SARS-CoV-2 Infection of Lung Epithelia Leads to an Increase in the Cleavage and Translocation of RNase-III Drosha; Loss of Drosha Is Associated with a Decrease in Viral Replication
et al., Genes, doi:10.3390/genes16101239, Oct 2025
In vitro study showing that SARS-CoV-2 infection leads to increased cleavage and cytoplasmic translocation of RNase-III Drosha, with loss of Drosha decreasing viral replication. The study suggests SARS-CoV-2 co-opts host RNA-processing enzymes to support viral replication through a non-canonical cytoplasmic function of Drosha distinct from its nuclear microRNA biogenesis role.
Winters et al., 20 Oct 2025, peer-reviewed, 11 authors.
Contact: ivmartinez@hsc.wvu.edu (corresponding author), mtwinters@mix.wvu.edu, twr0001@mix.wvu.edu, gms00019@mix.wvu.edu, orm0004@mix.wvu.edu, irw00003@mix.wvu.edu, emily.rice@emory.edu, nh00053@mix.wvu.edu, mlc00064@mix.wvu.edu, kjp0018@mix.wvu.edu, mrd00026@mix.wvu.edu.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
SARS-CoV-2 Infection of Lung Epithelia Leads to an Increase in the Cleavage and Translocation of RNase-III Drosha; Loss of Drosha Is Associated with a Decrease in Viral Replication
Genes, doi:10.3390/genes16101239
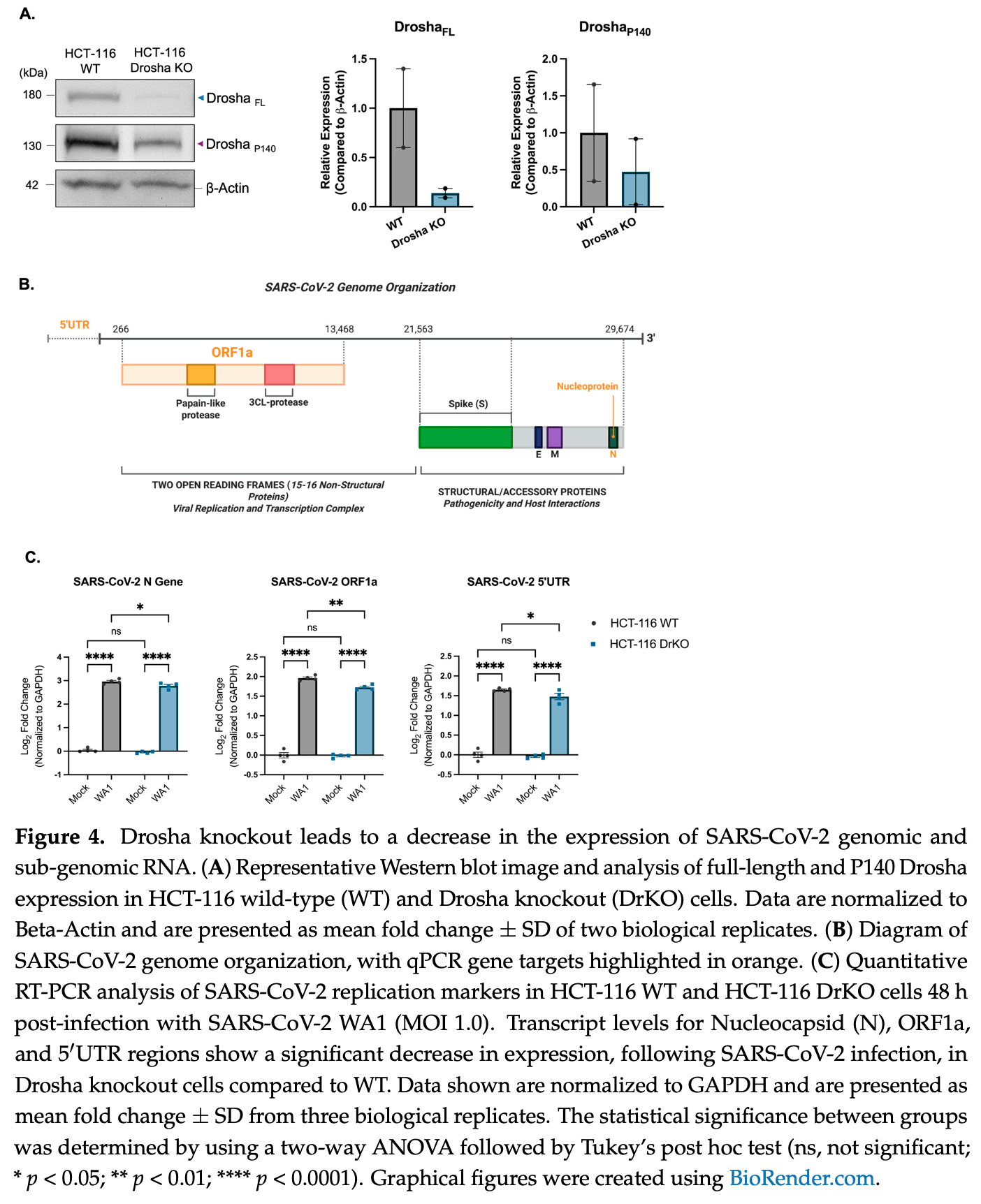
Background/Objectives: Since its emergence, COVID-19-caused by the novel coronavirus SARS-CoV-2-has affected millions globally and led to over 1.2 million deaths in the United States alone. This global impact, coupled with the emergence of five new human coronaviruses over the past two decades, underscores the urgency of understanding its pathogenic mechanisms at the molecular level-not only for managing the current pandemic but also preparing for future outbreaks. Small non-coding RNAs (sncRNAs) critically regulate host and viral gene expression, including antiviral responses. Among the molecular regulators implicated in antiviral defense, the microRNA-processing enzyme Drosha has emerged as a particularly intriguing factor. In addition to its canonical role, Drosha also exerts a non-canonical, interferon-independent antiviral function against several RNA viruses. Methods: To investigate this, we employed q/RT-PCR, Western blot, and immunocytochemistry/immunofluorescence in an immortalized normal human lung/bronchial epithelial cell line (NuLi-1), as well as a human colorectal carcinoma Drosha CRISPR knockout cell line. Results: In this study, we observed a striking shift in Drosha isoform expression following infection with multiple SARS-CoV-2 variants. This shift was absent following treatment with the viral mimetic poly (I:C) or infection with other RNA viruses, including the non-severe coronaviruses HCoV-OC43 and HCoV-229E. We also identified a distinct alteration in Drosha's cellular localization post SARS-CoV-2 infection. Moreover, Drosha ablation led to reduced expression of SARS-CoV-2 genomic and sub-genomic targets. Conclusions: Together, these observations not only elucidate a novel aspect of Drosha's antiviral role but also advance our understanding of SARS-CoV-2 host-pathogen interactions, highlighting potential therapeutic avenues for future human coronavirus infections.
Supplementary Materials: The following supporting information can be downloaded at: https:// www.mdpi.com/article/10.3390/genes16101239/s1 , Figure S1 : The expression of key proteins within the miRNA biogenesis pathway, at the mRNA and protein level, following SARS-CoV-2 infection. (A) Western Blot of four proteins within the miRNA biogenesis pathway in mock and SARS-CoV-2-infected (MOI 1.0) NuLi-1 cells. Dicer expression is significantly decreased following infection. (B) qRT-PCR of the mRNA transcripts of four key proteins within the miRNA biogenesis pathway. Following infection there is no significant change in expression at the transcript level. Data are normalized to Beta-Actin or GAPDH and are presented as mean fold change ± SD from at least three independent experiments. The statistical significance between groups was determined using a two-tailed students T test, or two-way ANOVA followed by Tukey's post hoc test (ns, not significant; * p < 0.05); Figure S2 . siRNA knockdown confirmation of Drosha p140 and p25 isoforms. (A) Western blot validation of Drosha isoforms by siRNA knockdown. NuLi-1 cells were transfected with 100 pmol Drosha-specific siRNA (siDrosha) or a non-targeting control siRNA (siControl). Western blotting shows a marked reduction in both FL and cleavage product (P140 and P25) bands in siDrosha-treated cells, confirming their identity as Drosha-specific. (B) qRT-PCR quantification showed a significant reduction in Drosha mRNA expression. Data..
References
Aguado, Schmid, May, Sabin, Panis et al., RNase III nucleases from diverse kingdoms serve as antiviral effectors, Nature, doi:10.1038/nature22990
Artimovič, Špaková, Macejková, Pribulová, Rabajdová et al., The ability of microRNAs to regulate the immune response in ischemia/reperfusion inflammatory pathways, Genes Immun, doi:10.1038/s41435-024-00283-6
Banerjee, Blanco, Bruce, Honson, Chen et al., SARS-CoV-2 Disrupts Splicing, Translation, and Protein Trafficking to Suppress Host Defenses, Cell, doi:10.1016/j.cell.2020.10.004
Bartel, MicroRNAs: Target Recognition and Regulatory Functions, Cell, doi:10.1016/j.cell.2009.01.002
Belshaw, Pybus, Rambaut, The evolution of genome compression and genomic novelty in RNA viruses, Genome Res, doi:10.1101/gr.6305707
Bouhaddou, Memon, Meyer, White, Rezelj et al., The Global Phosphorylation Landscape of SARS-CoV-2 Infection, Cell, doi:10.1016/j.cell.2020.06.034
Bruscella, Bottini, Baudesson, Pawlotsky, Feray et al., Viruses and miRNAs: More Friends than Foes, Front. Microbiol, doi:10.3389/fmicb.2017.00824
Cao, Cai, Xiao, Rao, Chen et al., The architecture of the SARS-CoV-2 RNA genome inside virion, Nat. Commun, doi:10.1038/s41467-021-22785-x
Case, Bailey, Kim, Chen, Diamond, Growth, detection, quantification, and inactivation of SARS-CoV-2, Virology, doi:10.1016/j.virol.2020.05.015
Chang, .-Y.; Rawlinson, Pitt, Taiaroa, Gleeson et al., Transcriptional and epi-transcriptional dynamics of SARS-CoV-2 during cellular infection, Cell Rep, doi:10.1016/j.celrep.2021.109108
Chatterjee, Thakur, SARS-CoV-2 Infection Triggers Phosphorylation: Potential Target for Anti-COVID-19 Therapeutics, Front. Immunol, doi:10.3389/fimmu.2022.829474
Cui, Li, Shi, Origin and evolution of pathogenic coronaviruses, Nat. Rev. Microbiol, doi:10.1038/s41579-018-0118-9
Dai, Chen, Youngren, Kulina, Yang et al., Cytoplasmic Drosha activity generated by alternative splicing, Nucleic Acids Res, doi:10.1093/nar/gkw668
Denli, Tops, Plasterk, Ketting, Hannon, Processing of primary microRNAs by the microprocessor complex, Nature, doi:10.1038/nature03049
Esteller, Non-coding RNAs in human disease, Nat. Rev. Genet, doi:10.1038/nrg3074
Faist, Schloer, Mecate-Zambrano, Janowski, Schreiber et al., Inhibition of p38 signaling curtails the SARS-CoV-2 induced inflammatory response but retains the IFN-dependent antiviral defense of the lung epithelial barrier, Antivir. Res, doi:10.1016/j.antiviral.2022.105475
Forster, Tate, Hertzog, MicroRNA as Type I Interferon-Regulated Transcripts and Modulators of the Innate Immune Response, Front. Immunol, doi:10.3389/fimmu.2015.00334
Girardi, López, Pfeffer, On the Importance of Host MicroRNAs During Viral Infection, Front. Genet, doi:10.3389/fgene.2018.00439
Gregory, Yan, Amuthan, Chendrimada, Doratotaj et al., The Microprocessor complex mediates the genesis of microRNAs, Nature, doi:10.1038/nature03120
Jopling, Yi, Lancaster, Lemon, Sarnow, Modulation of Hepatitis C Virus RNA Abundance by a Liver-Specific MicroRNA, Science, doi:10.1126/science.1113329
Kim, Lee, Yang, Kim, Kim et al., The Architecture of SARS-CoV-2 Transcriptome, Cell, doi:10.1016/j.cell.2020.04.011
Kincaid, Sullivan, Virus-Encoded microRNAs: An Overview and a Look to the Future, PLoS Pathog, doi:10.1371/journal.ppat.1003018
Lan, Allan, Malsick, Woo, Zhu et al., Secondary structural ensembles of the SARS-CoV-2 RNA genome in infected cells, Nat. Commun, doi:10.1038/s41467-022-28603-2
Lin, Sullivan, Expanding the role of Drosha to the regulation of viral gene expression, Proc. Natl. Acad. Sci, doi:10.1073/pnas.1105799108
Link, Grund, Diederichs, Alternative splicing affects the subcellular localization of Drosha, Nucleic Acids Res, doi:10.1093/nar/gkw400
Liu, Verma, Garcia, Ramage, Lucas et al., Targeting the coronavirus nucleocapsid protein through GSK-3 inhibition, Proc. Natl. Acad. Sci, doi:10.1073/pnas.2113401118
Lu, Liou, Zhou, Pinning down proline-directed phosphorylation signaling, Trends Cell Biol, doi:10.1016/S0962-8924(02)02253-5
Macveigh-Fierro, Rodriguez, Miles, Muller, Stealing the Show: KSHV Hijacks Host RNA Regulatory Pathways to Promote Infection, Viruses, doi:10.3390/v12091024
Martinez, Hayes, Barr, Harold, Xie et al., An Exportin-1dependent microRNA biogenesis pathway during human cell quiescence, Proc. Natl. Acad. Sci, doi:10.1073/pnas.1618732114
Minkoff, Tenoever, Innate immune evasion strategies of SARS-CoV-2, Nat. Rev. Microbiol, doi:10.1038/s41579-022-00839-1
Momen-Heravi, Bala, miRNA regulation of innate immunity, J. Leukoc. Biol, doi:10.1002/JLB.3MIR1117-459R
O'brien, Hayder, Zayed, Peng, Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation, Front. Endocrinol, doi:10.3389/fendo.2018.00402
Page, Génin, Baines, Hiscott, Interferon activation and innate immunity, Rev. Immunogenet
Park, Iwasaki, Type I and Type III Interferons-Induction, Signaling, Evasion, and Application to Combat COVID-19, Cell Host Microbe, doi:10.1016/j.chom.2020.05.008
Pawlica, Yario, White, Wang, Moss et al., SARS-CoV-2 expresses a microRNA-like small RNA able to selectively repress host genes, Proc. Natl. Acad. Sci, doi:10.1073/pnas.2116668118
Poggio, Vallese, Hartel, Morgenstern, Kanner et al., Perturbation of the host cell Ca 2+ homeostasis and ER-mitochondria contact sites by the SARS-CoV-2 structural proteins E and M, Cell Death Dis, doi:10.1038/s41419-023-05817-w
Prasad, Gour, Raghuvanshi, Kumar, The SARS-CoV-2 targeted human RNA binding proteins network biology to investigate COVID-19 associated manifestations, Int. J. Biol. Macromol, doi:10.1016/j.ijbiomac.2022.07.200
Shapiro, Langlois, Pham, Tenoever, Evidence for a cytoplasmic microprocessor of pri-miRNAs, RNA, doi:10.1261/rna.032268.112
Shapiro, Schmid, Aguado, Sabin, Yasunaga et al., Drosha as an interferon-independent antiviral factor, Proc. Natl. Acad. Sci, doi:10.1073/pnas.1319635111
Shemesh, Aktepe, Deerain, Mcauley, Audsley et al., SARS-CoV-2 suppresses IFNβ production mediated by NSP1, 5, 6, 15, ORF6 and ORF7b but does not suppress the effects of added interferon, PLoS Pathog, doi:10.1371/journal.ppat.1009800
Son, Kim, Yang, Kim, Role of the proline-rich disordered domain of DROSHA in intronic microRNA processing, Genes Dev, doi:10.1101/gad.350275.122
Tang, Li, Tucker, Ramratnam, Glycogen Synthase Kinase 3 Beta (GSK3β) Phosphorylates the RNAase III Enzyme Drosha at S300 and S302, PLoS ONE, doi:10.1371/journal.pone.0020391
Tang, Zhang, Tucker, Ramratnam, Phosphorylation of the RNase III enzyme Drosha at Serine300 or Serine302 is required for its nuclear localization, Nucleic Acids Res, doi:10.1093/nar/gkq547
Trobaugh, Klimstra, MicroRNA Regulation of RNA Virus Replication and Pathogenesis, Trends Mol. Med, doi:10.1016/j.molmed.2016.11.003
Tucker, Wong, Marri, Ali, Fedele et al., SARS-CoV-2 produces a microRNA CoV2-miR-O8 in patients with COVID-19 infection, iScience, doi:10.1016/j.isci.2023.108719
V'kovski, Kratzel, Steiner, Stalder, Thiel, Coronavirus biology and replication: Implications for SARS-CoV-2, Nat. Rev. Microbiol, doi:10.1038/s41579-020-00468-6
Yang, Li, She, Dou, Duong et al., Stress Induces p38 MAPK-Mediated Phosphorylation and Inhibition of Drosha-Dependent Cell Survival, Mol. Cell, doi:10.1016/j.molcel.2015.01.004
Ye, Yuan, Yuen, Fung, Chan et al., Zoonotic origins of human coronaviruses, Int. J. Biol. Sci, doi:10.7150/ijbs.45472
Yoo, Mitchison, Quantitative comparison of nuclear transport inhibition by SARS coronavirus ORF6 reveals the importance of oligomerization, Proc. Natl. Acad. Sci, doi:10.1073/pnas.2307997121
Zeid, Shanmugapriya, Rumney, Mosser, Caspase-mediated cleavage of miRNA processing proteins Drosha, DGCR8, Dicer, and TRBP2 in heat-shocked cells and its inhibition by HSP70 overexpression, Cell Stress Chaperones, doi:10.1007/s12192-021-01242-0
Zeng, Antia, Casorla-Perez, Puray-Chavez, Kutluay et al., Calpain-2 mediates SARS-CoV-2 entry via regulating ACE2 levels, mBio, doi:10.1128/mbio.02287-23
DOI record:
{
"DOI": "10.3390/genes16101239",
"ISSN": [
"2073-4425"
],
"URL": "http://dx.doi.org/10.3390/genes16101239",
"abstract": "<jats:p>Background/Objectives: Since its emergence, COVID-19—caused by the novel coronavirus SARS-CoV-2—has affected millions globally and led to over 1.2 million deaths in the United States alone. This global impact, coupled with the emergence of five new human coronaviruses over the past two decades, underscores the urgency of understanding its pathogenic mechanisms at the molecular level—not only for managing the current pandemic but also preparing for future outbreaks. Small non-coding RNAs (sncRNAs) critically regulate host and viral gene expression, including antiviral responses. Among the molecular regulators implicated in antiviral defense, the microRNA-processing enzyme Drosha has emerged as a particularly intriguing factor. In addition to its canonical role, Drosha also exerts a non-canonical, interferon-independent antiviral function against several RNA viruses. Methods: To investigate this, we employed q/RT-PCR, Western blot, and immunocytochemistry/immunofluorescence in an immortalized normal human lung/bronchial epithelial cell line (NuLi-1), as well as a human colorectal carcinoma Drosha CRISPR knockout cell line. Results: In this study, we observed a striking shift in Drosha isoform expression following infection with multiple SARS-CoV-2 variants. This shift was absent following treatment with the viral mimetic poly (I:C) or infection with other RNA viruses, including the non-severe coronaviruses HCoV-OC43 and HCoV-229E. We also identified a distinct alteration in Drosha’s cellular localization post SARS-CoV-2 infection. Moreover, Drosha ablation led to reduced expression of SARS-CoV-2 genomic and sub-genomic targets. Conclusions: Together, these observations not only elucidate a novel aspect of Drosha’s antiviral role but also advance our understanding of SARS-CoV-2 host–pathogen interactions, highlighting potential therapeutic avenues for future human coronavirus infections.</jats:p>",
"alternative-id": [
"genes16101239"
],
"author": [
{
"ORCID": "https://orcid.org/0000-0003-3532-928X",
"affiliation": [
{
"name": "Department of Microbiology, Immunology and Cell Biology, West Virginia University School of Medicine, West Virginia University, Morgantown, WV 26506, USA"
}
],
"authenticated-orcid": false,
"family": "Winters",
"given": "Michael T.",
"sequence": "first"
},
{
"ORCID": "https://orcid.org/0000-0001-6142-8033",
"affiliation": [
{
"name": "West Virginia University Cancer Institute, West Virginia University, Morgantown, WV 26506, USA"
}
],
"authenticated-orcid": false,
"family": "Westemeier-Rice",
"given": "Emily S.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Microbiology, Immunology and Cell Biology, West Virginia University School of Medicine, West Virginia University, Morgantown, WV 26506, USA"
}
],
"family": "Rawson",
"given": "Travis W.",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0009-0009-7984-3126",
"affiliation": [
{
"name": "West Virginia University School of Medicine, West Virginia University, Morgantown, WV 26506, USA"
}
],
"authenticated-orcid": false,
"family": "Patel",
"given": "Kiran J.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Microbiology, Immunology and Cell Biology, West Virginia University School of Medicine, West Virginia University, Morgantown, WV 26506, USA"
}
],
"family": "Sankey",
"given": "Gabriel M.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Biology, West Virginia University, Morgantown, WV 26506, USA"
}
],
"family": "Dixon-Gross",
"given": "Maya",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0009-0000-9684-4598",
"affiliation": [
{
"name": "Department of Microbiology, Immunology and Cell Biology, West Virginia University School of Medicine, West Virginia University, Morgantown, WV 26506, USA"
}
],
"authenticated-orcid": false,
"family": "McHugh",
"given": "Olivia R.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "West Virginia University Cancer Institute, West Virginia University, Morgantown, WV 26506, USA"
}
],
"family": "Hashemipour",
"given": "Nasrin",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0009-0007-4728-0846",
"affiliation": [
{
"name": "West Virginia University Cancer Institute, West Virginia University, Morgantown, WV 26506, USA"
}
],
"authenticated-orcid": false,
"family": "Carroll",
"given": "McKenna L.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Microbiology, Immunology and Cell Biology, West Virginia University School of Medicine, West Virginia University, Morgantown, WV 26506, USA"
}
],
"family": "Wilkerson",
"given": "Isabella R.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Microbiology, Immunology and Cell Biology, West Virginia University School of Medicine, West Virginia University, Morgantown, WV 26506, USA"
},
{
"name": "West Virginia University Cancer Institute, West Virginia University, Morgantown, WV 26506, USA"
}
],
"family": "Martinez",
"given": "Ivan",
"sequence": "additional"
}
],
"container-title": "Genes",
"container-title-short": "Genes",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2025,
10,
20
]
],
"date-time": "2025-10-20T10:44:10Z",
"timestamp": 1760957050000
},
"deposited": {
"date-parts": [
[
2025,
10,
22
]
],
"date-time": "2025-10-22T01:09:50Z",
"timestamp": 1761095390000
},
"funder": [
{
"DOI": "10.13039/100000048",
"award": [
"RSG-24-1039619-01-RMC"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/100000048",
"id-type": "DOI"
}
],
"name": "American Cancer Society"
},
{
"DOI": "10.13039/100000057",
"award": [
"5U54GM104942-03"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/100000057",
"id-type": "DOI"
}
],
"name": "IDeA CTR"
}
],
"indexed": {
"date-parts": [
[
2025,
10,
22
]
],
"date-time": "2025-10-22T21:32:52Z",
"timestamp": 1761168772792,
"version": "build-2065373602"
},
"is-referenced-by-count": 0,
"issue": "10",
"issued": {
"date-parts": [
[
2025,
10,
20
]
]
},
"journal-issue": {
"issue": "10",
"published-online": {
"date-parts": [
[
2025,
10
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
10,
20
]
],
"date-time": "2025-10-20T00:00:00Z",
"timestamp": 1760918400000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/2073-4425/16/10/1239/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "1239",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2025,
10,
20
]
]
},
"published-online": {
"date-parts": [
[
2025,
10,
20
]
]
},
"publisher": "MDPI AG",
"reference": [
{
"key": "ref_1",
"unstructured": "National Center for Health Statistics (2025, August 24). Centers for Disease Control and Prevention. Provisional COVID-19 Mortality Surveillance, Available online: https://www.cdc.gov/nchs/nvss/vsrr/covid19/."
},
{
"DOI": "10.1038/s41579-020-00468-6",
"article-title": "Coronavirus biology and replication: Implications for SARS-CoV-2",
"author": "Kratzel",
"doi-asserted-by": "crossref",
"first-page": "155",
"journal-title": "Nat. Rev. Microbiol.",
"key": "ref_2",
"volume": "19",
"year": "2021"
},
{
"DOI": "10.1038/s41579-018-0118-9",
"article-title": "Origin and evolution of pathogenic coronaviruses",
"author": "Cui",
"doi-asserted-by": "crossref",
"first-page": "181",
"journal-title": "Nat. Rev. Microbiol.",
"key": "ref_3",
"volume": "17",
"year": "2019"
},
{
"DOI": "10.1101/gr.6305707",
"article-title": "The evolution of genome compression and genomic novelty in RNA viruses",
"author": "Belshaw",
"doi-asserted-by": "crossref",
"first-page": "1496",
"journal-title": "Genome Res.",
"key": "ref_4",
"volume": "17",
"year": "2007"
},
{
"DOI": "10.3389/fgene.2018.00439",
"doi-asserted-by": "crossref",
"key": "ref_5",
"unstructured": "Girardi, E., López, P., and Pfeffer, S. (2018). On the Importance of Host MicroRNAs During Viral Infection. Front. Genet., 9."
},
{
"DOI": "10.1016/j.cell.2009.01.002",
"article-title": "MicroRNAs: Target Recognition and Regulatory Functions",
"author": "Bartel",
"doi-asserted-by": "crossref",
"first-page": "215",
"journal-title": "Cell",
"key": "ref_6",
"volume": "136",
"year": "2009"
},
{
"DOI": "10.1038/nrg3074",
"article-title": "Non-coding RNAs in human disease",
"author": "Esteller",
"doi-asserted-by": "crossref",
"first-page": "861",
"journal-title": "Nat. Rev. Genet.",
"key": "ref_7",
"volume": "12",
"year": "2011"
},
{
"DOI": "10.1126/science.1113329",
"article-title": "Modulation of Hepatitis C Virus RNA Abundance by a Liver-Specific MicroRNA",
"author": "Jopling",
"doi-asserted-by": "crossref",
"first-page": "1577",
"journal-title": "Science",
"key": "ref_8",
"volume": "309",
"year": "2005"
},
{
"DOI": "10.1038/s41435-024-00283-6",
"article-title": "The ability of microRNAs to regulate the immune response in ischemia/reperfusion inflammatory pathways",
"doi-asserted-by": "crossref",
"first-page": "277",
"journal-title": "Genes Immun.",
"key": "ref_9",
"volume": "25",
"year": "2024"
},
{
"DOI": "10.1002/JLB.3MIR1117-459R",
"article-title": "miRNA regulation of innate immunity",
"author": "Bala",
"doi-asserted-by": "crossref",
"first-page": "1205",
"journal-title": "J. Leukoc. Biol.",
"key": "ref_10",
"volume": "103",
"year": "2018"
},
{
"DOI": "10.3389/fimmu.2015.00334",
"doi-asserted-by": "crossref",
"key": "ref_11",
"unstructured": "Forster, S.C., Tate, M.D., and Hertzog, P.J. (2015). MicroRNA as Type I Interferon-Regulated Transcripts and Modulators of the Innate Immune Response. Front. Immunol., 6."
},
{
"DOI": "10.1016/j.cell.2020.10.004",
"article-title": "SARS-CoV-2 Disrupts Splicing, Translation, and Protein Trafficking to Suppress Host Defenses",
"author": "Banerjee",
"doi-asserted-by": "crossref",
"first-page": "1325",
"journal-title": "Cell",
"key": "ref_12",
"volume": "183",
"year": "2020"
},
{
"DOI": "10.1371/journal.ppat.1003018",
"doi-asserted-by": "crossref",
"key": "ref_13",
"unstructured": "Kincaid, R.P., and Sullivan, C.S. (2012). Virus-Encoded microRNAs: An Overview and a Look to the Future. PLoS Pathog., 8."
},
{
"DOI": "10.1016/j.molmed.2016.11.003",
"article-title": "MicroRNA Regulation of RNA Virus Replication and Pathogenesis",
"author": "Trobaugh",
"doi-asserted-by": "crossref",
"first-page": "80",
"journal-title": "Trends Mol. Med.",
"key": "ref_14",
"volume": "23",
"year": "2017"
},
{
"DOI": "10.1073/pnas.2116668118",
"article-title": "SARS-CoV-2 expresses a microRNA-like small RNA able to selectively repress host genes",
"author": "Pawlica",
"doi-asserted-by": "crossref",
"first-page": "e2116668118",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_15",
"volume": "118",
"year": "2021"
},
{
"DOI": "10.1016/j.isci.2023.108719",
"article-title": "SARS-CoV-2 produces a microRNA CoV2-miR-O8 in patients with COVID-19 infection",
"author": "Tucker",
"doi-asserted-by": "crossref",
"first-page": "108719",
"journal-title": "iScience",
"key": "ref_16",
"volume": "27",
"year": "2024"
},
{
"DOI": "10.3389/fmicb.2017.00824",
"doi-asserted-by": "crossref",
"key": "ref_17",
"unstructured": "Bruscella, P., Bottini, S., Baudesson, C., Pawlotsky, J.-M., Feray, C., and Trabucchi, M. (2017). Viruses and miRNAs: More Friends than Foes. Front. Microbiol., 8."
},
{
"DOI": "10.1038/nature03049",
"article-title": "Processing of primary microRNAs by the microprocessor complex",
"author": "Denli",
"doi-asserted-by": "crossref",
"first-page": "231",
"journal-title": "Nature",
"key": "ref_18",
"volume": "432",
"year": "2004"
},
{
"DOI": "10.1038/nature03120",
"article-title": "The Microprocessor complex mediates the genesis of microRNAs",
"author": "Gregory",
"doi-asserted-by": "crossref",
"first-page": "235",
"journal-title": "Nature",
"key": "ref_19",
"volume": "432",
"year": "2004"
},
{
"DOI": "10.3389/fendo.2018.00402",
"doi-asserted-by": "crossref",
"key": "ref_20",
"unstructured": "O’Brien, J., Hayder, H., Zayed, Y., and Peng, C. (2018). Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol., 9."
},
{
"article-title": "Interferon activation and innate immunity",
"author": "Baines",
"first-page": "374",
"journal-title": "Rev. Immunogenet.",
"key": "ref_21",
"volume": "2",
"year": "2000"
},
{
"DOI": "10.1371/journal.ppat.1010146",
"doi-asserted-by": "crossref",
"key": "ref_22",
"unstructured": "Shemesh, M., Aktepe, T.E., Deerain, J.M., McAuley, J.L., Audsley, M.D., David, C.T., Purcell, D.F.J., Urin, V., Hartmann, R., and Moseley, G.W. (2021). SARS-CoV-2 suppresses IFNβ production mediated by NSP1, 5, 6, 15, ORF6 and ORF7b but does not suppress the effects of added interferon. PLoS Pathog., 17."
},
{
"article-title": "Innate immune evasion strategies of SARS-CoV-2",
"author": "Minkoff",
"first-page": "178",
"journal-title": "Nat. Rev. Microbiol.",
"key": "ref_23",
"volume": "21",
"year": "2023"
},
{
"DOI": "10.1016/j.chom.2020.05.008",
"article-title": "Type I and Type III Interferons—Induction, Signaling, Evasion, and Application to Combat COVID-19",
"author": "Park",
"doi-asserted-by": "crossref",
"first-page": "870",
"journal-title": "Cell Host Microbe",
"key": "ref_24",
"volume": "27",
"year": "2020"
},
{
"DOI": "10.1101/gad.350275.122",
"article-title": "Role of the proline-rich disordered domain of DROSHA in intronic microRNA processing",
"author": "Son",
"doi-asserted-by": "crossref",
"first-page": "383",
"journal-title": "Genes Dev.",
"key": "ref_25",
"volume": "37",
"year": "2023"
},
{
"DOI": "10.1093/nar/gkw400",
"article-title": "Alternative splicing affects the subcellular localization of Drosha",
"author": "Link",
"doi-asserted-by": "crossref",
"first-page": "5330",
"journal-title": "Nucleic Acids Res.",
"key": "ref_26",
"volume": "44",
"year": "2016"
},
{
"article-title": "Cytoplasmic Drosha activity generated by alternative splicing",
"author": "Dai",
"first-page": "10454",
"journal-title": "Nucleic Acids Res.",
"key": "ref_27",
"volume": "44",
"year": "2016"
},
{
"DOI": "10.1073/pnas.1618732114",
"article-title": "An Exportin-1–dependent microRNA biogenesis pathway during human cell quiescence",
"author": "Martinez",
"doi-asserted-by": "crossref",
"first-page": "E4961",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_28",
"volume": "114",
"year": "2017"
},
{
"DOI": "10.1016/j.virol.2020.05.015",
"article-title": "Growth, detection, quantification, and inactivation of SARS-CoV-2",
"author": "Case",
"doi-asserted-by": "crossref",
"first-page": "39",
"journal-title": "Virology",
"key": "ref_29",
"volume": "548",
"year": "2020"
},
{
"DOI": "10.1261/rna.032268.112",
"article-title": "Evidence for a cytoplasmic microprocessor of pri-miRNAs",
"author": "Shapiro",
"doi-asserted-by": "crossref",
"first-page": "1338",
"journal-title": "RNA",
"key": "ref_30",
"volume": "18",
"year": "2012"
},
{
"DOI": "10.1073/pnas.1319635111",
"article-title": "Drosha as an interferon-independent antiviral factor",
"author": "Shapiro",
"doi-asserted-by": "crossref",
"first-page": "7108",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_31",
"volume": "111",
"year": "2014"
},
{
"DOI": "10.7150/ijbs.45472",
"article-title": "Zoonotic origins of human coronaviruses",
"author": "Ye",
"doi-asserted-by": "crossref",
"first-page": "1686",
"journal-title": "Int. J. Biol. Sci.",
"key": "ref_32",
"volume": "16",
"year": "2020"
},
{
"DOI": "10.1016/j.molcel.2015.01.004",
"article-title": "Stress Induces p38 MAPK-Mediated Phosphorylation and Inhibition of Drosha-Dependent Cell Survival",
"author": "Yang",
"doi-asserted-by": "crossref",
"first-page": "721",
"journal-title": "Mol. Cell",
"key": "ref_33",
"volume": "57",
"year": "2015"
},
{
"DOI": "10.1007/s12192-021-01242-0",
"article-title": "Caspase-mediated cleavage of miRNA processing proteins Drosha, DGCR8, Dicer, and TRBP2 in heat-shocked cells and its inhibition by HSP70 overexpression",
"author": "Zeid",
"doi-asserted-by": "crossref",
"first-page": "11",
"journal-title": "Cell Stress Chaperones",
"key": "ref_34",
"volume": "27",
"year": "2022"
},
{
"DOI": "10.1016/S0962-8924(02)02253-5",
"article-title": "Pinning down proline-directed phosphorylation signaling",
"author": "Lu",
"doi-asserted-by": "crossref",
"first-page": "164",
"journal-title": "Trends Cell Biol.",
"key": "ref_35",
"volume": "12",
"year": "2002"
},
{
"DOI": "10.1093/nar/gkq547",
"article-title": "Phosphorylation of the RNase III enzyme Drosha at Serine300 or Serine302 is required for its nuclear localization",
"author": "Tang",
"doi-asserted-by": "crossref",
"first-page": "6610",
"journal-title": "Nucleic Acids Res.",
"key": "ref_36",
"volume": "38",
"year": "2010"
},
{
"DOI": "10.1371/journal.pone.0020391",
"doi-asserted-by": "crossref",
"key": "ref_37",
"unstructured": "Tang, X., Li, M., Tucker, L., and Ramratnam, B. (2011). Glycogen Synthase Kinase 3 Beta (GSK3β) Phosphorylates the RNAase III Enzyme Drosha at S300 and S302. PLoS ONE, 6."
},
{
"DOI": "10.1016/j.cell.2020.06.034",
"article-title": "The Global Phosphorylation Landscape of SARS-CoV-2 Infection",
"author": "Bouhaddou",
"doi-asserted-by": "crossref",
"first-page": "685",
"journal-title": "Cell",
"key": "ref_38",
"volume": "182",
"year": "2020"
},
{
"DOI": "10.3389/fimmu.2022.829474",
"doi-asserted-by": "crossref",
"key": "ref_39",
"unstructured": "Chatterjee, B., and Thakur, S.S. (2022). SARS-CoV-2 Infection Triggers Phosphorylation: Potential Target for Anti-COVID-19 Therapeutics. Front. Immunol., 13."
},
{
"DOI": "10.1073/pnas.2307997121",
"article-title": "Quantitative comparison of nuclear transport inhibition by SARS coronavirus ORF6 reveals the importance of oligomerization",
"author": "Yoo",
"doi-asserted-by": "crossref",
"first-page": "e2307997121",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_40",
"volume": "121",
"year": "2024"
},
{
"DOI": "10.1038/nature22990",
"article-title": "RNase III nucleases from diverse kingdoms serve as antiviral effectors",
"author": "Aguado",
"doi-asserted-by": "crossref",
"first-page": "114",
"journal-title": "Nature",
"key": "ref_41",
"volume": "547",
"year": "2017"
},
{
"DOI": "10.1073/pnas.1105799108",
"article-title": "Expanding the role of Drosha to the regulation of viral gene expression",
"author": "Lin",
"doi-asserted-by": "crossref",
"first-page": "11229",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_42",
"volume": "108",
"year": "2011"
},
{
"DOI": "10.3390/v12091024",
"doi-asserted-by": "crossref",
"key": "ref_43",
"unstructured": "Macveigh-Fierro, D., Rodriguez, W., Miles, J., and Muller, M. (2020). Stealing the Show: KSHV Hijacks Host RNA Regulatory Pathways to Promote Infection. Viruses, 12."
},
{
"DOI": "10.1016/j.ijbiomac.2022.07.200",
"article-title": "The SARS-CoV-2 targeted human RNA binding proteins network biology to investigate COVID-19 associated manifestations",
"author": "Prasad",
"doi-asserted-by": "crossref",
"first-page": "853",
"journal-title": "Int. J. Biol. Macromol.",
"key": "ref_44",
"volume": "217",
"year": "2022"
},
{
"DOI": "10.1016/j.cell.2020.04.011",
"article-title": "The Architecture of SARS-CoV-2 Transcriptome",
"author": "Kim",
"doi-asserted-by": "crossref",
"first-page": "914",
"journal-title": "Cell",
"key": "ref_45",
"volume": "181",
"year": "2020"
},
{
"DOI": "10.1016/j.celrep.2021.109108",
"article-title": "Transcriptional and epi-transcriptional dynamics of SARS-CoV-2 during cellular infection",
"author": "Chang",
"doi-asserted-by": "crossref",
"first-page": "109108",
"journal-title": "Cell Rep.",
"key": "ref_46",
"volume": "35",
"year": "2021"
},
{
"DOI": "10.1128/mbio.02287-23",
"doi-asserted-by": "crossref",
"key": "ref_47",
"unstructured": "Zeng, Q., Antia, A., Casorla-Perez, L.A., Puray-Chavez, M., Kutluay, S.B., Ciorba, M.A., and Ding, S. (2024). Calpain-2 mediates SARS-CoV-2 entry via regulating ACE2 levels. mBio, 15."
},
{
"DOI": "10.1038/s41419-023-05817-w",
"article-title": "Perturbation of the host cell Ca2+ homeostasis and ER-mitochondria contact sites by the SARS-CoV-2 structural proteins E and M",
"author": "Poggio",
"doi-asserted-by": "crossref",
"first-page": "297",
"journal-title": "Cell Death Dis.",
"key": "ref_48",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.1016/j.antiviral.2022.105475",
"article-title": "Inhibition of p38 signaling curtails the SARS-CoV-2 induced inflammatory response but retains the IFN-dependent antiviral defense of the lung epithelial barrier",
"author": "Faist",
"doi-asserted-by": "crossref",
"first-page": "105475",
"journal-title": "Antivir. Res.",
"key": "ref_49",
"volume": "209",
"year": "2023"
},
{
"DOI": "10.1073/pnas.2113401118",
"article-title": "Targeting the coronavirus nucleocapsid protein through GSK-3 inhibition",
"author": "Liu",
"doi-asserted-by": "crossref",
"first-page": "e2113401118",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_50",
"volume": "118",
"year": "2021"
},
{
"DOI": "10.1038/s41467-021-22785-x",
"article-title": "The architecture of the SARS-CoV-2 RNA genome inside virion",
"author": "Cao",
"doi-asserted-by": "crossref",
"first-page": "3917",
"journal-title": "Nat. Commun.",
"key": "ref_51",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1038/s41467-022-28603-2",
"article-title": "Secondary structural ensembles of the SARS-CoV-2 RNA genome in infected cells",
"author": "Lan",
"doi-asserted-by": "crossref",
"first-page": "1128",
"journal-title": "Nat. Commun.",
"key": "ref_52",
"volume": "13",
"year": "2022"
}
],
"reference-count": 52,
"references-count": 52,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.mdpi.com/2073-4425/16/10/1239"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "SARS-CoV-2 Infection of Lung Epithelia Leads to an Increase in the Cleavage and Translocation of RNase-III Drosha; Loss of Drosha Is Associated with a Decrease in Viral Replication",
"type": "journal-article",
"update-policy": "https://doi.org/10.3390/mdpi_crossmark_policy",
"volume": "16"
}