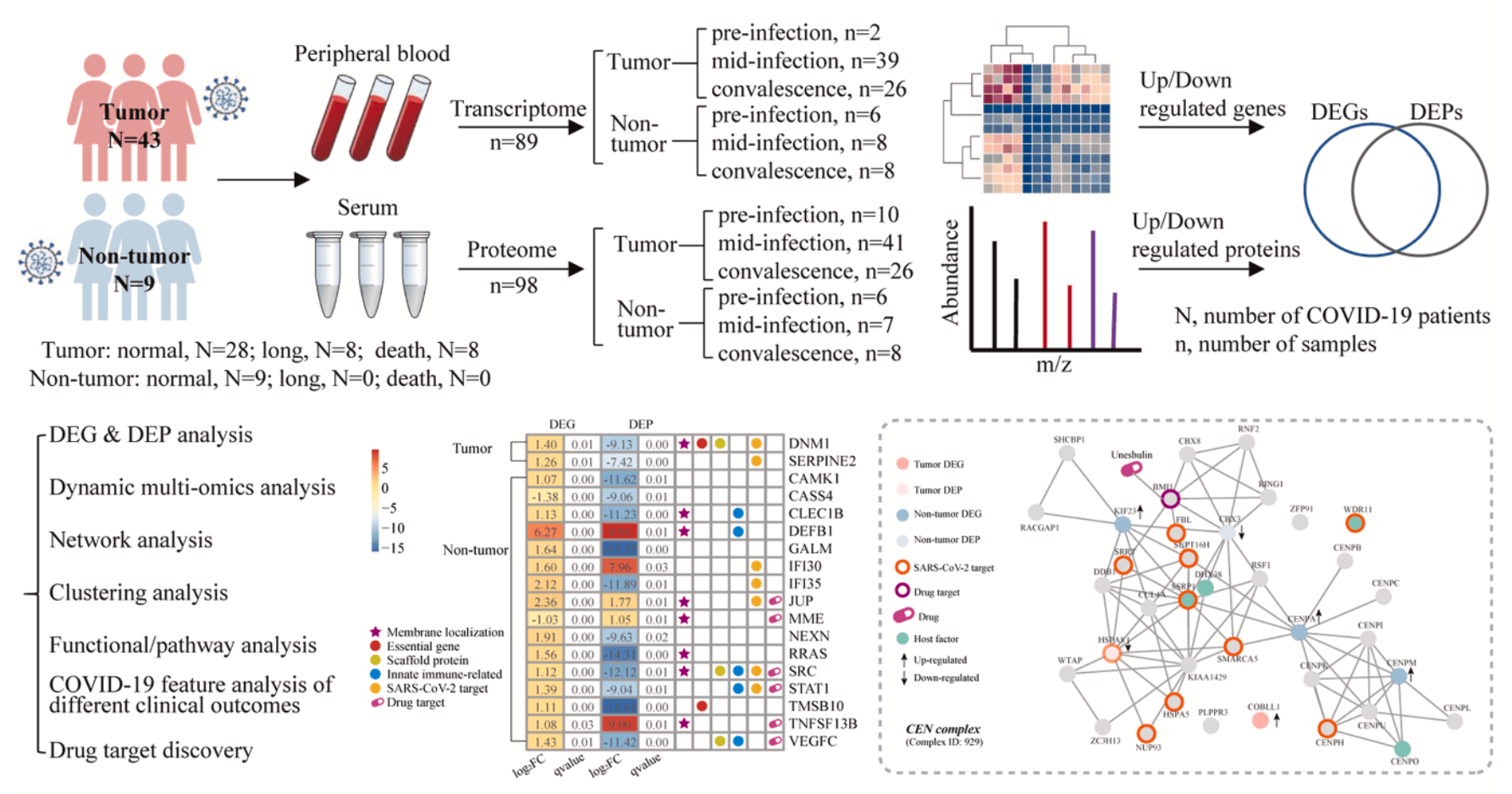
Multi-Omics Blood Atlas Reveals Host Immune Response Features of Immunocompromised Populations Following SARS-CoV-2 Infection
et al., Molecular & Cellular Proteomics, doi:10.1016/j.mcpro.2025.101068, Sep 2025
Multi-omics study analyzing blood samples from 52 COVID-19 patients showing distinct immune dysregulation patterns and identifying potential therapeutic targets.
Yang et al., 10 Sep 2025, peer-reviewed, 11 authors.
Contact: walzyaw@163.com, dongy@hsc.pku.edu.cn.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Multi-Omics Blood Atlas Reveals Host Immune Response Features of Immunocompromised Populations Following SARS-CoV-2 Infection
Molecular & Cellular Proteomics, doi:10.1016/j.mcpro.2025.101068
This multi-omics study reveals distinct immune dysregulation in hematological tumor patients with COVID-19, showing severe proteomic network perturbations and divergent immune signatures between long COVID (immunosuppression) and fatal cases (hyperactivation). Network analysis identifies potential therapeutic targets (e.g., HSPA8, SRC, and STAT1) and drug candidates, offering insights into COVID-19 mechanisms and tailored treatment strategies for high-risk patients.
DATA AVAILABILITY Data supporting the findings of this study have been deposited in iProX (integrated proteome resources) Proteo-meXchange under accession code PXD063172. The data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The data that support the findings of this study are available from the corresponding author upon reasonable request. Supplemental Data -This article contains supplemental data.
Ethics Approval and Consent to
Conflict of Interest - The authors declare that they do not have any conflicts of interest with the content of this article.
References
Ahern, Ai, Ainsworth, Allan, Allcock et al., A blood atlas of COVID-19 defines hallmarks of disease severity and specificity, Cell
Amani, Amani, Azvudine versus Paxlovid in COVID-19: a systematic review and meta-analysis, Rev. Med. Virol
Bader, Hogue, An automated method for finding molecular complexes in large protein interaction networks, BMC Bioinformatics
Banerjee, Blanco, Bruce, Honson, Chen et al., SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses, Cell
Bernal, Gomes Da Silva, Musungaie, Kovalchuk, Gonzalez et al., Molnupiravir for oral treatment of Covid-19 in nonhospitalized patients, N. Engl. J. Med
Bi, Liu, Ding, Liang, Zheng et al., Proteomic and metabolomic profiling of urine uncovers immune responses in patients with COVID-19, Cell Rep
Biji, Khatun, Swaraj, Narayan, Rajmani et al., Identification of COVID-19 prognostic markers and therapeutic targets through meta-analysis and validation of Omics data from nasopharyngeal samples, EBioMedicine
Blomen, Jae, Bigenzahn, Nieuwenhuis, Staring, Gene essentiality and synthetic lethality in haploid human cells, Science
Bouhaddou, Memon, Meyer, White, Rezelj et al., The global phosphorylation landscape of SARS-CoV-2 infection, Cell
Bouhaddou, Reuschl, Polacco, Thorne, Ummadi et al., SARS-CoV-2 variants evolve convergent strategies to remodel the host response, Cell
Breuer, Foroushani, Laird, Chen, Sribnaia et al., InnateDB: systems biology of innate immunity and beyondrecent updates and continuing curation, Nucleic Acids Res
Chen, Chen, Shi, Huang, Zhang et al., SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data, Gigascience
Chen, Zheng, Yu, Wang, Huang et al., Blood molecular markers associated with COVID-19 immunopathology and multi-organ damage, EMBO J
Cheong, Ravishankar, Sharma, Parkhurst, Grassmann et al., Epigenetic memory of coronavirus infection in innate immune cells and their progenitors, Cell
Daniloski, Jordan, Wessels, Hoagland, Kasela et al., Identification of required host factors for SARS-CoV-2 infection in human cells, Cell
Drew, Wallingford, Marcotte, MAP 2.0: integration of over 15,000 proteomic experiments builds a global compendium of human multiprotein assemblies, Mol. Syst. Biol
Filbin, Mehta, Schneider, Kays, Guess et al., Longitudinal proteomic analysis of severe COVID-19 reveals survival-associated signatures, tissue-specific cell death, and cell-cell interactions, Cell Rep. Med
Finkel, Gluck, Nachshon, Winkler, Fisher et al., SARS-CoV-2 uses a multipronged strategy to impede host protein synthesis, Nature
Foo, Cambou, Mok, Fajardo, Jung et al., The systemic inflammatory landscape of COVID-19 in pregnancy: extensive serum proteomic profiling of mother-infant dyads with in utero SARS-CoV-2, Cell Rep. Med
Fung, Babik, COVID-19 in immunocompromised hosts: what we know so far, Clin. Infect. Dis
Funk, Su, Ly, Feldman, Singh et al., The phenotypic landscape of essential human genes, Cell
Georg, Astaburuaga-García, Bonaguro, Brumhard, Michalick et al., Complement activation induces excessive T cell cytotoxicity in severe COVID-19, Cell
Geyer, Arend, Doll, Louiset, Virreira Winter et al., High-resolution serum proteome trajectories in COVID-19 reveal patient-specific seroconversion, EMBO Mol. Med
Gisby, Buang, Papadaki, Clarke, Malik et al., Multi-omics identify falling LRRC15 as a COVID-19 severity marker and persistent pro-thrombotic signals in convalescence, Nat. Commun
Han, Wang, Wang, Liu, Hu et al., ScaPD: a database for human scaffold proteins, BMC Bioinformatics
Huang, Zhou, Wernig, Südhof, ApoE2, ApoE3, and ApoE4 differentially stimulate APP transcription and Aβ secretion, Cell
Kanehisa, Furumichi, Sato, Kawashima, Ishiguro-Watanabe, KEGG for taxonomy-based analysis of pathways and genomes, Nucleic Acids Res
Kim, Langmead, Salzberg, HISAT: a fast spliced aligner with low memory requirements, Nat. Methods
Kim, Weller, Lin, Sheykhkarimli, Knapp et al., A proteome-scale map of the SARS-CoV-2-human contactome, Nat. Biotechnol
Lang, Armaos, Tartaglia, RNAct: Protein-RNA interaction predictions for model organisms with supporting experimental data, Nucleic Acids Res
Langerbeins, Hallek, COVID-19 in patients with hematologic malignancy, Blood
Lee, Shah, Hoyos, Solovyov, Douglas et al., Prolonged SARS-CoV-2 infection in patients with lymphoid malignancies, Cancer Discov
Leng, Ma, Zhang, Xu, Li et al., Spatial region-resolved proteome map reveals mechanism of COVID-19associated heart injury, Cell Rep
Li, Dewey, RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome, BMC Bioinformatics
Liu, Pan, Zhang, Li, Ma et al., Efficacy and safety of paxlovid in severe adult patients with SARS-Cov-2 infection: a multicenter randomized controlled study, Lancet Reg. Heal. West. Pac
Liu, Song, Zheng, Shi, Wu et al., A urinary proteomic landscape of COVID-19 progression identifies signaling pathways and therapeutic options, Sci. China Life Sci
Love, Huber, Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol
O'driscoll, Ribeiro Dos Santos, Wang, Cummings, Azman et al., Age-specific mortality and immunity patterns of SARS-CoV-2, Nature
Oughtred, Rust, Chang, Breitkreutz, Stark et al., The BioGRID database: a comprehensive biomedical resource of curated protein, genetic, and chemical interactions, Protein Sci
Pertea, Kim, Pertea, Leek, Salzberg, Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown, Nat. Protoc
Pinato, Zambelli, Aguilar-Company, Bower, Sng et al., Clinical portrait of the SARS-CoV-2 epidemic in European patients with cancer, Cancer Discov
Potts, Fletcher-Etherington, Nightingale, Mescia, Bergamaschi et al., Proteomic analysis of circulating immune cells identifies cellular phenotypes associated with COVID-19 severity, Cell Rep
Raman, Bluemke, Lüscher, Neubauer, Long COVID: post-acute sequelae of COVID-19 with a cardiovascular focus, Eur. Heart J
Ren, Wen, Fan, Hou, Su et al., COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas, Cell
Saravolatz, Depcinski, Sharma, Molnupiravir and nirmatrelvir-ritonavir: oral coronavirus disease 2019 antiviral drugs, Clin. Infect. Dis
Scherer, Babiker, Adelman, Allman, Key et al., SARS-CoV-2 evolution and immune escape in immunocompromised patients, N. Engl. J. Med
Schneider, Luna, Hoffmann, Ánchez-Rivera, Leal et al., Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks, Cell
Shannon, Markiel, Ozier, Baliga, Wang et al., Cytoscape: a software environment for integrated models of biomolecular interaction networks, Genome Res
Su, Chen, Yuan, Lausted, Choi et al., Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19, Cell
Thorne, Bouhaddou, Reuschl, Zuliani-Alvarez, Polacco et al., Evolution of enhanced innate immune evasion by SARS-CoV-2, Nature
Tidu, Janvier, Schaeffer, Sosnowski, Kuhn et al., The viral protein NSP1 acts as a ribosome gatekeeper for shutting down host translation and fostering SARS-CoV-2 translation, RNA
Tregoning, Flight, Higham, Wang, Pierce, Progress of the COVID-19 vaccine effort: viruses, vaccines and variants versus efficacy, effectiveness and escape, Nat. Rev. Immunol
Tsitsiridis, Steinkamp, Giurgiu, Brauner, Fobo et al., CORUM: the comprehensive resource of mammalian protein complexes-2022, Nucleic Acids Res
Uhl Én, Fagerberg, Hallström, Lindskog, Oksvold et al., Tissue-based map of the human proteome, Science
Valle, Kim-Schulze, Huang, Beckmann, Nirenberg et al., An inflammatory cytokine signature predicts COVID-19 severity and survival, Nat. Med
Van Phan, Tsitsiklis, Maguire, Haddad, Becker et al., Host-microbe multiomic profiling reveals age-dependent immune dysregulation associated with COVID-19 immunopathology, Sci. Transl. Med
Wang, Liu, Xie, Niu, Wang et al., Multiomics blood atlas reveals unique features of immune and platelet responses to SARS-CoV-2 Omicron breakthrough infection, Immunity
Wang, Yao, Ma, Ping, Fan et al., A singlecell transcriptomic landscape of the lungs of patients with COVID-19, Nat. Cell Biol
Wei, Alfajaro, Deweirdt, Hanna, Lu-Culligan et al., Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection, Cell
Weigang, Fuchs, Zimmer, Schnepf, Kern et al., Within-host evolution of SARS-CoV-2 in an immunosuppressed COVID-19 patient as a source of immune escape variants, Nat. Commun
Wilk, Rustagi, Zhao, Roque, Martínez-Col Ón et al., A single-cell atlas of the peripheral immune response in patients with severe COVID-19, Nat. Med
Wuchty, Siwo, Ferdig, Viral organization of human proteins, PLoS One
Yang, Fu, Lian, Dong, Zhang, Understanding human-virus protein-protein interactions using a human protein complex-based analysis framework, mSystems
Yang, Lian, Fu, Wuchty, Yang et al., HVIDB: a comprehensive database for human-virus protein-protein interactions, Brief. Bioinform
Yang, Zhu, Wang, Tang, Shen et al., Comparative analysis of dynamic transcriptomes reveals specific COVID-19 features and pathogenesis of immunocompromised populations, mSystems
Zaffagni, Harris, Patop, Pamudurti, Nguyen et al., SARS-CoV-2 Nsp14 mediates the effects of viral infection on the host cell transcriptome, Elife
Zhou, Liu, Gupta, Paramo, Hou et al., A comprehensive SARS-CoV-2-human protein-protein interactome reveals COVID-19 pathobiology and potential host therapeutic targets, Nat. Biotechnol
Zhou, Zhang, Zhao, Yu, Shen et al., TTD: therapeutic target database describing target druggability information, Nucleic Acids Res
DOI record:
{
"DOI": "10.1016/j.mcpro.2025.101068",
"ISSN": [
"1535-9476"
],
"URL": "http://dx.doi.org/10.1016/j.mcpro.2025.101068",
"alternative-id": [
"S1535947625001677"
],
"article-number": "101068",
"assertion": [
{
"label": "This article is maintained by",
"name": "publisher",
"value": "Elsevier"
},
{
"label": "Article Title",
"name": "articletitle",
"value": "Multi-Omics Blood Atlas Reveals Host Immune Response Features of Immunocompromised Populations Following SARS-CoV-2 Infection"
},
{
"label": "Journal Title",
"name": "journaltitle",
"value": "Molecular & Cellular Proteomics"
},
{
"label": "CrossRef DOI link to publisher maintained version",
"name": "articlelink",
"value": "https://doi.org/10.1016/j.mcpro.2025.101068"
},
{
"label": "Content Type",
"name": "content_type",
"value": "article"
},
{
"label": "Copyright",
"name": "copyright",
"value": "© 2025 The Authors. Published by Elsevier Inc on behalf of American Society for Biochemistry and Molecular Biologyé"
}
],
"author": [
{
"ORCID": "https://orcid.org/0000-0002-3229-5865",
"affiliation": [],
"authenticated-orcid": false,
"family": "Yang",
"given": "Xiaodi",
"sequence": "first"
},
{
"affiliation": [],
"family": "Shen",
"given": "Ye",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Tang",
"given": "Bo",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Zhu",
"given": "Jialin",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Wang",
"given": "Bingjie",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Wang",
"given": "Qingyun",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Tian",
"given": "Wenmin",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Wuchty",
"given": "Stefan",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Zhang",
"given": "Ziding",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Liang",
"given": "Zeyin",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Dong",
"given": "Yujun",
"sequence": "additional"
}
],
"container-title": "Molecular & Cellular Proteomics",
"container-title-short": "Molecular & Cellular Proteomics",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"mcponline.org",
"elsevier.com",
"sciencedirect.com"
]
},
"created": {
"date-parts": [
[
2025,
9,
11
]
],
"date-time": "2025-09-11T00:30:32Z",
"timestamp": 1757550632000
},
"deposited": {
"date-parts": [
[
2025,
10,
10
]
],
"date-time": "2025-10-10T03:27:29Z",
"timestamp": 1760066849000
},
"indexed": {
"date-parts": [
[
2025,
10,
11
]
],
"date-time": "2025-10-11T00:14:54Z",
"timestamp": 1760141694802,
"version": "build-2065373602"
},
"is-referenced-by-count": 0,
"issue": "10",
"issued": {
"date-parts": [
[
2025,
10
]
]
},
"journal-issue": {
"issue": "10",
"published-print": {
"date-parts": [
[
2025,
10
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://www.elsevier.com/tdm/userlicense/1.0/",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
10,
1
]
],
"date-time": "2025-10-01T00:00:00Z",
"timestamp": 1759276800000
}
},
{
"URL": "https://www.elsevier.com/legal/tdmrep-license",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
10,
1
]
],
"date-time": "2025-10-01T00:00:00Z",
"timestamp": 1759276800000
}
},
{
"URL": "http://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
9,
9
]
],
"date-time": "2025-09-09T00:00:00Z",
"timestamp": 1757376000000
}
}
],
"link": [
{
"URL": "https://api.elsevier.com/content/article/PII:S1535947625001677?httpAccept=text/xml",
"content-type": "text/xml",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://api.elsevier.com/content/article/PII:S1535947625001677?httpAccept=text/plain",
"content-type": "text/plain",
"content-version": "vor",
"intended-application": "text-mining"
}
],
"member": "78",
"original-title": [],
"page": "101068",
"prefix": "10.1016",
"published": {
"date-parts": [
[
2025,
10
]
]
},
"published-print": {
"date-parts": [
[
2025,
10
]
]
},
"publisher": "Elsevier BV",
"reference": [
{
"article-title": "Efficacy and safety of paxlovid in severe adult patients with SARS-Cov-2 infection: a multicenter randomized controlled study",
"author": "Liu",
"journal-title": "Lancet Reg. Heal. West. Pac.",
"key": "10.1016/j.mcpro.2025.101068_bib1",
"volume": "33",
"year": "2023"
},
{
"DOI": "10.1093/cid/ciac180",
"article-title": "Molnupiravir and nirmatrelvir-ritonavir: oral coronavirus disease 2019 antiviral drugs",
"author": "Saravolatz",
"doi-asserted-by": "crossref",
"first-page": "165",
"journal-title": "Clin. Infect. Dis.",
"key": "10.1016/j.mcpro.2025.101068_bib2",
"volume": "76",
"year": "2023"
},
{
"DOI": "10.1056/NEJMoa2116044",
"article-title": "Molnupiravir for oral treatment of Covid-19 in nonhospitalized patients",
"author": "Jayk Bernal",
"doi-asserted-by": "crossref",
"first-page": "509",
"journal-title": "N. Engl. J. Med.",
"key": "10.1016/j.mcpro.2025.101068_bib3",
"volume": "386",
"year": "2022"
},
{
"DOI": "10.1002/rmv.2551",
"article-title": "Azvudine versus Paxlovid in COVID-19: a systematic review and meta-analysis",
"author": "Amani",
"doi-asserted-by": "crossref",
"journal-title": "Rev. Med. Virol.",
"key": "10.1016/j.mcpro.2025.101068_bib4",
"volume": "34",
"year": "2024"
},
{
"DOI": "10.1038/s41577-021-00592-1",
"article-title": "Progress of the COVID-19 vaccine effort: viruses, vaccines and variants versus efficacy, effectiveness and escape",
"author": "Tregoning",
"doi-asserted-by": "crossref",
"first-page": "626",
"journal-title": "Nat. Rev. Immunol.",
"key": "10.1016/j.mcpro.2025.101068_bib5",
"volume": "21",
"year": "2021"
},
{
"DOI": "10.1038/s41586-020-2918-0",
"article-title": "Age-specific mortality and immunity patterns of SARS-CoV-2",
"author": "O'Driscoll",
"doi-asserted-by": "crossref",
"first-page": "140",
"journal-title": "Nature",
"key": "10.1016/j.mcpro.2025.101068_bib6",
"volume": "590",
"year": "2021"
},
{
"DOI": "10.1093/cid/ciaa863",
"article-title": "COVID-19 in immunocompromised hosts: what we know so far",
"author": "Fung",
"doi-asserted-by": "crossref",
"first-page": "340",
"journal-title": "Clin. Infect. Dis.",
"key": "10.1016/j.mcpro.2025.101068_bib7",
"volume": "72",
"year": "2021"
},
{
"DOI": "10.1182/blood.2021012251",
"article-title": "COVID-19 in patients with hematologic malignancy",
"author": "Langerbeins",
"doi-asserted-by": "crossref",
"first-page": "236",
"journal-title": "Blood",
"key": "10.1016/j.mcpro.2025.101068_bib8",
"volume": "140",
"year": "2022"
},
{
"DOI": "10.1158/2159-8290.CD-21-1033",
"article-title": "Prolonged SARS-CoV-2 infection in patients with lymphoid malignancies",
"author": "Lee",
"doi-asserted-by": "crossref",
"first-page": "62",
"journal-title": "Cancer Discov.",
"key": "10.1016/j.mcpro.2025.101068_bib9",
"volume": "12",
"year": "2022"
},
{
"DOI": "10.1016/j.celrep.2022.110955",
"article-title": "Spatial region-resolved proteome map reveals mechanism of COVID-19-associated heart injury",
"author": "Leng",
"doi-asserted-by": "crossref",
"journal-title": "Cell Rep.",
"key": "10.1016/j.mcpro.2025.101068_bib10",
"volume": "39",
"year": "2022"
},
{
"DOI": "10.1016/j.celrep.2021.110271",
"article-title": "Proteomic and metabolomic profiling of urine uncovers immune responses in patients with COVID-19",
"author": "Bi",
"doi-asserted-by": "crossref",
"journal-title": "Cell Rep.",
"key": "10.1016/j.mcpro.2025.101068_bib11",
"volume": "38",
"year": "2022"
},
{
"DOI": "10.1016/j.cell.2021.01.053",
"article-title": "COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas",
"author": "Ren",
"doi-asserted-by": "crossref",
"first-page": "1895",
"journal-title": "Cell",
"key": "10.1016/j.mcpro.2025.101068_bib12",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1038/s41586-021-04352-y",
"article-title": "Evolution of enhanced innate immune evasion by SARS-CoV-2",
"author": "Thorne",
"doi-asserted-by": "crossref",
"first-page": "487",
"journal-title": "Nature",
"key": "10.1016/j.mcpro.2025.101068_bib13",
"volume": "602",
"year": "2022"
},
{
"article-title": "The systemic inflammatory landscape of COVID-19 in pregnancy: extensive serum proteomic profiling of mother-infant dyads with in utero SARS-CoV-2",
"author": "Foo",
"journal-title": "Cell Rep. Med.",
"key": "10.1016/j.mcpro.2025.101068_bib14",
"volume": "2",
"year": "2021"
},
{
"DOI": "10.7554/eLife.71945",
"article-title": "SARS-CoV-2 Nsp14 mediates the effects of viral infection on the host cell transcriptome",
"author": "Zaffagni",
"doi-asserted-by": "crossref",
"journal-title": "Elife",
"key": "10.1016/j.mcpro.2025.101068_bib15",
"volume": "11",
"year": "2022"
},
{
"DOI": "10.1016/j.cell.2023.07.019",
"article-title": "Epigenetic memory of coronavirus infection in innate immune cells and their progenitors",
"author": "Cheong",
"doi-asserted-by": "crossref",
"first-page": "3882",
"journal-title": "Cell",
"key": "10.1016/j.mcpro.2025.101068_bib16",
"volume": "186",
"year": "2023"
},
{
"DOI": "10.1007/s11427-021-2070-y",
"article-title": "A urinary proteomic landscape of COVID-19 progression identifies signaling pathways and therapeutic options",
"author": "Liu",
"doi-asserted-by": "crossref",
"first-page": "1866",
"journal-title": "Sci. China Life Sci.",
"key": "10.1016/j.mcpro.2025.101068_bib17",
"volume": "65",
"year": "2022"
},
{
"DOI": "10.1016/j.celrep.2023.112613",
"article-title": "Proteomic analysis of circulating immune cells identifies cellular phenotypes associated with COVID-19 severity",
"author": "Potts",
"doi-asserted-by": "crossref",
"journal-title": "Cell Rep.",
"key": "10.1016/j.mcpro.2025.101068_bib18",
"volume": "42",
"year": "2023"
},
{
"DOI": "10.1038/s41587-022-01475-z",
"article-title": "A proteome-scale map of the SARS-CoV-2–human contactome",
"author": "Kim",
"doi-asserted-by": "crossref",
"first-page": "140",
"journal-title": "Nat. Biotechnol.",
"key": "10.1016/j.mcpro.2025.101068_bib19",
"volume": "41",
"year": "2023"
},
{
"DOI": "10.1038/s41587-022-01474-0",
"article-title": "A comprehensive SARS-CoV-2–human protein–protein interactome reveals COVID-19 pathobiology and potential host therapeutic targets",
"author": "Zhou",
"doi-asserted-by": "crossref",
"first-page": "128",
"journal-title": "Nat. Biotechnol.",
"key": "10.1016/j.mcpro.2025.101068_bib20",
"volume": "41",
"year": "2023"
},
{
"DOI": "10.15252/emmm.202114167",
"article-title": "High-resolution serum proteome trajectories in COVID-19 reveal patient-specific seroconversion",
"author": "Geyer",
"doi-asserted-by": "crossref",
"journal-title": "EMBO Mol. Med.",
"key": "10.1016/j.mcpro.2025.101068_bib21",
"volume": "13",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2020.10.037",
"article-title": "Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19",
"author": "Su",
"doi-asserted-by": "crossref",
"first-page": "1479",
"journal-title": "Cell",
"key": "10.1016/j.mcpro.2025.101068_bib22",
"volume": "183",
"year": "2020"
},
{
"DOI": "10.1093/nar/gkac1015",
"article-title": "CORUM: the comprehensive resource of mammalian protein complexes-2022",
"author": "Tsitsiridis",
"doi-asserted-by": "crossref",
"first-page": "D539",
"journal-title": "Nucleic Acids Res.",
"key": "10.1016/j.mcpro.2025.101068_bib23",
"volume": "51",
"year": "2023"
},
{
"DOI": "10.15252/msb.202010016",
"article-title": "hu.MAP 2.0: integration of over 15,000 proteomic experiments builds a global compendium of human multiprotein assemblies",
"author": "Drew",
"doi-asserted-by": "crossref",
"journal-title": "Mol. Syst. Biol.",
"key": "10.1016/j.mcpro.2025.101068_bib24",
"volume": "17",
"year": "2021"
},
{
"DOI": "10.1093/bib/bbaa425",
"article-title": "HVIDB: a comprehensive database for human-virus protein-protein interactions",
"author": "Yang",
"doi-asserted-by": "crossref",
"first-page": "832",
"journal-title": "Brief. Bioinform.",
"key": "10.1016/j.mcpro.2025.101068_bib25",
"volume": "22",
"year": "2021"
},
{
"DOI": "10.1128/mSystems.00303-18",
"article-title": "Understanding human-virus protein-protein interactions using a human protein complex-based analysis framework",
"author": "Yang",
"doi-asserted-by": "crossref",
"journal-title": "mSystems",
"key": "10.1016/j.mcpro.2025.101068_bib26",
"volume": "4",
"year": "2019"
},
{
"DOI": "10.1016/j.cell.2022.01.012",
"article-title": "A blood atlas of COVID-19 defines hallmarks of disease severity and specificity",
"author": "Ahern",
"doi-asserted-by": "crossref",
"first-page": "916",
"journal-title": "Cell",
"key": "10.1016/j.mcpro.2025.101068_bib27",
"volume": "185",
"year": "2022"
},
{
"article-title": "Longitudinal proteomic analysis of severe COVID-19 reveals survival-associated signatures, tissue-specific cell death, and cell-cell interactions",
"author": "Filbin",
"journal-title": "Cell Rep. Med.",
"key": "10.1016/j.mcpro.2025.101068_bib28",
"volume": "2",
"year": "2021"
},
{
"DOI": "10.1158/2159-8290.CD-20-0773",
"article-title": "Clinical portrait of the SARS-CoV-2 epidemic in European patients with cancer",
"author": "Pinato",
"doi-asserted-by": "crossref",
"first-page": "1465",
"journal-title": "Cancer Discov.",
"key": "10.1016/j.mcpro.2025.101068_bib29",
"volume": "10",
"year": "2020"
},
{
"key": "10.1016/j.mcpro.2025.101068_bib30",
"series-title": "COVID-19 rapid guideline: managing the long-term effects of COVID-19",
"year": "2020"
},
{
"DOI": "10.1093/eurheartj/ehac031",
"article-title": "Long COVID: post-acute sequelae of COVID-19 with a cardiovascular focus",
"author": "Raman",
"doi-asserted-by": "crossref",
"first-page": "1157",
"journal-title": "Eur. Heart J.",
"key": "10.1016/j.mcpro.2025.101068_bib31",
"volume": "43",
"year": "2022"
},
{
"DOI": "10.1128/msystems.01385-23",
"article-title": "Comparative analysis of dynamic transcriptomes reveals specific COVID-19 features and pathogenesis of immunocompromised populations",
"author": "Yang",
"doi-asserted-by": "crossref",
"journal-title": "mSystems",
"key": "10.1016/j.mcpro.2025.101068_bib32",
"volume": "9",
"year": "2024"
},
{
"DOI": "10.1093/gigascience/gix120",
"article-title": "SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data",
"author": "Chen",
"doi-asserted-by": "crossref",
"first-page": "1",
"journal-title": "Gigascience",
"key": "10.1016/j.mcpro.2025.101068_bib33",
"volume": "7",
"year": "2018"
},
{
"DOI": "10.1038/nmeth.3317",
"article-title": "HISAT: a fast spliced aligner with low memory requirements",
"author": "Kim",
"doi-asserted-by": "crossref",
"first-page": "357",
"journal-title": "Nat. Methods",
"key": "10.1016/j.mcpro.2025.101068_bib34",
"volume": "12",
"year": "2015"
},
{
"DOI": "10.1038/nprot.2016.095",
"article-title": "Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown",
"author": "Pertea",
"doi-asserted-by": "crossref",
"first-page": "1650",
"journal-title": "Nat. Protoc.",
"key": "10.1016/j.mcpro.2025.101068_bib35",
"volume": "11",
"year": "2016"
},
{
"DOI": "10.1186/1471-2105-12-323",
"article-title": "RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "323",
"journal-title": "BMC Bioinformatics",
"key": "10.1016/j.mcpro.2025.101068_bib36",
"volume": "12",
"year": "2011"
},
{
"DOI": "10.1186/s13059-014-0550-8",
"article-title": "Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2",
"author": "Love",
"doi-asserted-by": "crossref",
"first-page": "550",
"journal-title": "Genome Biol.",
"key": "10.1016/j.mcpro.2025.101068_bib37",
"volume": "15",
"year": "2014"
},
{
"DOI": "10.15252/embj.2020105896",
"article-title": "Blood molecular markers associated with COVID-19 immunopathology and multi-organ damage",
"author": "Chen",
"doi-asserted-by": "crossref",
"journal-title": "EMBO J.",
"key": "10.1016/j.mcpro.2025.101068_bib38",
"volume": "39",
"year": "2020"
},
{
"DOI": "10.1016/j.ebiom.2021.103525",
"article-title": "Identification of COVID-19 prognostic markers and therapeutic targets through meta-analysis and validation of Omics data from nasopharyngeal samples",
"author": "Biji",
"doi-asserted-by": "crossref",
"journal-title": "EBioMedicine",
"key": "10.1016/j.mcpro.2025.101068_bib39",
"volume": "70",
"year": "2021"
},
{
"DOI": "10.1038/s41556-021-00796-6",
"article-title": "A single-cell transcriptomic landscape of the lungs of patients with COVID-19",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "1314",
"journal-title": "Nat. Cell Biol.",
"key": "10.1016/j.mcpro.2025.101068_bib40",
"volume": "23",
"year": "2021"
},
{
"DOI": "10.1016/j.immuni.2023.05.007",
"article-title": "Multi-omics blood atlas reveals unique features of immune and platelet responses to SARS-CoV-2 Omicron breakthrough infection",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "1410",
"journal-title": "Immunity",
"key": "10.1016/j.mcpro.2025.101068_bib41",
"volume": "56",
"year": "2023"
},
{
"DOI": "10.1038/s41467-022-35454-4",
"article-title": "Multi-omics identify falling LRRC15 as a COVID-19 severity marker and persistent pro-thrombotic signals in convalescence",
"author": "Gisby",
"doi-asserted-by": "crossref",
"first-page": "7775",
"journal-title": "Nat. Commun.",
"key": "10.1016/j.mcpro.2025.101068_bib42",
"volume": "13",
"year": "2022"
},
{
"article-title": "Host-microbe multiomic profiling reveals age-dependent immune dysregulation associated with COVID-19 immunopathology",
"author": "van Phan",
"journal-title": "Sci. Transl. Med.",
"key": "10.1016/j.mcpro.2025.101068_bib43",
"volume": "16",
"year": "2024"
},
{
"DOI": "10.1002/pro.3978",
"article-title": "The BioGRID database: a comprehensive biomedical resource of curated protein, genetic, and chemical interactions",
"author": "Oughtred",
"doi-asserted-by": "crossref",
"first-page": "187",
"journal-title": "Protein Sci.",
"key": "10.1016/j.mcpro.2025.101068_bib44",
"volume": "30",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2022.10.017",
"article-title": "The phenotypic landscape of essential human genes",
"author": "Funk",
"doi-asserted-by": "crossref",
"first-page": "4634",
"journal-title": "Cell",
"key": "10.1016/j.mcpro.2025.101068_bib45",
"volume": "185",
"year": "2022"
},
{
"DOI": "10.1126/science.aac7557",
"article-title": "Gene essentiality and synthetic lethality in haploid human cells",
"author": "Blomen",
"doi-asserted-by": "crossref",
"first-page": "1092",
"journal-title": "Science",
"key": "10.1016/j.mcpro.2025.101068_bib46",
"volume": "350",
"year": "2015"
},
{
"DOI": "10.1186/s12859-017-1806-6",
"article-title": "ScaPD: a database for human scaffold proteins",
"author": "Han",
"doi-asserted-by": "crossref",
"first-page": "386",
"journal-title": "BMC Bioinformatics",
"key": "10.1016/j.mcpro.2025.101068_bib47",
"volume": "18",
"year": "2017"
},
{
"DOI": "10.1016/j.cell.2020.10.028",
"article-title": "Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection",
"author": "Wei",
"doi-asserted-by": "crossref",
"first-page": "76",
"journal-title": "Cell",
"key": "10.1016/j.mcpro.2025.101068_bib48",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2020.10.030",
"article-title": "Identification of required host factors for SARS-CoV-2 infection in human cells",
"author": "Daniloski",
"doi-asserted-by": "crossref",
"first-page": "92",
"journal-title": "Cell",
"key": "10.1016/j.mcpro.2025.101068_bib49",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2020.12.006",
"article-title": "Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks",
"author": "Schneider",
"doi-asserted-by": "crossref",
"first-page": "120",
"journal-title": "Cell",
"key": "10.1016/j.mcpro.2025.101068_bib50",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1093/nar/gks1147",
"article-title": "InnateDB: systems biology of innate immunity and beyond—recent updates and continuing curation",
"author": "Breuer",
"doi-asserted-by": "crossref",
"first-page": "D1228",
"journal-title": "Nucleic Acids Res.",
"key": "10.1016/j.mcpro.2025.101068_bib51",
"volume": "41",
"year": "2013"
},
{
"DOI": "10.1101/gr.1239303",
"article-title": "Cytoscape: a software environment for integrated models of biomolecular interaction networks",
"author": "Shannon",
"doi-asserted-by": "crossref",
"first-page": "2498",
"journal-title": "Genome Res.",
"key": "10.1016/j.mcpro.2025.101068_bib52",
"volume": "13",
"year": "2003"
},
{
"DOI": "10.1186/1471-2105-4-2",
"article-title": "An automated method for finding molecular complexes in large protein interaction networks",
"author": "Bader",
"doi-asserted-by": "crossref",
"first-page": "2",
"journal-title": "BMC Bioinformatics",
"key": "10.1016/j.mcpro.2025.101068_bib53",
"volume": "4",
"year": "2003"
},
{
"DOI": "10.1093/nar/gky1055",
"article-title": "The gene ontology resource: 20 years and still GOing strong",
"doi-asserted-by": "crossref",
"first-page": "D330",
"journal-title": "Nucleic Acids Res.",
"key": "10.1016/j.mcpro.2025.101068_bib54",
"volume": "47",
"year": "2019"
},
{
"DOI": "10.1093/nar/gkac963",
"article-title": "KEGG for taxonomy-based analysis of pathways and genomes",
"author": "Kanehisa",
"doi-asserted-by": "crossref",
"first-page": "D587",
"journal-title": "Nucleic Acids Res.",
"key": "10.1016/j.mcpro.2025.101068_bib55",
"volume": "51",
"year": "2023"
},
{
"DOI": "10.1126/science.1260419",
"article-title": "Tissue-based map of the human proteome",
"author": "Uhlén",
"doi-asserted-by": "crossref",
"journal-title": "Science",
"key": "10.1016/j.mcpro.2025.101068_bib56",
"volume": "347",
"year": "2015"
},
{
"DOI": "10.1093/nar/gkad751",
"article-title": "TTD: therapeutic target database describing target druggability information",
"author": "Zhou",
"doi-asserted-by": "crossref",
"first-page": "D1465",
"journal-title": "Nucleic Acids Res.",
"key": "10.1016/j.mcpro.2025.101068_bib57",
"volume": "52",
"year": "2024"
},
{
"DOI": "10.1261/rna.078121.120",
"article-title": "The viral protein NSP1 acts as a ribosome gatekeeper for shutting down host translation and fostering SARS-CoV-2 translation",
"author": "Tidu",
"doi-asserted-by": "crossref",
"first-page": "253",
"journal-title": "RNA",
"key": "10.1016/j.mcpro.2025.101068_bib58",
"volume": "27",
"year": "2021"
},
{
"DOI": "10.1371/journal.pone.0011796",
"article-title": "Viral organization of human proteins",
"author": "Wuchty",
"doi-asserted-by": "crossref",
"journal-title": "PLoS One",
"key": "10.1016/j.mcpro.2025.101068_bib59",
"volume": "5",
"year": "2010"
},
{
"DOI": "10.1093/nar/gky967",
"article-title": "RNAct: Protein-RNA interaction predictions for model organisms with supporting experimental data",
"author": "Lang",
"doi-asserted-by": "crossref",
"first-page": "D601",
"journal-title": "Nucleic Acids Res.",
"key": "10.1016/j.mcpro.2025.101068_bib60",
"volume": "47",
"year": "2019"
},
{
"DOI": "10.1016/j.cell.2016.12.044",
"article-title": "ApoE2, ApoE3, and ApoE4 differentially stimulate APP transcription and Aβ secretion",
"author": "Huang",
"doi-asserted-by": "crossref",
"first-page": "427",
"journal-title": "Cell",
"key": "10.1016/j.mcpro.2025.101068_bib61",
"volume": "168",
"year": "2017"
},
{
"DOI": "10.1016/j.cell.2021.12.040",
"article-title": "Complement activation induces excessive T cell cytotoxicity in severe COVID-19",
"author": "Georg",
"doi-asserted-by": "crossref",
"first-page": "493",
"journal-title": "Cell",
"key": "10.1016/j.mcpro.2025.101068_bib62",
"volume": "185",
"year": "2022"
},
{
"DOI": "10.1016/j.cell.2020.10.004",
"article-title": "SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses",
"author": "Banerjee",
"doi-asserted-by": "crossref",
"first-page": "1325",
"journal-title": "Cell",
"key": "10.1016/j.mcpro.2025.101068_bib64",
"volume": "183",
"year": "2020"
},
{
"DOI": "10.1038/s41586-021-03610-3",
"article-title": "SARS-CoV-2 uses a multipronged strategy to impede host protein synthesis",
"author": "Finkel",
"doi-asserted-by": "crossref",
"first-page": "240",
"journal-title": "Nature",
"key": "10.1016/j.mcpro.2025.101068_bib65",
"volume": "594",
"year": "2021"
},
{
"DOI": "10.1038/s41591-020-0944-y",
"article-title": "A single-cell atlas of the peripheral immune response in patients with severe COVID-19",
"author": "Wilk",
"doi-asserted-by": "crossref",
"first-page": "1070",
"journal-title": "Nat. Med.",
"key": "10.1016/j.mcpro.2025.101068_bib66",
"volume": "26",
"year": "2020"
},
{
"DOI": "10.1038/s41467-021-26602-3",
"article-title": "Within-host evolution of SARS-CoV-2 in an immunosuppressed COVID-19 patient as a source of immune escape variants",
"author": "Weigang",
"doi-asserted-by": "crossref",
"first-page": "6405",
"journal-title": "Nat. Commun.",
"key": "10.1016/j.mcpro.2025.101068_bib67",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1056/NEJMc2202861",
"article-title": "SARS-CoV-2 evolution and immune escape in immunocompromised patients",
"author": "Scherer",
"doi-asserted-by": "crossref",
"first-page": "2436",
"journal-title": "N. Engl. J. Med.",
"key": "10.1016/j.mcpro.2025.101068_bib68",
"volume": "386",
"year": "2022"
},
{
"DOI": "10.1038/s41591-020-1051-9",
"article-title": "An inflammatory cytokine signature predicts COVID-19 severity and survival",
"author": "Del Valle",
"doi-asserted-by": "crossref",
"first-page": "1636",
"journal-title": "Nat. Med.",
"key": "10.1016/j.mcpro.2025.101068_bib69",
"volume": "26",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2023.08.026",
"article-title": "SARS-CoV-2 variants evolve convergent strategies to remodel the host response",
"author": "Bouhaddou",
"doi-asserted-by": "crossref",
"first-page": "4597",
"journal-title": "Cell",
"key": "10.1016/j.mcpro.2025.101068_bib70",
"volume": "186",
"year": "2023"
},
{
"DOI": "10.1016/j.cell.2020.06.034",
"article-title": "The global phosphorylation landscape of SARS-CoV-2 infection",
"author": "Bouhaddou",
"doi-asserted-by": "crossref",
"first-page": "685",
"journal-title": "Cell",
"key": "10.1016/j.mcpro.2025.101068_bib71",
"volume": "182",
"year": "2020"
}
],
"reference-count": 70,
"references-count": 70,
"relation": {},
"resource": {
"primary": {
"URL": "https://linkinghub.elsevier.com/retrieve/pii/S1535947625001677"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Multi-Omics Blood Atlas Reveals Host Immune Response Features of Immunocompromised Populations Following SARS-CoV-2 Infection",
"type": "journal-article",
"update-policy": "https://doi.org/10.1016/elsevier_cm_policy",
"volume": "24"
}