Comparison of the Neutralization Power of Sotrovimab Against SARS-CoV-2 Variants: Development of a Rapid Computational Method
et al., JMIR Bioinformatics and Biotechnology, doi:10.2196/58018, Oct 2024
Sotrovimab for COVID-19
45th treatment shown to reduce risk in
August 2022, now with p = 0.00048 from 29 studies, recognized in 42 countries.
Efficacy is variant dependent.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
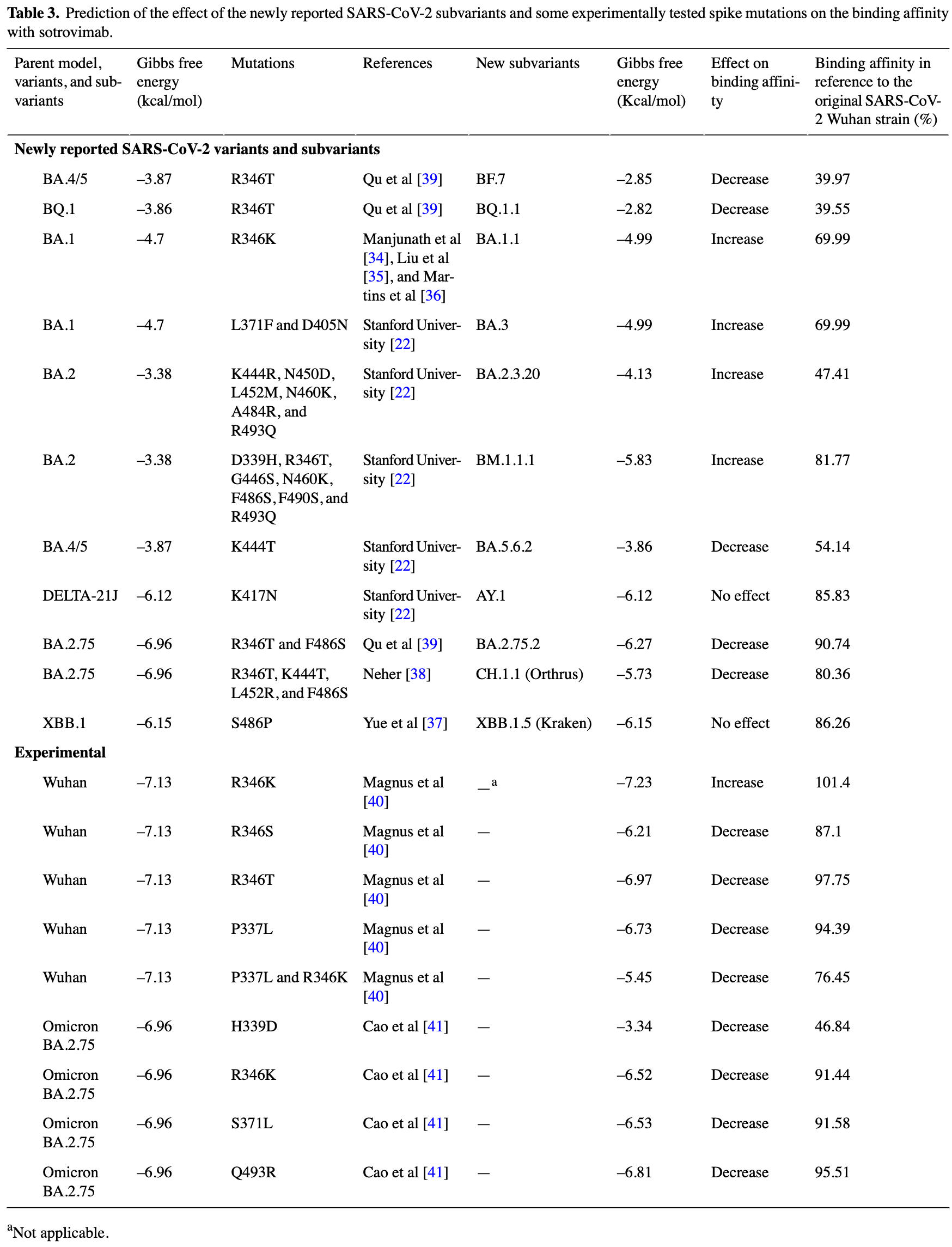
In silico computational method developed to predict the neutralization power of monoclonal antibodies against new SARS-CoV-2 variants. The method evaluates binding affinity of antibodies based on molecular interactions and Gibbs free energy compared to a reference model. Analysis of sotrovimab (S309) shows decreased binding affinity to delta and omicron variants compared to the original Wuhan strain. The G339H and G339D mutations are found to play a key role in sotrovimab's reduced neutralization of omicron subvariants, which aligns with published clinical reports.
Ashoor et al., 10 Oct 2024, peer-reviewed, 3 authors.
Contact: danana@agu.edu.bh.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Comparison of the Neutralization Power of Sotrovimab Against SARS-CoV-2 Variants: Development of a Rapid Computational Method
JMIR Bioinformatics and Biotechnology, doi:10.2196/58018
Background: The rapid evolution of SARS-CoV-2 imposed a huge challenge on disease control. Immune evasion caused by genetic variations of the SARS-CoV-2 spike protein's immunogenic epitopes affects the efficiency of monoclonal antibody-based therapy of COVID-19. Therefore, a rapid method is needed to evaluate the efficacy of the available monoclonal antibodies against the new emerging variants or potential novel variants.
Objective: The aim of this study is to develop a rapid computational method to evaluate the neutralization power of anti-SARS-CoV-2 monoclonal antibodies against new SARS-CoV-2 variants and other potential new mutations.
Methods: The amino acid sequence of the extracellular domain of the spike proteins of the severe acute respiratory syndrome coronavirus (GenBank accession number YP_009825051.1) and SARS-CoV-2 (GenBank accession number YP_009724390.1) were used to create computational 3D models for the native spike proteins. Specific mutations were introduced to the curated sequence to generate the different variant spike models. The neutralization potential of sotrovimab (S309) against these variants was evaluated based on its molecular interactions and Gibbs free energy in comparison to a reference model after molecular replacement of the reference receptor-binding domain with the variant's receptor-binding domain.
Results: Our results show a loss in the binding affinity of the neutralizing antibody S309 with both SARS-CoV and SARS-CoV-2. The binding affinity of S309 was greater to the Alpha, Beta, Gamma, and Kappa variants than to the original Wuhan strain of SARS-CoV-2. However, S309 showed a substantially decreased binding affinity to the Delta and Omicron variants. Based on the mutational profile of Omicron subvariants, our data describe the effect of the G339H and G339D mutations and their role in escaping antibody neutralization, which is in line with published clinical reports. Conclusions: This method is rapid, applicable, and of interest to adapt the use of therapeutic antibodies to the treatment of emerging variants. It could be applied to antibody-based treatment of other viral infections.
Authors' Contributions DA carried out the in silico analysis, designed the methodology, curated the data, and drafted and edited the manuscript. MM designed the illustrations and figures. M-DF conceptualized the study, analyzed the data, drafted and edited the manuscript, and supervised the study.
Conflicts of Interest None declared.
Multimedia
References
Aggarwal, Beaty, Bennett, Carlson, Mayer et al., Change in effectiveness of sotrovimab for preventing hospitalization and mortality for at-risk COVID-19 outpatients during an Omicron BA.1 and BA.1.1-predominant phase, Int J Infect Dis, doi:10.1016/j.ijid.2022.10.002
Amicone, Borges, Alves, Isidro, Zé-Zé et al., Mutation rate of SARS-CoV-2 and emergence of mutators during experimental evolution, Evol Med Public Health, doi:10.1093/emph/eoac010
Arora, Kempf, Nehlmeier, Schulz, Cossmann et al., Augmented neutralisation resistance of emerging omicron subvariants BA.2.12, Lancet Infect Dis, doi:10.1016/s1473-3099(22)00422-4
Ashoor, Khalaf, Marzouq, Jarjanazi, Chlif et al., A computational approach to evaluate the combined effect of SARS-CoV-2 RBD mutations and ACE2 receptor genetic variants on infectivity: the COVID-19 host-pathogen nexus, Front Cell Infect Microbiol, doi:10.3389/fcimb.2021.707194
Ashoor, Marzouq, Trabelsi, Chlif, Abotalib et al., How concerning is a SARS-CoV-2 variant of concern? Computational predictions and the variants labeling system, Front Cell Infect Microbiol, doi:10.3389/fcimb.2022.868205
Asif, Ilyas, Abdullah, Sarfraz, Mustafa et al., The comparison of mutational progression in SARS-CoV-2: a short updated overview, JMP. Oct, doi:10.3390/jmp3040018
Barnes, Jette, Abernathy, Dam, Esswein et al., SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies, Nature. Dec, doi:10.1038/s41586-020-2852-1
Benkert, Biasini, Schwede, Toward the estimation of the absolute quality of individual protein structure models, Bioinformatics, doi:10.1093/bioinformatics/btq662
Cao, Wang, Jian, Song, Yisimayi, Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies, Nature. Feb, doi:10.1038/s41586-021-04385-3
Cheng, Li, Zhan, Zhao, Xue et al., Clinical application of antibody immunity against SARS-CoV-2: comprehensive review on immunoassay and immunotherapy, Clin Rev Allergy Immunol, doi:10.1007/s12016-021-08912-y
Cox, Peacock, Harvey, Hughes, Wright et al., SARS-CoV-2 variant evasion of monoclonal antibodies based on in vitro studies, Nat Rev Microbiol, doi:10.1038/s41579-022-00809-7
Dejnirattisai, Zhou, Ginn, Duyvesteyn, Supasa et al., The antigenic anatomy of SARS-CoV-2 receptor binding domain, Cell. Apr, doi:10.1016/j.cell.2021.02.032
Delano, The PyMOL Molecular Graphics System, PyMOL
Focosi, Quiroga, Mcconnell, Johnson, Casadevall, Convergent evolution in SARS-CoV-2 spike creates a variant soup from which new COVID-19 waves emerge, Int J Mol Sci. Jan, doi:10.3390/ijms24032264
Guex, Peitsch, SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling, Electrophoresis. Dec, doi:10.1002/elps.1150181505
Gupta, Gonzalez-Rojas, Juarez, Casal, Moya et al., Early treatment for Covid-19 with SARS-CoV-2 neutralizing antibody sotrovimab, N Engl J Med. Nov, doi:10.1056/nejmoa2107934
Gupta, Gonzalez-Rojas, Juarez, Casal, Moya et al., Effect of sotrovimab on hospitalization or death among high-risk patients with mild to moderate COVID-19: a randomized clinical trial, JAMA. Apr, doi:10.1001/jama.2022.2832
Hastie, Li, Bedinger, Schendel, Dennison et al., Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: a global consortium study, Science, doi:10.1126/science.abh2315
Heo, Sotrovimab: first approval, Drugs. Mar, doi:10.1007/s40265-022-01690-7
Hodcroft, Covariants, CoVariants
Huang, Wu, Zheng, Xie, He et al., Atlas of currently available human neutralizing antibodies against SARS-CoV-2 and escape by Omicron sub-variants BA, BA.3. Immunity, doi:10.1016/j.immuni.2022.06.005
Imai, Ito, Kiso, Yamayoshi, Uraki et al., Efficacy of antiviral agents against omicron subvariants BQ.1.1 and XBB, N Engl J Med. Jan, doi:10.1056/nejmc2214302
Kumar, Karuppanan, Subramaniam, Omicron (BA.1) and sub-variants (BA.1.1, BA.2, and BA.3) of SARS-CoV-2 spike infectivity and pathogenicity: a comparative sequence and structural-based computational assessment, J Med Virol, doi:10.1002/jmv.27927
Laskowski, Swindells, LigPlot+: multiple ligand-protein interaction diagrams for drug discovery, J Chem Inf Model, doi:10.1021/ci200227u
Liu, Xiong, Sun, Hu, Thilakavathy et al., Omicron: a chimera of two early SARS-CoV-2 lineages, Signal Transduct Target Ther. Mar, doi:10.1038/s41392-022-00949-5
Lv, Deng, Ye, Cao, Sun et al., Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody, Science. Sep, doi:10.1126/science.abc5881
Magnus, Hiergeist, Schuster, Rohrhofer, Medenbach et al., Targeted escape of SARS-CoV-2 from monoclonal antibody S309, the precursor of sotrovimab, Front Immunol, doi:10.3389/fimmu.2022.966236
Manjunath, Gaonkar, Saleh, Husain, A comprehensive review on Covid-19 Omicron (B.1.1.529) variant, Saudi J Biol Sci, doi:10.1016/j.sjbs.2022.103372
Mannar, Saville, Sun, Zhu, Marti et al., SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization, Nat Commun. Aug, doi:10.1038/s41467-022-32262-8
Martins, Do Nascimento, Nooruzzaman, Yuan, Chen et al., The omicron variant BA.1.1 presents a lower pathogenicity than B.1 D614G and delta variants in a feline model of SARS-CoV-2 infection, J Virol, doi:10.1128/jvi.00961-22
Mccallum, Czudnochowski, Rosen, Zepeda, Bowen et al., Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement, Science. Feb, doi:10.1126/science.abn8652
Mittal, Khattri, Verma, Structural and antigenic variations in the spike protein of emerging SARS-CoV-2 variants, PLoS Pathog, doi:10.1371/journal.ppat.1010260
Mohapatra, Mahal, Kutikuppala, Pal, Kandi et al., Renewed global threat by the novel SARS-CoV-2 variants, Front Virol, doi:10.3389/fviro.2022.1077155
Myung, Pires, Ascher, CSM-AB: graph-based antibody-antigen binding affinity prediction and docking scoring function, Bioinformatics. Jan, doi:10.1093/bioinformatics/btab762
Neher, Variant report 2022-12-22
Piccoli, Park, Tortorici, Czudnochowski, Walls et al., Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology, Cell. Nov, doi:10.1016/j.cell.2020.09.037
Pinto, Park, Beltramello, Walls, Tortorici et al., Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody, Nature. Jul, doi:10.1038/s41586-020-2349-y
Qu, Evans, Faraone, Zheng, Carlin et al., Enhanced neutralization resistance of SARS-CoV-2 Omicron subvariants BQ.1, Cell Host Microbe, doi:10.1016/j.chom.2022.11.012
Saville, Mannar, Zhu, Srivastava, Berezuk et al., Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants, Nat Commun. Feb, doi:10.1038/s41467-022-28324-6
Song, Gui, Wang, Xiang, Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2, PLoS Pathog, doi:10.1371/journal.ppat.1007236
Studer, Rempfer, Waterhouse, Gumienny, Haas et al., QMEANDisCo-distance constraints applied on model quality estimation, Bioinformatics. Mar, doi:10.1093/bioinformatics/btz828
Van De Veerdonk, Bourboulis, Pickkers, Derde, Leavis et al., A guide to immunotherapy for COVID-19, Nat Med. Jan, doi:10.1038/s41591-021-01643-9
Van Dorp, Acman, Richard, Shaw, Ford et al., Emergence of genomic diversity and recurrent mutations in SARS-CoV-2, Infect Genet Evol, doi:10.1016/j.meegid.2020.104351
Wang, Liu, Zhang, Wang, Hong et al., Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies, Nat Commun. Feb, doi:10.1038/s41467-022-28528-w
Waterhouse, Bertoni, Bienert, Studer, Tauriello et al., SWISS-MODEL: homology modelling of protein structures and complexes, Nucleic Acids Res, doi:10.1093/nar/gky427
Willett, Grove, Maclean, Wilkie, Lorenzo et al., SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway, Nat Microbiol, doi:10.1038/s41564-022-01143-7
Yue, Song, Wang, Jian, Chen et al., Enhanced transmissibility of XBB.1.5 is contributed by both strong ACE2 binding and antibody evasion, bioRxiv, doi:10.1101/2023.01.03.522427
Zhao, Zhou, Tian, Huang, Liu et al., Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape, JMIR Bioinform Biotech, doi:10.1038/s41467-022-32665-7
DOI record:
{
"DOI": "10.2196/58018",
"ISSN": [
"2563-3570"
],
"URL": "http://dx.doi.org/10.2196/58018",
"abstract": "<jats:sec>\n <jats:title>Background</jats:title>\n <jats:p>The rapid evolution of SARS-CoV-2 imposed a huge challenge on disease control. Immune evasion caused by genetic variations of the SARS-CoV-2 spike protein’s immunogenic epitopes affects the efficiency of monoclonal antibody–based therapy of COVID-19. Therefore, a rapid method is needed to evaluate the efficacy of the available monoclonal antibodies against the new emerging variants or potential novel variants.</jats:p>\n </jats:sec>\n <jats:sec>\n <jats:title>Objective</jats:title>\n <jats:p>The aim of this study is to develop a rapid computational method to evaluate the neutralization power of anti–SARS-CoV-2 monoclonal antibodies against new SARS-CoV-2 variants and other potential new mutations.</jats:p>\n </jats:sec>\n <jats:sec>\n <jats:title>Methods</jats:title>\n <jats:p>The amino acid sequence of the extracellular domain of the spike proteins of the severe acute respiratory syndrome coronavirus (GenBank accession number YP_009825051.1) and SARS-CoV-2 (GenBank accession number YP_009724390.1) were used to create computational 3D models for the native spike proteins. Specific mutations were introduced to the curated sequence to generate the different variant spike models. The neutralization potential of sotrovimab (S309) against these variants was evaluated based on its molecular interactions and Gibbs free energy in comparison to a reference model after molecular replacement of the reference receptor-binding domain with the variant’s receptor-binding domain.</jats:p>\n </jats:sec>\n <jats:sec>\n <jats:title>Results</jats:title>\n <jats:p>Our results show a loss in the binding affinity of the neutralizing antibody S309 with both SARS-CoV and SARS-CoV-2. The binding affinity of S309 was greater to the Alpha, Beta, Gamma, and Kappa variants than to the original Wuhan strain of SARS-CoV-2. However, S309 showed a substantially decreased binding affinity to the Delta and Omicron variants. Based on the mutational profile of Omicron subvariants, our data describe the effect of the G339H and G339D mutations and their role in escaping antibody neutralization, which is in line with published clinical reports.</jats:p>\n </jats:sec>\n <jats:sec>\n <jats:title>Conclusions</jats:title>\n <jats:p>This method is rapid, applicable, and of interest to adapt the use of therapeutic antibodies to the treatment of emerging variants. It could be applied to antibody-based treatment of other viral infections.</jats:p>\n </jats:sec>",
"author": [
{
"ORCID": "http://orcid.org/0000-0002-0376-6859",
"affiliation": [],
"authenticated-orcid": false,
"family": "Ashoor",
"given": "Dana",
"sequence": "first"
},
{
"ORCID": "http://orcid.org/0000-0001-9881-0412",
"affiliation": [],
"authenticated-orcid": false,
"family": "Marzouq",
"given": "Maryam",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0001-8834-7460",
"affiliation": [],
"authenticated-orcid": false,
"family": "Fathallah",
"given": "M-Dahmani",
"sequence": "additional"
}
],
"container-title": "JMIR Bioinformatics and Biotechnology",
"container-title-short": "JMIR Bioinform Biotech",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2024,
4,
19
]
],
"date-time": "2024-04-19T04:15:54Z",
"timestamp": 1713500154000
},
"deposited": {
"date-parts": [
[
2024,
10,
10
]
],
"date-time": "2024-10-10T14:00:43Z",
"timestamp": 1728568843000
},
"indexed": {
"date-parts": [
[
2024,
10,
11
]
],
"date-time": "2024-10-11T04:17:57Z",
"timestamp": 1728620277646
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2024,
10,
10
]
]
},
"language": "en",
"member": "1010",
"original-title": [],
"page": "e58018",
"prefix": "10.2196",
"published": {
"date-parts": [
[
2024,
10,
10
]
]
},
"published-online": {
"date-parts": [
[
2024,
10,
10
]
]
},
"publisher": "JMIR Publications Inc.",
"reference": [
{
"DOI": "10.1016/j.meegid.2020.104351",
"doi-asserted-by": "publisher",
"key": "ref1"
},
{
"DOI": "10.1093/emph/eoac010",
"doi-asserted-by": "publisher",
"key": "ref2"
},
{
"DOI": "10.3390/ijms24032264",
"doi-asserted-by": "publisher",
"key": "ref3"
},
{
"key": "ref4",
"unstructured": "Emergency Use AuthorizationFood and Drug Administration2024-09-17https://www.fda.gov/emergency-preparedness-and-response/mcm-legal-regulatory-and-policy-framework/emergency-use-authorization#coviddrugs"
},
{
"key": "ref5",
"unstructured": "COVID-19 medicinesEuropean Medicines Agency2024-09-17https://www.ema.europa.eu/en/human-regulatory/overview/public-health-threats/coronavirus-disease-covid-19/treatments-vaccines/covid-19-treatments"
},
{
"DOI": "10.1126/science.abh2315",
"doi-asserted-by": "publisher",
"key": "ref6"
},
{
"DOI": "10.1371/journal.ppat.1010260",
"doi-asserted-by": "publisher",
"key": "ref7"
},
{
"DOI": "10.1038/s41586-020-2852-1",
"doi-asserted-by": "publisher",
"key": "ref8"
},
{
"DOI": "10.1016/j.cell.2020.09.037",
"doi-asserted-by": "publisher",
"key": "ref9"
},
{
"DOI": "10.1038/s41586-020-2349-y",
"doi-asserted-by": "publisher",
"key": "ref10"
},
{
"DOI": "10.1002/jmv.27927",
"doi-asserted-by": "publisher",
"key": "ref11"
},
{
"DOI": "10.1126/science.abn8652",
"doi-asserted-by": "publisher",
"key": "ref12"
},
{
"DOI": "10.1007/s40265-022-01690-7",
"doi-asserted-by": "publisher",
"key": "ref13"
},
{
"DOI": "10.3389/fcimb.2021.707194",
"doi-asserted-by": "publisher",
"key": "ref14"
},
{
"DOI": "10.1038/s41467-022-32665-7",
"doi-asserted-by": "publisher",
"key": "ref15"
},
{
"DOI": "10.1126/science.abc5881",
"doi-asserted-by": "publisher",
"key": "ref16"
},
{
"DOI": "10.1056/nejmoa2107934",
"doi-asserted-by": "publisher",
"key": "ref17"
},
{
"DOI": "10.1001/jama.2022.2832",
"doi-asserted-by": "publisher",
"key": "ref18"
},
{
"DOI": "10.3389/fcimb.2022.868205",
"doi-asserted-by": "publisher",
"key": "ref19"
},
{
"key": "ref20",
"unstructured": "Protein Data BankRCSB2024-09-17https://www.rcsb.org/"
},
{
"key": "ref21",
"unstructured": "HodcroftECoVariantsCoVariants2024-09-17https://covariants.org/"
},
{
"key": "ref22",
"unstructured": "SARS-CoV-2 VariantsStanford University: Coronavirus Antiviral & Resistance Database2024-09-17https://covdb.stanford.edu/variants/omicron_ba_1_3/"
},
{
"DOI": "10.1093/nar/gky427",
"doi-asserted-by": "publisher",
"key": "ref23"
},
{
"key": "ref24",
"unstructured": "DeLanoWThe PyMOL Molecular Graphics SystemPyMOL20022024-09-17http://www.pymol.org/"
},
{
"DOI": "10.1371/journal.ppat.1007236",
"doi-asserted-by": "publisher",
"key": "ref25"
},
{
"DOI": "10.1016/j.cell.2021.02.032",
"doi-asserted-by": "publisher",
"key": "ref26"
},
{
"DOI": "10.1038/s41467-022-32262-8",
"doi-asserted-by": "publisher",
"key": "ref27"
},
{
"DOI": "10.1038/s41467-022-28528-w",
"doi-asserted-by": "publisher",
"key": "ref28"
},
{
"DOI": "10.1038/s41467-022-28324-6",
"doi-asserted-by": "publisher",
"key": "ref29"
},
{
"key": "ref30",
"unstructured": "The GROMOS Software for (Bio)Molecular Simulation. Volume 1: About the GROMOS package: OverviewThe GROMOS Software for (Bio)Molecular Simulation. Volume 1: About the GROMOS package: Overview20232024-09-17https://www.gromos.net/gromos11_pdf_manuals/vol1.pdf"
},
{
"DOI": "10.1002/elps.1150181505",
"doi-asserted-by": "publisher",
"key": "ref31"
},
{
"DOI": "10.1021/ci200227u",
"doi-asserted-by": "publisher",
"key": "ref32"
},
{
"DOI": "10.1093/bioinformatics/btab762",
"doi-asserted-by": "publisher",
"key": "ref33"
},
{
"DOI": "10.1016/j.sjbs.2022.103372",
"doi-asserted-by": "publisher",
"key": "ref34"
},
{
"DOI": "10.1038/s41392-022-00949-5",
"doi-asserted-by": "publisher",
"key": "ref35"
},
{
"DOI": "10.1128/jvi.00961-22",
"doi-asserted-by": "publisher",
"key": "ref36"
},
{
"DOI": "10.1101/2023.01.03.522427",
"doi-asserted-by": "publisher",
"key": "ref37"
},
{
"key": "ref38",
"unstructured": "NeherRVariant report 2022-12-22GitHub2024-09-17https://github.com/neherlab/SARS-CoV-2_variant-reports/blob/d2d531c6deb12e52e5a6fde9af25f2cce023302b/reports/variant_report_2022-12-22.md"
},
{
"DOI": "10.1016/j.chom.2022.11.012",
"doi-asserted-by": "publisher",
"key": "ref39"
},
{
"DOI": "10.3389/fimmu.2022.966236",
"doi-asserted-by": "publisher",
"key": "ref40"
},
{
"DOI": "10.1038/s41586-021-04385-3",
"doi-asserted-by": "publisher",
"key": "ref41"
},
{
"DOI": "10.1093/bioinformatics/btq662",
"doi-asserted-by": "publisher",
"key": "ref42"
},
{
"DOI": "10.1093/bioinformatics/btz828",
"doi-asserted-by": "publisher",
"key": "ref43"
},
{
"DOI": "10.3390/jmp3040018",
"doi-asserted-by": "publisher",
"key": "ref44"
},
{
"DOI": "10.1016/j.immuni.2022.06.005",
"doi-asserted-by": "publisher",
"key": "ref45"
},
{
"DOI": "10.1038/s41591-021-01643-9",
"doi-asserted-by": "publisher",
"key": "ref46"
},
{
"DOI": "10.1007/s12016-021-08912-y",
"doi-asserted-by": "publisher",
"key": "ref47"
},
{
"DOI": "10.1016/j.ijid.2022.10.002",
"doi-asserted-by": "publisher",
"key": "ref48"
},
{
"DOI": "10.1016/s1473-3099(22)00422-4",
"doi-asserted-by": "publisher",
"key": "ref49"
},
{
"DOI": "10.1056/nejmc2214302",
"doi-asserted-by": "publisher",
"key": "ref50"
},
{
"DOI": "10.1038/s41579-022-00809-7",
"doi-asserted-by": "publisher",
"key": "ref51"
},
{
"key": "ref52",
"unstructured": "COVID-19 weekly epidemiological update, edition 119, 23 November 2022World Health Organization20222024-09-17https://iris.who.int/handle/10665/364724"
},
{
"DOI": "10.1038/s41564-022-01143-7",
"doi-asserted-by": "publisher",
"key": "ref53"
},
{
"DOI": "10.3389/fviro.2022.1077155",
"doi-asserted-by": "publisher",
"key": "ref54"
}
],
"reference-count": 54,
"references-count": 54,
"relation": {
"has-preprint": [
{
"asserted-by": "object",
"id": "10.2196/preprints.58018",
"id-type": "doi"
}
]
},
"resource": {
"primary": {
"URL": "https://bioinform.jmir.org/2024/1/e58018"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Comparison of the Neutralization Power of Sotrovimab Against SARS-CoV-2 Variants: Development of a Rapid Computational Method",
"type": "journal-article",
"volume": "5"
}