: a multicentre, prospective analysis, Lancet Microbe, doi:10.1016/S2666-5247(23)00336-1
Subong, Ozawa, Bio-Chemoinformatics-Driven analysis of nsp7 and nsp8 mutations and their effects on viral replication protein complex stability, Curr Issues Mol Biol, doi:10.3390/cimb46030165
Sun, Wang, Yang, Mutation N856K in Spike reduces fusogenicity and infectivity of Omicron BA.1, Sig Transduct Target Ther, doi:10.1038/s41392-022-01281-8
Tegally, Moir, Everatt, Giovanetti, Scheepers et al., Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa, Nat Med, doi:10.1038/s41591-022-01911-2
Wilkinson, Richter, Casey, Osman, Mirza et al., Recurrent SARS-CoV-2 mutations in immunodeficient patients, Virus Evol, doi:10.1093/ve/veac050
Wu, Guo, Yuan, Cao, Wang et al., Duration of viable virus shedding and polymerase chain reaction positivity of the SARS-CoV-2 Omicron variant in the upper respiratory tract: a systematic review and metaanalysis, Int J Infect Dis, doi:10.1016/j.ijid.2023.02.011
DOI record:
{
"DOI": "10.1186/s12879-025-10740-w",
"ISSN": [
"1471-2334"
],
"URL": "http://dx.doi.org/10.1186/s12879-025-10740-w",
"alternative-id": [
"10740"
],
"article-number": "359",
"assertion": [
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Received",
"name": "received",
"order": 1,
"value": "24 August 2024"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "3 March 2025"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "First Online",
"name": "first_online",
"order": 3,
"value": "13 March 2025"
},
{
"group": {
"label": "Declarations",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 1
},
{
"group": {
"label": "Ethics approval and consent to participate",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 2,
"value": "The study protocol has been approved by the Territorial Ethical Committee Lombardy 3 (5164_Case.Report_11.09.2024_P). This study was conducted following the principles of the 1964 Declaration of Helsinki. The data used in this study were anonymized before use."
},
{
"group": {
"label": "Consent for publication",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 3,
"value": "A written informed consent was obtained by the patient."
},
{
"group": {
"label": "Competing interests",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 4,
"value": "The authors declare no competing interests."
}
],
"author": [
{
"affiliation": [],
"family": "Bolis",
"given": "Matteo",
"sequence": "first"
},
{
"affiliation": [],
"family": "Uceda Renteria",
"given": "Sara",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Alagna",
"given": "Laura",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Liparoti",
"given": "Arianna",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Passerini",
"given": "Beatrice Zita",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Pastena",
"given": "Andrea",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Parisi",
"given": "Alessandra",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Callegaro",
"given": "Annapaola",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Bandera",
"given": "Alessandra",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Muscatello",
"given": "Antonio",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Alteri",
"given": "Claudia",
"sequence": "additional"
}
],
"container-title": "BMC Infectious Diseases",
"container-title-short": "BMC Infect Dis",
"content-domain": {
"crossmark-restriction": false,
"domain": [
"link.springer.com"
]
},
"created": {
"date-parts": [
[
2025,
3,
13
]
],
"date-time": "2025-03-13T16:58:28Z",
"timestamp": 1741885108000
},
"deposited": {
"date-parts": [
[
2025,
3,
13
]
],
"date-time": "2025-03-13T16:58:30Z",
"timestamp": 1741885110000
},
"funder": [
{
"DOI": "10.13039/501100009702",
"award": [
"STOP-COVID"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100009702",
"id-type": "DOI"
}
],
"name": "Fondazione IRCCS Ca' Granda Ospedale Maggiore Policlinico"
}
],
"indexed": {
"date-parts": [
[
2025,
3,
14
]
],
"date-time": "2025-03-14T04:25:50Z",
"timestamp": 1741926350760,
"version": "3.38.0"
},
"is-referenced-by-count": 0,
"issue": "1",
"issued": {
"date-parts": [
[
2025,
3,
13
]
]
},
"journal-issue": {
"issue": "1",
"published-online": {
"date-parts": [
[
2025,
12
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by-nc-nd/4.0",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
3,
13
]
],
"date-time": "2025-03-13T00:00:00Z",
"timestamp": 1741824000000
}
},
{
"URL": "https://creativecommons.org/licenses/by-nc-nd/4.0",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
3,
13
]
],
"date-time": "2025-03-13T00:00:00Z",
"timestamp": 1741824000000
}
}
],
"link": [
{
"URL": "https://link.springer.com/content/pdf/10.1186/s12879-025-10740-w.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://link.springer.com/article/10.1186/s12879-025-10740-w/fulltext.html",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://link.springer.com/content/pdf/10.1186/s12879-025-10740-w.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "297",
"original-title": [],
"prefix": "10.1186",
"published": {
"date-parts": [
[
2025,
3,
13
]
]
},
"published-online": {
"date-parts": [
[
2025,
3,
13
]
]
},
"publisher": "Springer Science and Business Media LLC",
"reference": [
{
"DOI": "10.1093/infdis/jiaa666",
"author": "JH Baang",
"doi-asserted-by": "publisher",
"first-page": "23",
"issue": "1",
"journal-title": "J Infect Dis",
"key": "10740_CR1",
"unstructured": "Baang JH, Smith C, Mirabelli C, Valesano AL, Manthei DM, Bachman MA, Wobus CE, Adams M, Washer L, Martin ET, Lauring AS. Prolonged severe acute respiratory syndrome coronavirus 2 replication in an immunocompromised patient. J Infect Dis. 2021;223(1):23–7. https://doi.org/10.1093/infdis/jiaa666.",
"volume": "223",
"year": "2021"
},
{
"DOI": "10.1056/NEJMc2031364",
"author": "B Choi",
"doi-asserted-by": "publisher",
"first-page": "2291",
"issue": "23",
"journal-title": "N Engl J Med",
"key": "10740_CR2",
"unstructured": "Choi B, Choudhary MC, Regan J, Sparks JA, Padera RF, Qiu X, Solomon IH, Kuo HH, Boucau J, Bowman K, Adhikari UD, Winkler ML, Mueller AA, Hsu TY, Desjardins M, Baden LR, Chan BT, Walker BD, Lichterfeld M, Brigl M, Kwon DS, Kanjilal S, Richardson ET, Jonsson AH, Alter G, Barczak AK, Hanage WP, Yu XG, Gaiha GD, Seaman MS, Cernadas M, Li JZ. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med. 2020;383(23):2291–3. https://doi.org/10.1056/NEJMc2031364.",
"volume": "383",
"year": "2020"
},
{
"DOI": "10.1016/j.xcrm.2023.100943",
"author": "C Chaguza",
"doi-asserted-by": "publisher",
"first-page": "100943",
"issue": "2",
"journal-title": "Cell Rep Med",
"key": "10740_CR3",
"unstructured": "Chaguza C, Hahn AM, Petrone ME, Zhou S, Ferguson D, Breban MI, Pham K, Peña-Hernández MA, Castaldi C, Hill V, Yale, Schulz W, Swanstrom RI, Roberts SC, Grubaugh ND. Accelerated SARS-CoV-2 intrahost evolution leading to distinct genotypes during chronic infection. Cell Rep Med. 2023;4(2):100943. https://doi.org/10.1016/j.xcrm.2023.100943.",
"volume": "4",
"year": "2023"
},
{
"DOI": "10.1093/ve/veac050",
"author": "SAJ Wilkinson",
"doi-asserted-by": "publisher",
"first-page": "veac050",
"issue": "2",
"journal-title": "Virus Evol",
"key": "10740_CR4",
"unstructured": "Wilkinson SAJ, Richter A, Casey A, Osman H, Mirza JD, Stockton J, Quick J, Ratcliffe L, Sparks N, Cumley N, Poplawski R, Nicholls SN, Kele B, Harris K, Peacock TP, Loman NJ. Recurrent SARS-CoV-2 mutations in immunodeficient patients. Virus Evol. 2022;8(2):veac050. https://doi.org/10.1093/ve/veac050.",
"volume": "8",
"year": "2022"
},
{
"DOI": "10.1016/S2666-5247(23)00336-1",
"author": "Z Raglow",
"doi-asserted-by": "publisher",
"first-page": "e235",
"issue": "3",
"journal-title": "Lancet Microbe",
"key": "10740_CR5",
"unstructured": "Raglow Z, Surie D, Chappell JD, Zhu Y, Martin ET, Kwon JH, Frosch AE, Mohamed A, Gilbert J, Bendall EE, Bahr A, Halasa N, Talbot HK, Grijalva CG, Baughman A, Womack KN, Johnson C, Swan SA, Koumans E, McMorrow ML, Harcourt JL, Atherton LJ, Burroughs A, Thornburg NJ, Self WH, Lauring AS. Investigating respiratory viruses in the acutely ill (IVY) network. SARS-CoV-2 shedding and evolution in patients who were immunocompromised during the Omicron period: a multicentre, prospective analysis. Lancet Microbe. 2024;5(3):e235–46. https://doi.org/10.1016/S2666-5247(23)00336-1.",
"volume": "5",
"year": "2024"
},
{
"DOI": "10.1128/mbio.00110-24",
"author": "AD Marques",
"doi-asserted-by": "publisher",
"first-page": "e0011024",
"issue": "3",
"journal-title": "mBio",
"key": "10740_CR6",
"unstructured": "Marques AD, Graham-Wooten J, Fitzgerald AS, Sobel Leonard A, Cook EJ, Everett JK, Rodino KG, Moncla LH, Kelly BJ, Collman RG, Bushman FD. SARS-CoV-2 evolution during prolonged infection in immunocompromised patients. mBio. 2024;15(3):e0011024. https://doi.org/10.1128/mbio.00110-24.",
"volume": "15",
"year": "2024"
},
{
"DOI": "10.1093/cid/ciac769",
"author": "JI Hogan",
"doi-asserted-by": "publisher",
"first-page": "342",
"issue": "2",
"journal-title": "Clin Infect Dis",
"key": "10740_CR7",
"unstructured": "Hogan JI, Duerr R, Dimartino D, Marier C, Hochman SE, Mehta S, Wang G, Heguy A. Remdesivir resistance in transplant recipients with persistent coronavirus disease 2019. Clin Infect Dis. 2023;76(2):342–5. https://doi.org/10.1093/cid/ciac769.",
"volume": "76",
"year": "2023"
},
{
"DOI": "10.1016/S1473-3099(23)00815-0",
"doi-asserted-by": "publisher",
"key": "10740_CR8",
"unstructured": "Machkovech HM, Hahn AM, Garonzik Wang J, Grubaugh ND, Halfmann PJ, Johnson MC, Lemieux JE, O’Connor DH, Piantadosi A, Wei W, Friedrich TC. Persistent SARS-CoV-2 infection: significance and implications. Lancet Infect Dis 2024 Feb 7:S1473-3099(23)00815-0. doi: https://doi.org/10.1016/S1473-3099(23)00815-0."
},
{
"DOI": "10.3389/fviro.2022.942555",
"author": "M Ghafari",
"doi-asserted-by": "publisher",
"first-page": "942555",
"journal-title": "Front Virol",
"key": "10740_CR9",
"unstructured": "Ghafari M, Liu Q, Dhillon A, Katzourakis A, Weissman DB. Investigating the evolutionary origins of the first three SARS-CoV-2 variants of concern. Front Virol. 2022;2:942555.",
"volume": "2",
"year": "2022"
},
{
"DOI": "10.1038/s41591-022-01911-2",
"doi-asserted-by": "publisher",
"key": "10740_CR10",
"unstructured": "Tegally H, Moir M, Everatt J, Giovanetti M, Scheepers C, Wilkinson E, Subramoney K, Makatini Z, Moyo S, Amoako DG, Baxter C, Althaus CL, Anyaneji UJ, Kekana D, Viana R, Giandhari J, Lessells RJ, Maponga T, Maruapula D, Choga W, Matshaba M, Mbulawa MB, Msomi N, NGS -SA consortium;, Naidoo Y, Pillay S, Sanko TJ, San JE, Scott L, Singh L, Magini NA, Smith-Lawrence P, Stevens W, Dor G, Tshiabuila D, Wolter N, Preiser W, Treurnicht FK, Venter M, Chiloane G, McIntyre C, O’Toole A, Ruis C, Peacock TP, Roemer C, Kosakovsky Pond SL, Williamson C, Pybus OG, Bhiman JN, Glass A, Martin DP, Jackson B, Rambaut A, Laguda-Akingba O, Gaseitsiwe S, von Gottberg A, de Oliveira T. Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nat Med. 2022;28(9):1785–1790. https://doi.org/10.1038/s41591-022-01911-2"
},
{
"DOI": "10.1016/j.jiph.2022.11.024",
"author": "K Dhama",
"doi-asserted-by": "publisher",
"first-page": "4",
"issue": "1",
"journal-title": "J Infect Public Health",
"key": "10740_CR11",
"unstructured": "Dhama K, Nainu F, Frediansyah A, Yatoo MI, Mohapatra RK, Chakraborty S, Zhou H, Islam MR, Mamada SS, Kusuma HI, Rabaan AA, Alhumaid S, Mutair AA, Iqhrammullah M, Al-Tawfiq JA, Mohaini MA, Alsalman AJ, Tuli HS, Chakraborty C, Harapan H. Global emerging Omicron variant of SARS-CoV-2: impacts, challenges and strategies. J Infect Public Health. 2023;16(1):4–14. https://doi.org/10.1016/j.jiph.2022.11.024.",
"volume": "16",
"year": "2023"
},
{
"DOI": "10.1038/s41579-023-00878-2",
"author": "PV Markov",
"doi-asserted-by": "publisher",
"first-page": "361",
"journal-title": "Nat Rev Microbiol",
"key": "10740_CR12",
"unstructured": "Markov PV, Ghafari M, Beer M, et al. The evolution of SARS-CoV-2. Nat Rev Microbiol. 2023;21:361–79.",
"volume": "21",
"year": "2023"
},
{
"DOI": "10.1016/j.xcrm.2023.100943",
"author": "C Chaguza",
"doi-asserted-by": "publisher",
"first-page": "100943",
"journal-title": "Cell Rep Med",
"key": "10740_CR13",
"unstructured": "Chaguza C, Hahn AM, Petrone ME, et al. Accelerated SARS-CoV-2 intrahost evolution leading to distinct genotypes during chronic infection. Cell Rep Med. 2023;4:100943.",
"volume": "4",
"year": "2023"
},
{
"key": "10740_CR14",
"unstructured": "https://covdb.stanford.edu/drms, last access 05/02/2025."
},
{
"DOI": "10.1038/s41392-022-01281-8",
"author": "C Sun",
"doi-asserted-by": "publisher",
"first-page": "75",
"journal-title": "Sig Transduct Target Ther",
"key": "10740_CR15",
"unstructured": "Sun C, Wang H, Yang J, et al. Mutation N856K in Spike reduces fusogenicity and infectivity of Omicron BA.1. Sig Transduct Target Ther. 2023;8:75. https://doi.org/10.1038/s41392-022-01281-8.",
"volume": "8",
"year": "2023"
},
{
"DOI": "10.1056/NEJMc2031670",
"author": "T Aydillo",
"doi-asserted-by": "publisher",
"first-page": "2586",
"issue": "26",
"journal-title": "N Engl J Med",
"key": "10740_CR16",
"unstructured": "Aydillo T, Gonzalez-Reiche AS, Aslam S, van de Guchte A, Khan Z, Obla A, Dutta J, van Bakel H, Aberg J, García-Sastre A, Shah G, Hohl T, Papanicolaou G, Perales MA, Sepkowitz K, Babady NE, Kamboj M. Shedding of viable SARS-CoV-2 after immunosuppressive therapy for Cancer. N Engl J Med. 2020;383(26):2586–8. https://doi.org/10.1056/NEJMc2031670.",
"volume": "383",
"year": "2020"
},
{
"DOI": "10.1016/j.ijid.2023.02.011",
"author": "Y Wu",
"doi-asserted-by": "publisher",
"first-page": "228",
"journal-title": "Int J Infect Dis",
"key": "10740_CR17",
"unstructured": "Wu Y, Guo Z, Yuan J, Cao G, Wang Y, Gao P, Liu J, Liu M. Duration of viable virus shedding and polymerase chain reaction positivity of the SARS-CoV-2 Omicron variant in the upper respiratory tract: a systematic review and meta-analysis. Int J Infect Dis. 2023;129:228–35. https://doi.org/10.1016/j.ijid.2023.02.011.",
"volume": "129",
"year": "2023"
},
{
"DOI": "10.1016/j.jinf.2023.01.024",
"author": "SW Kang",
"doi-asserted-by": "publisher",
"first-page": "412",
"issue": "4",
"journal-title": "J Infect",
"key": "10740_CR18",
"unstructured": "Kang SW, Kim JW, Kim JY, Lim SY, Jang CY, Chang E, Yang JS, Kim KC, Jang HC, Kim D, Shin Y, Lee JY, Kim SH. Characteristics and risk factors of prolonged viable virus shedding in immunocompromised patients with COVID-19: a prospective cohort study. J Infect. 2023;86(4):412–4. https://doi.org/10.1016/j.jinf.2023.01.024.",
"volume": "86",
"year": "2023"
},
{
"DOI": "10.1038/s41467-024-47941-x",
"author": "H Igari",
"doi-asserted-by": "publisher",
"first-page": "3604",
"journal-title": "Nat Commun",
"key": "10740_CR19",
"unstructured": "Igari H, Sakao S, Ishige T, et al. Dynamic diversity of SARS-CoV-2 genetic mutations in a lung transplantation patient with persistent COVID-19. Nat Commun. 2024;15:3604. https://doi.org/10.1038/s41467-024-47941-x.",
"volume": "15",
"year": "2024"
},
{
"DOI": "10.1128/aac.00266-23",
"author": "R Palomino-Cabrera",
"doi-asserted-by": "publisher",
"first-page": "e0026623",
"issue": "7",
"journal-title": "Antimicrob Agents Chemother",
"key": "10740_CR20",
"unstructured": "Palomino-Cabrera R, Tejerina F, Molero-Salinas A, Ferris M, Veintimilla C, Catalán P, Rodríguez Macias G, Alonso R, Muñoz P, García de Viedma D, Pérez-Lago L. Gregorio Marañón Microbiology-ID COVID 19 study group. Frequent emergence of resistance mutations following complex Intra-Host genomic dynamics in SARS-CoV-2 patients receiving Sotrovimab. Antimicrob Agents Chemother. 2023;67(7):e0026623. https://doi.org/10.1128/aac.00266-23.",
"volume": "67",
"year": "2023"
},
{
"DOI": "10.1038/s42003-023-04923-x",
"author": "SB Park",
"doi-asserted-by": "publisher",
"first-page": "556",
"journal-title": "Commun Biol",
"key": "10740_CR21",
"unstructured": "Park SB, Khan M, Chiliveri SC, et al. SARS-CoV-2 Omicron variants harbor Spike protein mutations responsible for their attenuated fusogenic phenotype. Commun Biol. 2023;6:556. https://doi.org/10.1038/s42003-023-04923-x.",
"volume": "6",
"year": "2023"
},
{
"DOI": "10.1080/22221751.2021",
"author": "A Baj",
"doi-asserted-by": "publisher",
"first-page": "2010",
"issue": "1",
"journal-title": "Emerg Microbes Infect",
"key": "10740_CR22",
"unstructured": "Baj A, Novazzi F, Drago Ferrante F, Genoni A, Tettamanzi E, Catanoso G, Dalla Gasperina D, Dentali F, Focosi D, Maggi F. Spike protein evolution in the SARS-CoV-2 Delta variant of concern: a case series from Northern Lombardy. Emerg Microbes Infect. 2021;10(1):2010–5. https://doi.org/10.1080/22221751.2021.",
"volume": "10",
"year": "2021"
},
{
"DOI": "10.1007/s13337-021-00747-7",
"author": "D Paul",
"doi-asserted-by": "publisher",
"first-page": "703",
"issue": "4",
"journal-title": "Virus Disease",
"key": "10740_CR23",
"unstructured": "Paul D, Pyne N, Paul S. Mutation profile of SARS-CoV-2 Spike protein and identification of potential multiple epitopes within Spike protein for vaccine development against SARS-CoV-2. Virus Disease. 2021;32(4):703–26. https://doi.org/10.1007/s13337-021-00747-7.",
"volume": "32",
"year": "2021"
},
{
"DOI": "10.2217/fvl-2022-0181",
"doi-asserted-by": "publisher",
"key": "10740_CR24",
"unstructured": "Ciccozzi M, Pascarella S. Two sides of the same coin: the N-terminal and the receptor binding domains of SARS-CoV-2 Spike. Future Virol. 2023 Feb. https://doi.org/10.2217/fvl-2022-0181."
},
{
"DOI": "10.3390/cimb46030165",
"author": "BJJ Subong",
"doi-asserted-by": "publisher",
"first-page": "2598",
"issue": "3",
"journal-title": "Curr Issues Mol Biol",
"key": "10740_CR25",
"unstructured": "Subong BJJ, Ozawa T. Bio-Chemoinformatics-Driven analysis of nsp7 and nsp8 mutations and their effects on viral replication protein complex stability. Curr Issues Mol Biol. 2024;46(3):2598–619. https://doi.org/10.3390/cimb46030165.",
"volume": "46",
"year": "2024"
},
{
"DOI": "10.1126/science.abg0821",
"author": "KA Lythgoe",
"doi-asserted-by": "publisher",
"first-page": "eabg0821",
"issue": "6539",
"journal-title": "Science",
"key": "10740_CR26",
"unstructured": "Lythgoe KA, Hall M, Ferretti L, de Cesare M, MacIntyre-Cockett G, Trebes A, Andersson M, Otecko N, Wise EL, Moore N, Lynch J, Kidd S, Cortes N, Mori M, Williams R, Vernet G, Justice A, Green A, Nicholls SM, Ansari MA, Abeler-Dörner L, Moore CE, Peto TEA, Eyre DW, Shaw R, Simmonds P, Buck D, Todd JA, Oxford Virus Sequencing Analysis Group (OVSG), Connor TR, Ashraf S, da Silva Filipe A, Shepherd J, Thomson EC, COVID-19 Genomics UK (COG-UK) Consortium, Bonsall D, Fraser C, Golubchik T. SARS-CoV-2 within-host diversity and transmission. Science. 2021;372(6539):eabg0821. https://doi.org/10.1126/science.abg0821.",
"volume": "372",
"year": "2021"
},
{
"DOI": "10.1016/j.celrep.2021.110205",
"author": "J Li",
"doi-asserted-by": "publisher",
"first-page": "110205",
"issue": "2",
"journal-title": "Cell Rep",
"key": "10740_CR27",
"unstructured": "Li J, Du P, Yang L, Zhang J, Song C, Chen D, Song Y, Ding N, Hua M, Han K, Song R, Xie W, Chen Z, Wang X, Liu J, Xu Y, Gao G, Wang Q, Pu L, Di L, Li J, Yue J, Han J, Zhao X, Yan Y, Yu F, Wu AR, Zhang F, Gao YQ, Huang Y, Wang J, Zeng H, Chen C. Two-step fitness selection for intra-host variations in SARS-CoV-2. Cell Rep. 2022;38(2):110205. https://doi.org/10.1016/j.celrep.2021.110205.",
"volume": "38",
"year": "2022"
},
{
"DOI": "10.1093/molbev/msad204",
"author": "M Hou",
"doi-asserted-by": "publisher",
"first-page": "msad204",
"issue": "9",
"journal-title": "Mol Biol Evol",
"key": "10740_CR28",
"unstructured": "Hou M, Shi J, Gong Z, Wen H, Lan Y, Deng X, Fan Q, Li J, Jiang M, Tang X, Wu CI, Li F, Ruan Y. Intra- vs. Interhost evolution of SARS-CoV-2 driven by uncorrelated Selection-The evolution thwarted. Mol Biol Evol. 2023;40(9):msad204. https://doi.org/10.1093/molbev/msad204.",
"volume": "40",
"year": "2023"
},
{
"DOI": "10.1038/s41586-021-03291-y",
"author": "SA Kemp",
"doi-asserted-by": "publisher",
"first-page": "277",
"journal-title": "Nature",
"key": "10740_CR29",
"unstructured": "Kemp SA, Collier DA, Datir RP, Ferreira I, Gayed S, Jahun A, Hosmillo M, Rees-Spear C, Mlcochova P, Lumb IU, et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592:277–82.",
"volume": "592",
"year": "2021"
},
{
"DOI": "10.1128/jvi.01618-23",
"author": "M Farjo",
"doi-asserted-by": "publisher",
"first-page": "e0161823",
"issue": "1",
"journal-title": "J Virol",
"key": "10740_CR30",
"unstructured": "Farjo M, Koelle K, Martin MA, Gibson LL, Walden KKO, Rendon G, Fields CJ, Alnaji FG, Gallagher N, Luo CH, Mostafa HH, Manabe YC, Pekosz A, Smith RL, McManus DD, Brooke CB. Within-host evolutionary dynamics and tissue compartmentalization during acute SARS-CoV-2 infection. J Virol. 2024;98(1):e0161823. https://doi.org/10.1128/jvi.01618-23.",
"volume": "98",
"year": "2024"
},
{
"DOI": "10.1016/S1473-3099(24)00353-0",
"author": "MH Choi",
"doi-asserted-by": "publisher",
"first-page": "1213",
"issue": "11",
"journal-title": "Lancet Infect Dis",
"key": "10740_CR31",
"unstructured": "Choi MH, Wan EYF, Wong ICK, Chan EWY, Chu WM, Tam AR, Yuen KY, Hung IFN. Comparative effectiveness of combination therapy with nirmatrelvir-ritonavir and Remdesivir versus monotherapy with Remdesivir or nirmatrelvir-ritonavir in patients hospitalised with COVID-19: a target trial emulation study. Lancet Infect Dis. 2024;24(11):1213–24. https://doi.org/10.1016/S1473-3099(24)00353-0.",
"volume": "24",
"year": "2024"
},
{
"DOI": "10.3389/fphar.2024.1401658",
"author": "Y Qiu",
"doi-asserted-by": "publisher",
"first-page": "1401658",
"journal-title": "Front Pharmacol",
"key": "10740_CR32",
"unstructured": "Qiu Y, Wen H, Wang H, Sun W, Li G, Li S, Wang Y, Zhai J, Zhan Y, Su Y, Long Z, Li Z, Ye F. Real-world effectiveness and safety of nirmatrelvir-ritonavir (Paxlovid)-treated for COVID-19 patients with onset of more than 5 days: a retrospective cohort study. Front Pharmacol. 2024;15:1401658. https://doi.org/10.3389/fphar.2024.1401658.",
"volume": "15",
"year": "2024"
}
],
"reference-count": 32,
"references-count": 32,
"relation": {},
"resource": {
"primary": {
"URL": "https://bmcinfectdis.biomedcentral.com/articles/10.1186/s12879-025-10740-w"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "SARS-CoV-2 genomic evolution during a severe and long-lasting omicron infection under antiviral therapy",
"type": "journal-article",
"update-policy": "https://doi.org/10.1007/springer_crossmark_policy",
"volume": "25"
}