Decoding the genome of SARS-CoV-2: a pathway to drug development through translation inhibition
et al., RNA Biology, doi:10.1080/15476286.2024.2433830, Dec 2024
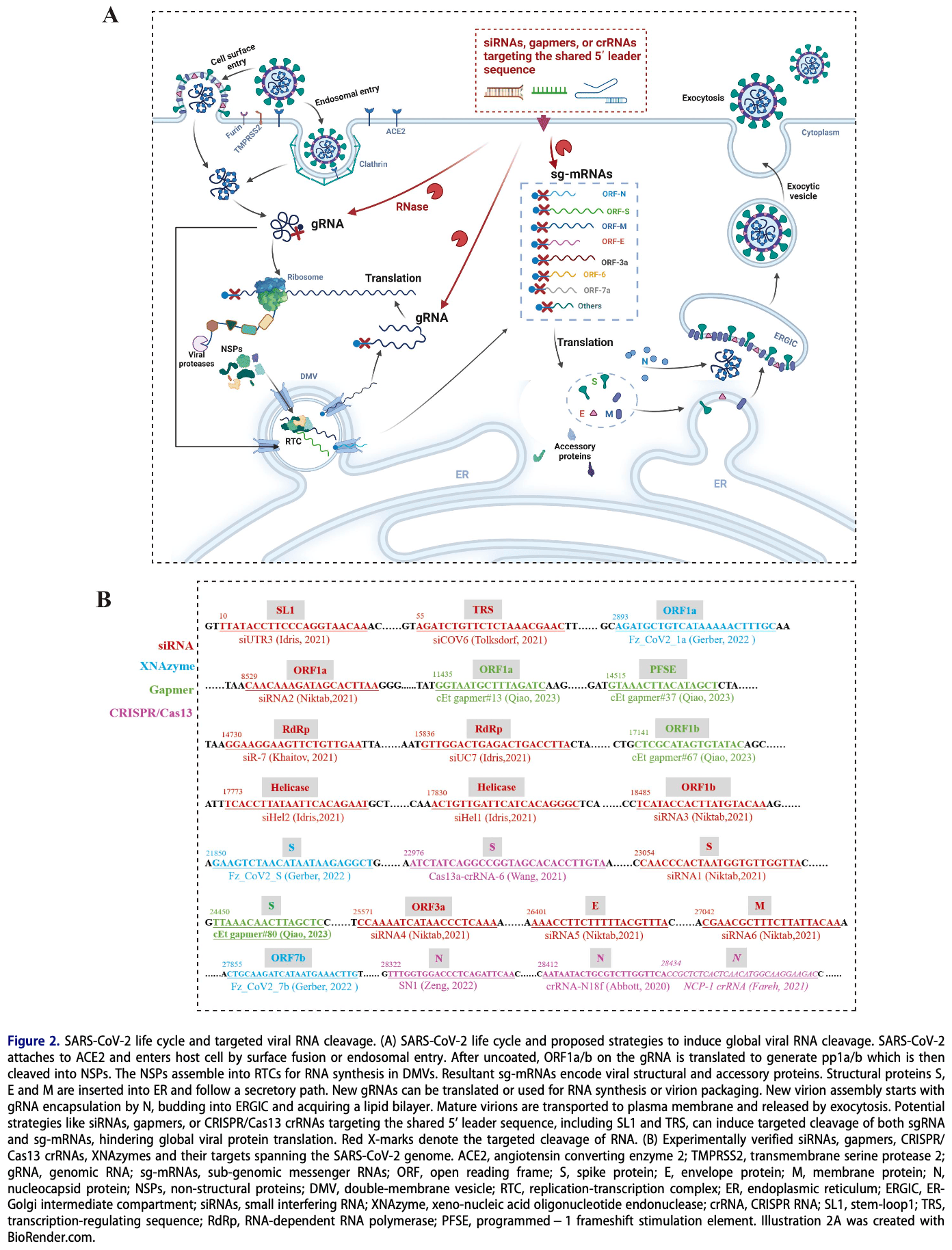
Review of the mechanism of SARS-CoV-2 protein translation and potential therapeutic strategies targeting viral RNA for inhibition. Protein translation is crucial for the entire intracellular life cycle of SARS-CoV-2, with genomic RNA immediately translated upon release into the cytoplasm to produce early non-structural proteins, followed by translation of subgenomic mRNAs to obtain structural proteins for virion assembly in the later stage. Authors discuss the current understanding of SARS-CoV-2 translation initiation, noting the roles of conserved RNA elements and untranslated regions in regulating translation. Strategies for blocking translation through targeted viral RNA cleavage (using antisense oligonucleotides, siRNA, CRISPR-Cas13) or inhibiting viral RNA element functions (using small molecule inhibitors) are highlighted.
Wu et al., 4 Dec 2024, peer-reviewed, 4 authors.
Contact: t-lxh@163.com.
Decoding the genome of SARS-CoV-2: a pathway to drug development through translation inhibition
RNA Biology, doi:10.1080/15476286.2024.2433830
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes the coronavirus disease 2019 (COVID-19) pandemic and is continuously spreading globally. The continuous emergence of new SARS-CoV-2 variants keeps posing threats, highlighting the need for fast-acting, mutation-resistant broadspectrum therapeutics. Protein translation is vital for SARS-CoV-2 replication, producing early nonstructural proteins for RNA replication and transcription, and late structural proteins for virion assembly. Targeted blocking of viral protein translation is thus a potential approach to developing effective anti-SARS-CoV-2 drugs. SARS-CoV-2, as an obligate parasite, utilizes the host's translation machinery. Translation-blocking strategies that target the SARS-CoV-2 mRNA, especially those that target its conserved elements are generally preferred. In this review, we discuss the current understanding of SARS-CoV-2 translation, highlighting the important conserved motifs and structures involved in its regulation. We also discuss the current strategies for blocking SARS-CoV-2 translation through viral RNA degradation or RNA element dysfunction.
References
Abbott, Dhamdhere, Liu, Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza, Cell, doi:10.1016/j.cell.2020.04.020
Aldhumani, Hossain, Fairchild, RNA sequence and ligand binding alter conformational profile of SARS-CoV-2 stem loop II motif, Biochem Biophys Res Commun, doi:10.1016/j.bbrc.2021.01.013
Anokhina, Miller, Targeting ribosomal frameshifting as an antiviral strategy: from HIV-1 to SARS-CoV-2, Acc Chem Res, doi:10.1021/acs.accounts.1c00316
Babendure, Babendure, Ding, Control of mammalian translation by mRNA structure near caps, RNA, doi:10.1261/rna.2309906
Baldassarre, Paolini, Bruno, Potential use of noncoding RNAs and innovative therapeutic strategies to target the 5'UTR of SARS-CoV-2, Epigenomics, doi:10.2217/epi-2020-0162
Banerjee, Blanco, Bruce, SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses, Cell, doi:10.1016/j.cell.2020.10.004
Baranov, Henderson, Anderson, Programmed ribosomal frameshifting in decoding the SARS-CoV genome, Virology, doi:10.1016/j.virol.2004.11.038
Berry, Waghray, Mortimer, Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning, Structure, doi:10.1016/j.str.2011.08.002
Bhatt, Scaiola, Loughran, Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome, Science, doi:10.1126/science.abf3546
Brant, Tian, Majerciak, SARS-CoV-2: from its discovery to genome structure, transcription, and replication, Cell Biosci, doi:10.1186/s13578-021-00643-z
Chamond, Deforges, Ulryck, 40S recruitment in the absence of eIF4G/4A by EMCV IRES refines the model for translation initiation on the archetype of type II IRESs, Nucleic Acids Res, doi:10.1093/nar/gku720
Chen, Tarn, uORF-mediated translational control: recently elucidated mechanisms and implications in cancer, RNA Biol, doi:10.1080/15476286.2019.1632634
Condé, Allatif, Ohlmann, Translation of SARS-CoV-2 gRNA is extremely efficient and competitive despite a High degree of secondary structures and the presence of an uORF, Viruses, doi:10.3390/v14071505
Cortese, Lee, Cerikan, Integrative imaging reveals SARS-CoV-2-induced reshaping of subcellular morphologies, Cell Host Microbe, doi:10.1016/j.chom.2020.11.003
Coutard, Valle, De Lamballerie, The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade, Antiviral Res, doi:10.1016/j.antiviral.2020.104742
Crooke, Baker, Crooke, Antisense technology: an overview and prospectus, Nat Rev Drug Discov, doi:10.1038/s41573-021-00162-z
Crooke, Liang, Baker, Antisense technology: a review, J Biol Chem, doi:10.1016/j.jbc.2021.100416
De Breyne, Yu, Unbehaun, Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites, Proc Natl Acad Sci, doi:10.1073/pnas.0900153106
Dey, Yan, Schlick, Abolished frameshifting for predicted structure-stabilizing SARS-CoV-2 mutants: implications to alternative conformations and their statistical structural analyses, RNA, doi:10.1261/rna.080035.124
Dhorne-Pollet, Fitzpatrick, Costa, Antisense oligonucleotides targeting ORF1b block replication of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), Front Microbiol, doi:10.3389/fmicb.2022.915202
Dinman, Mechanisms and implications of programmed translational frameshifting, Wiley Interdiscip Rev RNA, doi:10.1002/wrna.1126
Embarc-Buh, Francisco-Velilla, Martinez-Salas, RNA-Binding proteins at the host-pathogen interface targeting viral regulatory elements, Viruses, doi:10.3390/v13060952
Fareh, Zhao, Hu, Reprogrammed CRISPR-Cas13b suppresses SARS-CoV-2 replication and circumvents its mutational escape through mismatch tolerance, Nat Commun, doi:10.1038/s41467-021-24577-9
Fernández-Miragall, Salas, Structural organization of a viral IRES depends on the integrity of the GNRA motif, RNA, doi:10.1261/rna.5950603
Fields, Howley, De, Coronaviruses
Finkel, Gluck, Nachshon, SARS-CoV-2 uses a multipronged strategy to impede host protein synthesis, Nature, doi:10.1038/s41586-021-03610-3
Finkel, Mizrahi, Nachshon, The coding capacity of SARS-CoV-2, Nature, doi:10.1038/s41586-020-2739-1
Firth, Brierley, Non-canonical translation in RNA viruses, J Gen Virol, doi:10.1099/vir.0.042499-0
Franch, Gerdes, U-turns and regulatory RNAs, Curr Opin Microbiol, doi:10.1016/S1369-5274(00)00069-2
Frye, Cunningham, Mihailescu, Characterization of the SARS-CoV-2 genome 3 0 -untranslated region interactions with Host MicroRNAs, ACS Omega, doi:10.1021/acsomega.4c01050
Ganser, Kelly, Herschlag, The roles of structural dynamics in the cellular functions of RNAs, Nat Rev Mol Cell Biol, doi:10.1038/s41580-019-0136-0
Gerber, Donde, Matheson, Xnazymes targeting the SARS-CoV-2 genome inhibit viral infection, Nat Commun, doi:10.1038/s41467-022-34339-w
Gerresheim, Dünnes, Nieder-Röhrmann, microRNA-122 target sites in the hepatitis C virus RNA NS5B coding region and 3 0 untranslated region: function in replication and influence of RNA secondary structure, Cell Mol Life Sci, doi:10.1007/s00018-016-2377-9
Goebel, Miller, Bennett, A hypervariable region within the 3 0 cis -acting element of the murine coronavirus genome is nonessential for RNA synthesis but affects pathogenesis, J Virol, doi:10.1128/JVI.00803-06
Gorbalenya, Baker, Baric, The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2, Nat Microbiol, doi:10.1038/s41564-020-0695-z
Gross, Vicens, Einhorn, The IRES5 0 UTR of the dicistrovirus cricket paralysis virus is a type III IRES containing an essential pseudoknot structure, Nucleic Acids Res, doi:10.1093/nar/gkx622
Gutell, Cannone, Konings, Predicting U-turns in ribosomal RNA with comparative sequence analysis, J Mol Biol, doi:10.1006/jmbi.2000.3900
Hahn, Hahn, Rice, Conserved elements in the 3 0 untranslated region of flavivirus RNAs and potential cyclization sequences, J Mol Biol, doi:10.1016/0022-2836(87)90455-4
Haniff, Tong, Liu, Targeting the SARS-CoV-2 RNA genome with small molecule binders and ribonuclease targeting chimera (RIBOTAC) degraders, ACS Cent Sci, doi:10.1021/acscentsci.0c00984
Hartenian, Nandakumar, The molecular virology of coronaviruses, J Biol Chem, doi:10.1074/jbc.REV120.013930
Hashem, Des Georges, Dhote, Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit, Nature, doi:10.1038/nature12658
Hegde, Tang, Zhao, Inhibition of SARS-CoV-2 by targeting conserved viral RNA structures and sequences, Front Chem, doi:10.3389/fchem.2021.802766
Hilgenfeld, Peiris, From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses, Antiviral Res, doi:10.1016/j.antiviral.2013.08.015
Hinnebusch, Ivanov, Sonenberg, Translational control by 5 0 -untranslated regions of eukaryotic mRNAs, Science, doi:10.1126/science.aad9868
Hoang, Luong, Ayun, A novel approach of antiviral drugs targeting viral genomes, Microorganisms, doi:10.3390/microorganisms10081552
Huang, Lokugamage, Rozovics, SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRnas: viral mRNAs are resistant to nsp1-induced RNA cleavage, PloS Pathog, doi:10.1371/journal.ppat.1002433
Huston, Wan, Strine, Comprehensive in vivo secondary structure of the SARS-CoV-2 genome reveals novel regulatory motifs and mechanisms, Mol Cell, doi:10.1016/j.molcel.2020.12.041
Idris, Davis, Supramaniam, A SARS-CoV-2 targeted siRNA-nanoparticle therapy for COVID-19, Mol Ther, doi:10.1016/j.ymthe.2021.05.004
Imperatore, Cunningham, Pellegrene, Highly conserved s2m element of SARS-CoV-2 dimerizes via a kissing complex and interacts with host miRNA-1307-3p, Nucleic Acids Res, doi:10.1093/nar/gkab1226
Jaafar, Kieft, Viral RNA structure-based strategies to manipulate translation, Nat Rev Microbiol, doi:10.1038/s41579-018-0117-x
Jiang, Joshi, Gan, The highly conserved stem-loop II motif is dispensable for SARS-CoV-2, J Virol, doi:10.1128/jvi.00635-23
Johnstone, Bazzini, Giraldez, Upstream ORFs are prevalent translational repressors in vertebrates, Embo J, doi:10.15252/embj.201592759
Kean, The role of mRNA 5 0 -noncoding and 3 0 -end sequences on 40S ribosomal subunit recruitment, and how RNA viruses successfully compete with cellular mRNAs to ensure their own protein synthesis, Biol Cell, doi:10.1016/S0248-4900(03)00030-3
Kelly, Olson, Neupane, Structural and functional conservation of the programmed -1 ribosomal frameshift signal of SARS coronavirus 2 (SARS-CoV-2), J Biol Chem, doi:10.1074/jbc.AC120.013449
Khaitov, Nikonova, Shilovskiy, Silencing of SARS-CoV-2 with modified siRNA-peptide dendrimer formulation, Allergy, doi:10.1111/all.14850
Khan, Terenzi, Liu, A viral pan-end RNA element and host complex define a SARS-CoV-2 regulon, Nat Commun, doi:10.1038/s41467-023-39091-3
Kim, Kim, Park, A high-resolution temporal atlas of the SARS-CoV-2 translatome and transcriptome, Nat Commun, doi:10.1038/s41467-021-25361-5
Kim, Lee, Yang, The architecture of SARS-CoV-2 transcriptome, Cell, doi:10.1016/j.cell.2020.04.011
Kozak, Initiation of translation in prokaryotes and eukaryotes, Gene, doi:10.1016/S0378-1119(99)00210-3
Kozak, Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes, Cell, doi:10.1016/0092-8674(86)90762-2
Kozak, Pushing the limits of the scanning mechanism for initiation of translation, Gene, doi:10.1016/S0378-1119(02)01056-9
Kung, Lee, Chiang, Molecular virology of SARS-CoV-2 and related coronaviruses, Microbiol Mol Biol Rev, doi:10.1128/mmbr.00026-21
Kwan, Thompson, Noncanonical translation initiation in eukaryotes, Cold Spring Harb Perspect Biol, doi:10.1101/cshperspect.a032672
Lan, Allan, Malsick, Secondary structural ensembles of the SARS-CoV-2 RNA genome in infected cells, Nat Commun, doi:10.1038/s41467-022-28603-2
Lapointe, Grosely, Johnson, Dynamic competition between SARS-CoV-2 NSP1 and mRNA on the human ribosome inhibits translation initiation, Proc Natl Acad Sci, doi:10.1073/pnas.2017715118
Lee, Budhathoki, Lee, Broad-spectrum antiviral activity of 3D8, a nucleic acid-hydrolyzing single-chain variable fragment (scFv), targeting SARS-CoV-2 and multiple coronaviruses in vitro, Viruses, doi:10.3390/v13040650
Lei, Cheng, Wang, The influence of host miRNA binding to RNA within RNA viruses on virus multiplication, Front Cell Infect Microbiol, doi:10.3389/fcimb.2022.802149
Leppek, Das, Barna, Functional 5 0 UTR mRNA structures in eukaryotic translation regulation and how to find them, Nat Rev Mol Cell Biol, doi:10.1038/nrm.2017.103
Li, Callahan, Phadke, Automated flow synthesis of peptide-pna conjugates, ACS Cent Sci, doi:10.1021/acscentsci.1c01019
Li, Hilgenfeld, Whitley, Therapeutic strategies for COVID-19: progress and lessons learned, Nat Rev Drug Discov, doi:10.1038/s41573-023-00672-y
Li, Kang, Liu, Structural lability in stem-loop 1 drives a 5 0 UTR-3 0 UTR interaction in coronavirus replication, J Mol Biol, doi:10.1016/j.jmb.2008.01.068
Li, Sczepanski, Targeting a conserved structural element from the SARS-CoV-2 genome using l-DNA aptamers, RSC Chem Biol, doi:10.1039/D1CB00172H
Li, Structure-based design of antisense oligonucleotides that inhibit SARS-CoV-2 replication, bioRxiv
Li, Tang, Atlas of interactions between SARS-CoV-2 macromolecules and host proteins, Cell Insight, doi:10.1016/j.cellin.2022.100068
Li, Zhang, Zhang, LinearTurboFold: linear-time global prediction of conserved structures for RNA homologs with applications to SARS-CoV-2, Proc Natl Acad Sci U S A, doi:10.1073/pnas.2116269118
Lukavsky, Structure and function of HCV IRES domains, Virus Res, doi:10.1016/j.virusres.2008.06.004
Lulla, Wandel, Bandyra, Targeting the conserved stem loop 2 motif in the SARS-CoV-2 genome, J Virol, doi:10.1128/JVI.00663-21
Malone, Urakova, Snijder, Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design, Nat Rev Mol Cell Biol, doi:10.1038/s41580-021-00432-z
Manfredonia, Nithin, Ponce-Salvatierra, Genome-wide mapping of SARS-CoV-2 RNA structures identifies therapeutically-relevant elements, Nucleic Acids Res, doi:10.1093/nar/gkaa1053
Martinez-Salas, Velilla, Fernandez-Chamorro, Insights into structural and mechanistic features of viral IRES elements, Front Microbiol, doi:10.3389/fmicb.2017.02629
Mathez, Cagno, Small molecules targeting viral RNA, Int J Mol Sci, doi:10.3390/ijms241713500
Mayr, What are 3 0 UTRs doing? Cold Spring, Harb Perspect Biol, doi:10.1101/cshperspect.a034728
Miao, Tidu, Eriani, Secondary structure of the SARS-CoV-2 5'-UTR, RNA Biology, doi:10.1080/15476286.2020.1814556
Morandi, Manfredonia, Simon, Genome-scale deconvolution of RNA structure ensembles, Nat Methods, doi:10.1038/s41592-021-01075-w
Nabiabad, Amini, Demirdas, Specific delivering of RNAi using spike's aptamer-functionalized lipid nanoparticles for targeting SARS-CoV-2: a strong anti-covid drug in a clinical case study, Chem Biol Drug Des, doi:10.1111/cbdd.13978
Niktab, Haghparast, Beigi, Design of advanced siRNA therapeutics for the treatment of COVID-19, Meta Gene, doi:10.1016/j.mgene.2021.100910
Ou, Liu, Lei, Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV, Nat Commun, doi:10.1038/s41467-020-15562-9
Pfafenrot, Schneider, Müller, Inhibition of SARS-CoV-2 coronavirus proliferation by designer antisense-circRNAs, Nucleic Acids Res, doi:10.1093/nar/gkab1096
Plant, Pérez-Alvarado, Jacobs, A three-stemmed mRNA pseudoknot in the SARS coronavirus frameshift signal, PLOS Biol, doi:10.1371/journal.pbio.0030172
Preiss, Hentze, From factors to mechanisms: translation and translational control in eukaryotes, Curr Opin Genet Dev, doi:10.1016/S0959-437X(99)00005-2
Qiao, Wotring, Zhang, Antisense oligonucleotides to therapeutically target SARS-CoV-2 infection, PLOS ONE, doi:10.1371/journal.pone.0281281
Ramos-Lorente, Berzal-Herranz, Romero-López, Recruitment of the 40S ribosomal subunit by the West Nile virus 3 0 UTR promotes the cross-talk between the viral genomic ends for translation regulation, Virus Res, doi:10.1016/j.virusres.2024.199340
Rasekhian, Roohvand, Habtemariam, The role of 3'UTR of RNA viruses on mRNA stability and translation enhancement, MRMC, doi:10.2174/1389557521666210217092305
Roberts, Langer, Wood, Advances in oligonucleotide drug delivery, Nat Rev Drug Discov, doi:10.1038/s41573-020-0075-7
Robertson, Igel, Baertsch, The structure of a rigorously conserved RNA element within the SARS virus genome, PLOS Biol, doi:10.1371/journal.pbio.0030005
Roman, Lewicka, Koirala, The SARS-CoV-2 programmed -1 ribosomal frameshifting element crystal structure solved to 2.09 Å using chaperone-assisted RNA crystallography, ACS Chem Biol, doi:10.1021/acschembio.1c00324
Rosenke, Leventhal, Moulton, Inhibition of SARS-CoV-2 in Vero cell cultures by peptide-conjugated morpholino oligomers, J Antimicrob Chemother, doi:10.1093/jac/dkaa460
Sajjanar, Raina, Untranslated regions (UTRs) orchestrate translation reprogramming in cellular stress responses, J Therm Biol, doi:10.1016/j.jtherbio.2017.02.006
Schlick, Zhu, Dey, To knot or not to knot: multiple conformations of the SARS-CoV-2 frameshifting RNA element, J Am Chem Soc, doi:10.1021/jacs.1c03003
Schoggins, Wilson, Panis, A diverse range of gene products are effectors of the type I interferon antiviral response, Nature, doi:10.1038/nature09907
Schubert, Karousis, Jomaa, SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation, Nat Struct Mol Biol, doi:10.1038/s41594-020-0511-8
Shang, Wan, Luo, Cell entry mechanisms of SARS-CoV-2, Proc Natl Acad Sci, doi:10.1073/pnas.2003138117
Slobodin, Sehrawat, Cap-independent translation and a precisely located RNA sequence enable SARS-CoV-2 to control host translation and escape anti-viral response, Nucleic Acids Res, doi:10.1093/nar/gkac615
Snijder, Bredenbeek, Dobbe, Unique and conserved features of genome and proteome of sars-coronavirus, an early split-off from the coronavirus group 2 lineage, J Mol Biol, doi:10.1016/S0022-2836(03)00865-9
Snijder, Limpens, De Wilde, A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis, PLoS Biol, doi:10.1371/journal.pbio.3000715
Sola, Almazán, Zúñiga, Continuous and discontinuous RNA synthesis in coronaviruses, Annu Rev Virol, doi:10.1146/annurev-virology-100114-055218
Sorokin, Vassilenko, Terenin, Non-canonical translation initiation mechanisms employed by Eukaryotic viral mRNAs, Biochemistry (Mosc), doi:10.1134/S0006297921090042
Spahn, Mulder, Cryo-em visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor, Cell, doi:10.1016/j.cell.2004.08.001
Stern-Ginossar, Thompson, Mathews, Translational control in virus-infected cells, Cold Spring Harb Perspect Biol, doi:10.1101/cshperspect.a033001
Su, Ma, Feng, Efficient inhibition of SARS-CoV-2 using chimeric antisense oligonucleotides through RNase L activation*, Angew Chem Int Ed Engl, doi:10.1002/anie.202105942
Sun, Abriola, Niederer, Restriction of SARS-CoV-2 replication by targeting programmed -1 ribosomal frameshifting, Proc Natl Acad Sci, doi:10.1073/pnas.2023051118
Sun, Li, Ju, In vivo structural characterization of the SARS-CoV-2 RNA genome identifies host proteins vulnerable to repurposed drugs, Cell, doi:10.1016/j.cell.2021.02.008
Szczesniak, Baliga-Gil, Jarmolowicz, Structural and functional RNA motifs of SARS-CoV-2 and influenza a virus as a target of viral inhibitors, Int J Mol Sci, doi:10.3390/ijms24021232
Tan, Yin, RNAi, a new therapeutic strategy against viral infection, Cell Res, doi:10.1038/sj.cr.7290248
Tengs, Kristoffersen, Bachvaroff, A mobile genetic element with unknown function found in distantly related viruses, Virol J, doi:10.1186/1743-422X-10-132
Thoms, Buschauer, Ameismeier, Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2, Science, doi:10.1126/science.abc8665
Tidu, Janvier, Schaeffer, The viral protein NSP1 acts as a ribosome gatekeeper for shutting down host translation and fostering SARS-CoV-2 translation, RNA, doi:10.1261/rna.078121.120
Tolksdorf, Nie, Niemeyer, Inhibition of SARS-CoV-2 replication by a small interfering RNA targeting the leader sequence, Viruses, doi:10.3390/v13102030
Varricchio, Mathez, Pillonel, Geneticin shows selective antiviral activity against SARS-CoV-2 by interfering with programmed -1 ribosomal frameshifting, Antiviral Res, doi:10.1016/j.antiviral.2022.105452
Vora, Fontana, Mao, Targeting stem-loop 1 of the SARS-CoV-2 5 0 UTR to suppress viral translation and Nsp1 evasion, Proc Natl Acad Sci, doi:10.1073/pnas.2117198119
Wacker, Weigand, Akabayov, Secondary structure determination of conserved SARS-CoV-2 RNA elements by NMR spectroscopy, Nucleic Acids Res, doi:10.1093/nar/gkaa1013
Wang, Zhou, Wang, Rapid design and development of CRISPR-Cas13a targeting SARS-CoV-2 spike protein, Theranostics, doi:10.7150/thno.51479
Weinlich, Hüttelmaier, Schierhorn, IGF2BP1 enhances HCV ires-mediated translation initiation via the 3 0 UTR, RNA, doi:10.1261/rna.1578409
Wethmar, The regulatory potential of upstream open reading frames in eukaryotic gene expression, Wiley Interdiscip Rev RNA, doi:10.1002/wrna.1245
Wu, Luo, Developing effective siRNAs to reduce the expression of key viral genes of COVID-19, Int J Biol Sci, doi:10.7150/ijbs.59151
Yang, Olatunji, Rhodes, Discovery of small molecules targeting the frameshifting element RNA in SARS-CoV-2 viral genome, ACS Med Chem Lett, doi:10.1021/acsmedchemlett.3c00051
Yao, Sun, Chen, RBM24 inhibits the translation of SARS-CoV-2 polyproteins by targeting the 5 0 -untranslated region, Antiviral Res, doi:10.1016/j.antiviral.2022.105478
Yokoyama, Machida, Iwasaki, HCV IRES captures an actively translating 80S ribosome, Mol Cell, doi:10.1016/j.molcel.2019.04.022
Zafferani, Haddad, Luo, Amilorides inhibit SARS-CoV-2 replication in vitro by targeting RNA structures, Sci Adv, doi:10.1126/sciadv.abl6096
Zamore, Haley, Ribo-gnome: the big world of small RNAs, Science, doi:10.1126/science.1111444
Zeng, Liu, Nguyenla, Broad-spectrum crispr-mediated inhibition of SARS-CoV-2 variants and endemic coronaviruses in vitro, Nat Commun, doi:10.1038/s41467-022-30546-7
Zhang, Almazi, Ong, Nanoparticle delivery platforms for RNAi therapeutics targeting COVID-19 disease in the respiratory tract, Int J Mol Sci, doi:10.3390/ijms23052408
Zhang, Huang, Ren, Comparison of viral RNA-host protein interactomes across pathogenic RNA viruses informs rapid antiviral drug discovery for SARS-CoV-2, Cell Res, doi:10.1038/s41422-021-00581-y
Zhang, Huang, Xie, In vivo structure and dynamics of the SARS-CoV-2 RNA genome, Nat Commun, doi:10.1038/s41467-021-25999-1
Zhang, Zheludev, Hagey, Cryo-em and antisense targeting of the 28-kDa frameshift stimulation element from the SARS-CoV-2 RNA genome, Nat Struct Mol Biol, doi:10.1038/s41594-021-00653-y
Zhou, Chen, Wang, Advances of CRISPR-Cas13 system in COVID-19 diagnosis and treatment, Genes Dis, doi:10.1016/j.gendis.2022.11.016
Zhu, Lee, Woo, An intranasal ASO therapeutic targeting SARS-CoV-2, Nat Commun, doi:10.1038/s41467-022-32216-0
Ziebuhr, Snijder, Gorbalenya, Virus-encoded proteinases and proteolytic processing in the Nidovirales, J Gen Virol, doi:10.1099/0022-1317-81-4-853
Ziv, Gabryelska, Lun, COMRADES determines in vivo RNA structures and interactions, Nat Methods, doi:10.1038/s41592-018-0121-0
Ziv, Price, Shalamova, The short-and long-range RNA-RNA interactome of SARS-CoV-2, Mol Cell, doi:10.1016/j.molcel.2020.11.004
DOI record:
{
"DOI": "10.1080/15476286.2024.2433830",
"ISSN": [
"1547-6286",
"1555-8584"
],
"URL": "http://dx.doi.org/10.1080/15476286.2024.2433830",
"alternative-id": [
"10.1080/15476286.2024.2433830"
],
"assertion": [
{
"label": "Peer Review Statement",
"name": "peerreview_statement",
"order": 1,
"value": "The publishing and review policy for this title is described in its Aims & Scope."
},
{
"URL": "http://www.tandfonline.com/action/journalInformation?show=aimsScope&journalCode=krnb20",
"label": "Aim & Scope",
"name": "aims_and_scope_url",
"order": 2,
"value": "http://www.tandfonline.com/action/journalInformation?show=aimsScope&journalCode=krnb20"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Revised",
"name": "revised",
"order": 1,
"value": "2024-11-16"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "2024-11-19"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Published",
"name": "published",
"order": 3,
"value": "2024-12-04"
}
],
"author": [
{
"affiliation": [
{
"name": "Department of Pharmaceutics, Key Laboratory of Drug-Targeting and Drug Delivery System of the Education Ministry and Sichuan Province, Sichuan Engineering Laboratory for Plant-Sourced Drug and Sichuan Research Center for Drug Precision Industrial Technology, West China School of Pharmacy, Sichuan University, Chengdu, China"
}
],
"family": "Wu",
"given": "Shan-Na",
"sequence": "first"
},
{
"affiliation": [
{
"name": "Key Laboratory of Birth Defects and Related Diseases of Women and Children, Children’s Medicine Key Laboratory of Sichuan Province, Department of Pharmacy/Evidence-Based Pharmacy Center, West China Second University Hospital, Sichuan University, Chengdu, China"
}
],
"family": "Xiao",
"given": "Ting",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Key Laboratory of Birth Defects and Related Diseases of Women and Children, Children’s Medicine Key Laboratory of Sichuan Province, Department of Pharmacy/Evidence-Based Pharmacy Center, West China Second University Hospital, Sichuan University, Chengdu, China"
}
],
"family": "Chen",
"given": "Hui",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Pharmaceutics, Key Laboratory of Drug-Targeting and Drug Delivery System of the Education Ministry and Sichuan Province, Sichuan Engineering Laboratory for Plant-Sourced Drug and Sichuan Research Center for Drug Precision Industrial Technology, West China School of Pharmacy, Sichuan University, Chengdu, China"
}
],
"family": "Li",
"given": "Xiao-Hong",
"sequence": "additional"
}
],
"container-title": "RNA Biology",
"container-title-short": "RNA Biology",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"www.tandfonline.com"
]
},
"created": {
"date-parts": [
[
2024,
12,
4
]
],
"date-time": "2024-12-04T15:38:17Z",
"timestamp": 1733326697000
},
"deposited": {
"date-parts": [
[
2024,
12,
12
]
],
"date-time": "2024-12-12T14:41:42Z",
"timestamp": 1734014502000
},
"funder": [
{
"DOI": "10.13039/501100018542",
"award": [
"2022NSFSC0722"
],
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/501100018542",
"id-type": "DOI"
}
],
"name": "Natural Science Foundation of Sichuan Province"
}
],
"indexed": {
"date-parts": [
[
2024,
12,
12
]
],
"date-time": "2024-12-12T15:10:22Z",
"timestamp": 1734016222442,
"version": "3.30.2"
},
"is-referenced-by-count": 0,
"issue": "1",
"issued": {
"date-parts": [
[
2024,
12,
4
]
]
},
"journal-issue": {
"issue": "1",
"published-print": {
"date-parts": [
[
2024,
12,
31
]
]
}
},
"language": "en",
"license": [
{
"URL": "http://creativecommons.org/licenses/by-nc/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
12,
4
]
],
"date-time": "2024-12-04T00:00:00Z",
"timestamp": 1733270400000
}
}
],
"link": [
{
"URL": "https://www.tandfonline.com/doi/pdf/10.1080/15476286.2024.2433830",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "301",
"original-title": [],
"page": "1290-1307",
"prefix": "10.1080",
"published": {
"date-parts": [
[
2024,
12,
4
]
]
},
"published-online": {
"date-parts": [
[
2024,
12,
4
]
]
},
"published-print": {
"date-parts": [
[
2024,
12,
31
]
]
},
"publisher": "Informa UK Limited",
"reference": [
{
"DOI": "10.1038/s41564-020-0695-z",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_2_1"
},
{
"DOI": "10.1016/j.antiviral.2013.08.015",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_3_1"
},
{
"DOI": "10.1038/s41573-023-00672-y",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_4_1"
},
{
"DOI": "10.1128/mmbr.00026-21",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_5_1"
},
{
"DOI": "10.1007/978-3-030-85109-5_2",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_6_1"
},
{
"DOI": "10.3390/v13060952",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_7_1"
},
{
"DOI": "10.1038/s41580-019-0136-0",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_8_1"
},
{
"DOI": "10.1016/j.cell.2020.04.011",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_9_1"
},
{
"DOI": "10.1073/pnas.2003138117",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_10_1"
},
{
"DOI": "10.1038/s41467-020-15562-9",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_11_1"
},
{
"DOI": "10.1016/j.antiviral.2020.104742",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_12_1"
},
{
"DOI": "10.1099/0022-1317-81-4-853",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_13_1"
},
{
"DOI": "10.1016/S0022-2836(03)00865-9",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_14_1"
},
{
"DOI": "10.1126/science.abc8665",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_15_1"
},
{
"DOI": "10.1038/s41594-020-0511-8",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_16_1"
},
{
"DOI": "10.1038/s41580-021-00432-z",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_17_1"
},
{
"DOI": "10.1371/journal.pbio.3000715",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_18_1"
},
{
"DOI": "10.1016/j.chom.2020.11.003",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_19_1"
},
{
"DOI": "10.1074/jbc.REV120.013930",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_20_1"
},
{
"DOI": "10.1146/annurev-virology-100114-055218",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_21_1"
},
{
"author": "Fields BN",
"key": "e_1_3_4_22_1",
"unstructured": "Fields BN, Howley PM, Griffin DE. Coronaviruses, in fields virology, H. KV, Editor. PA (USA): Lippincott Williams & Wilkins; 2001.",
"volume-title": "Coronaviruses, in fields virology",
"year": "2001"
},
{
"DOI": "10.1186/s13578-021-00643-z",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_23_1"
},
{
"DOI": "10.1371/journal.pbio.0030172",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_24_1"
},
{
"DOI": "10.1126/science.abf3546",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_25_1"
},
{
"DOI": "10.1021/acs.accounts.1c00316",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_26_1"
},
{
"DOI": "10.1016/j.cell.2020.10.004",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_27_1"
},
{
"DOI": "10.1073/pnas.2017715118",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_28_1"
},
{
"DOI": "10.1038/s41586-021-03610-3",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_29_1"
},
{
"DOI": "10.1261/rna.078121.120",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_30_1"
},
{
"DOI": "10.1371/journal.ppat.1002433",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_31_1"
},
{
"DOI": "10.1073/pnas.2117198119",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_32_1"
},
{
"DOI": "10.1016/S0378-1119(99)00210-3",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_33_1"
},
{
"DOI": "10.3390/v14071505",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_34_1"
},
{
"DOI": "10.1038/nature09907",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_35_1"
},
{
"DOI": "10.1126/science.aad9868",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_36_1"
},
{
"DOI": "10.1261/rna.2309906",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_37_1"
},
{
"DOI": "10.1080/15476286.2019.1632634",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_38_1"
},
{
"DOI": "10.15252/embj.201592759",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_39_1"
},
{
"DOI": "10.1002/wrna.1245",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_40_1"
},
{
"DOI": "10.1080/15476286.2020.1814556",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_41_1"
},
{
"DOI": "10.1016/j.cell.2021.02.008",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_42_1"
},
{
"DOI": "10.1093/nar/gkaa1053",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_43_1"
},
{
"DOI": "10.1016/j.molcel.2020.12.041",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_44_1"
},
{
"DOI": "10.1038/s41467-021-25361-5",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_45_1"
},
{
"DOI": "10.1038/s41586-020-2739-1",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_46_1"
},
{
"DOI": "10.1093/nar/gkac615",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_47_1"
},
{
"DOI": "10.2174/1389557521666210217092305",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_48_1"
},
{
"DOI": "10.1016/j.virusres.2024.199340",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_49_1"
},
{
"DOI": "10.1016/S0378-1119(02)01056-9",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_50_1"
},
{
"DOI": "10.1016/0092-8674(86)90762-2",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_51_1"
},
{
"DOI": "10.1134/S0006297921090042",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_52_1"
},
{
"DOI": "10.1099/vir.0.042499-0",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_53_1"
},
{
"DOI": "10.1101/cshperspect.a032672",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_54_1"
},
{
"DOI": "10.1073/pnas.0900153106",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_55_1"
},
{
"DOI": "10.1016/j.cell.2004.08.001",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_56_1"
},
{
"DOI": "10.1038/nature12658",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_57_1"
},
{
"DOI": "10.1093/nar/gku720",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_58_1"
},
{
"DOI": "10.3389/fmicb.2017.02629",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_59_1"
},
{
"DOI": "10.1016/j.virusres.2008.06.004",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_60_1"
},
{
"DOI": "10.1093/nar/gkx622",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_61_1"
},
{
"DOI": "10.1261/rna.5950603",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_62_1"
},
{
"DOI": "10.1016/j.str.2011.08.002",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_63_1"
},
{
"DOI": "10.1016/S1369-5274(00)00069-2",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_64_1"
},
{
"DOI": "10.1006/jmbi.2000.3900",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_65_1"
},
{
"DOI": "10.1016/j.molcel.2019.04.022",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_66_1"
},
{
"DOI": "10.1016/j.jtherbio.2017.02.006",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_67_1"
},
{
"DOI": "10.1101/cshperspect.a033001",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_68_1"
},
{
"DOI": "10.1038/s41579-018-0117-x",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_69_1"
},
{
"DOI": "10.1038/s41467-021-25999-1",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_70_1"
},
{
"DOI": "10.1093/nar/gkaa1013",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_71_1"
},
{
"DOI": "10.1128/JVI.00803-06",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_72_1"
},
{
"DOI": "10.1016/j.molcel.2020.11.004",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_73_1"
},
{
"DOI": "10.1038/s41592-021-01075-w",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_74_1"
},
{
"DOI": "10.1093/nar/gkab1226",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_75_1"
},
{
"DOI": "10.1101/cshperspect.a034728",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_76_1"
},
{
"DOI": "10.1021/acsomega.4c01050",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_77_1"
},
{
"key": "e_1_3_4_78_1",
"unstructured": "Predicted targets of hsa-miR-1307-3p mature miRNA. Available from: https://www.mirbase.org/mature/MIMAT0005951"
},
{
"DOI": "10.3389/fcimb.2022.802149",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_79_1"
},
{
"DOI": "10.1007/s00018-016-2377-9",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_80_1"
},
{
"DOI": "10.1371/journal.pbio.0030005",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_81_1"
},
{
"DOI": "10.1038/s41592-018-0121-0",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_82_1"
},
{
"DOI": "10.1016/0022-2836(87)90455-4",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_83_1"
},
{
"DOI": "10.1073/pnas.2116269118",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_84_1"
},
{
"DOI": "10.1016/j.jmb.2008.01.068",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_85_1"
},
{
"DOI": "10.1016/S0959-437X(99)00005-2",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_86_1"
},
{
"DOI": "10.1016/S0248-4900(03)00030-3",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_87_1"
},
{
"DOI": "10.1038/s41422-021-00581-y",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_88_1"
},
{
"DOI": "10.1016/j.cellin.2022.100068",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_89_1"
},
{
"DOI": "10.1261/rna.1578409",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_90_1"
},
{
"DOI": "10.1038/s41467-023-39091-3",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_91_1"
},
{
"DOI": "10.1038/s41594-021-00653-y",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_92_1"
},
{
"DOI": "10.1021/acschembio.1c00324",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_93_1"
},
{
"DOI": "10.1021/jacs.1c03003",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_94_1"
},
{
"DOI": "10.1261/rna.080035.124",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_95_1"
},
{
"DOI": "10.1038/s41467-022-28603-2",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_96_1"
},
{
"DOI": "10.3390/microorganisms10081552",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_97_1"
},
{
"DOI": "10.1016/j.jbc.2021.100416",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_98_1"
},
{
"DOI": "10.1038/s41573-021-00162-z",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_99_1"
},
{
"DOI": "10.1038/s41573-020-0075-7",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_100_1"
},
{
"DOI": "10.2217/epi-2020-0162",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_101_1"
},
{
"DOI": "10.3389/fmicb.2022.915202",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_102_1"
},
{
"DOI": "10.1371/journal.pone.0281281",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_103_1"
},
{
"DOI": "10.1002/anie.202105942",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_104_1"
},
{
"DOI": "10.1126/science.1111444",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_105_1"
},
{
"DOI": "10.1038/sj.cr.7290248",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_106_1"
},
{
"DOI": "10.3390/ijms23052408",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_107_1"
},
{
"DOI": "10.1016/j.mgene.2021.100910",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_108_1"
},
{
"DOI": "10.1111/all.14850",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_109_1"
},
{
"DOI": "10.7150/ijbs.59151",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_110_1"
},
{
"DOI": "10.3390/v13102030",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_111_1"
},
{
"DOI": "10.1016/j.ymthe.2021.05.004",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_112_1"
},
{
"DOI": "10.1111/cbdd.13978",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_113_1"
},
{
"DOI": "10.1016/j.gendis.2022.11.016",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_114_1"
},
{
"DOI": "10.1016/j.cell.2020.04.020",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_115_1"
},
{
"DOI": "10.7150/thno.51479",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_116_1"
},
{
"DOI": "10.1038/s41467-021-24577-9",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_117_1"
},
{
"DOI": "10.1038/s41467-022-30546-7",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_118_1"
},
{
"DOI": "10.1038/s41467-022-34339-w",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_119_1"
},
{
"DOI": "10.3390/v13040650",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_120_1"
},
{
"DOI": "10.3389/fchem.2021.802766",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_121_1"
},
{
"DOI": "10.1038/s41467-022-32216-0",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_122_1"
},
{
"DOI": "10.1093/jac/dkaa460",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_123_1"
},
{
"DOI": "10.1021/acscentsci.1c01019",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_124_1"
},
{
"DOI": "10.1093/nar/gkab1096",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_125_1"
},
{
"DOI": "10.1126/sciadv.abl6096",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_126_1"
},
{
"DOI": "10.1016/j.antiviral.2022.105478",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_127_1"
},
{
"DOI": "10.1101/2021.08.23.457434",
"doi-asserted-by": "crossref",
"key": "e_1_3_4_128_1",
"unstructured": "Li Y. Structure-based design of antisense oligonucleotides that inhibit SARS-CoV-2 replication. bioRxiv 2021."
},
{
"DOI": "10.1074/jbc.AC120.013449",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_129_1"
},
{
"DOI": "10.1073/pnas.2023051118",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_130_1"
},
{
"DOI": "10.1016/j.antiviral.2022.105452",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_131_1"
},
{
"DOI": "10.1021/acsmedchemlett.3c00051",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_132_1"
},
{
"DOI": "10.1021/acscentsci.0c00984",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_133_1"
},
{
"DOI": "10.1016/j.bbrc.2021.01.013",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_134_1"
},
{
"DOI": "10.1128/JVI.00663-21",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_135_1"
},
{
"DOI": "10.1039/D1CB00172H",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_136_1"
},
{
"DOI": "10.3390/ijms24021232",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_137_1"
},
{
"DOI": "10.1038/nrm.2017.103",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_138_1"
},
{
"DOI": "10.1002/wrna.1126",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_139_1"
},
{
"DOI": "10.1016/j.virol.2004.11.038",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_140_1"
},
{
"DOI": "10.3390/ijms241713500",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_141_1"
},
{
"DOI": "10.1128/jvi.00635-23",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_142_1"
},
{
"DOI": "10.1186/1743-422X-10-132",
"doi-asserted-by": "publisher",
"key": "e_1_3_4_143_1"
}
],
"reference-count": 142,
"references-count": 142,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.tandfonline.com/doi/full/10.1080/15476286.2024.2433830"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Decoding the genome of SARS-CoV-2: a pathway to drug development through translation inhibition",
"type": "journal-article",
"update-policy": "https://doi.org/10.1080/tandf_crossmark_01",
"volume": "21"
}