In Silico Analysis of Probiotic Bacteria Changes Across COVID-19 Severity Stages
et al., Microorganisms, doi:10.3390/microorganisms12112353, Nov 2024
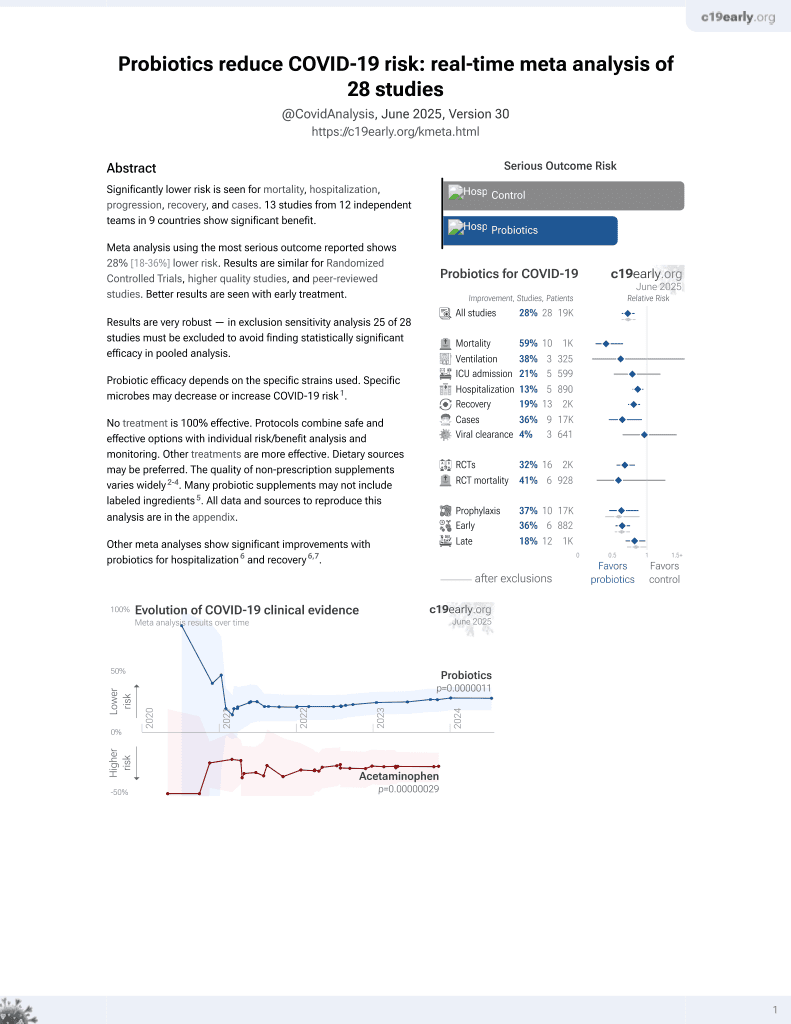
Probiotics for COVID-19
20th treatment shown to reduce risk in
March 2021, now with p = 0.00000044 from 29 studies.
No treatment is 100% effective. Protocols
combine treatments.
6,400+ studies for
210+ treatments. c19early.org
|
Reanalysis of publicly available microbiome datasets from 7 studies totaling 581 COVID-19 patients and controls, showing significant differential abundance of beneficial bacterial genera, particularly Bifidobacterium and Bacteroides, across disease severity stages. The consistent presence of Bifidobacterium in milder cases suggests a potential protective role influencing immune modulation. Bacteroides showed a complex pattern, with increased abundance in moderate cases but lower in mild and severe, possibly related to inflammation. The findings emphasize the potential of specific bacterial genera as biomarkers of COVID-19 severity and therapeutic targets.
Probiotic efficacy depends on the specific strains used. Specific microbes may decrease or increase COVID-19 risk1.
Taufer et al., 18 Nov 2024, peer-reviewed, 3 authors.
Contact: prampelotto@hcpa.edu.br (corresponding author), clarissa.taufer@gmail.com, juliana.silva@unilasalle.edu.br.
In Silico Analysis of Probiotic Bacteria Changes Across COVID-19 Severity Stages
Microorganisms, doi:10.3390/microorganisms12112353
The gut microbiota plays a crucial role in modulating the immune response during COVID-19, with several studies reporting significant alterations in specific bacterial genera, including Akkermansia, Bacteroides, Bifidobacterium, Faecalibacterium, Lactobacillus, Oscillospira, and Ruminococcus. These genera are symbionts of the gut microbiota and contribute to host health. However, comparing results across studies is challenging due to differences in analysis methods and reference databases. We screened 16S rRNA raw datasets available in public databases on COVID-19, focusing on the V3-V4 region of the bacterial genome. In total, seven studies were included. All samples underwent the same bioinformatics pipeline, evaluating the differential abundance of these seven bacterial genera at each level of severity. The reanalysis identified significant changes in differential abundance. Bifidobacterium emerged as a potential biomarker of disease severity and a therapeutic target. Bacteroides presented a complex pattern, possibly related to disease-associated inflammation or opportunistic pathogen growth. Lactobacillus showed significant changes in abundance across the COVID-19 stages. On the other hand, Akkermansia and Faecalibacterium did not show significant differences, while Oscillospira and Ruminococcus produced statistically significant results but with limited relevance to COVID-19 severity. Our findings reveal new insights into the differential abundance of key bacterial genera in COVID-19, particularly Bifidobacterium and Bacteroides.
Conflicts of Interest: The authors declare no conflicts of interest.
References
Abellan-Schneyder, Matchado, Reitmeier, Sommer, Sewald et al., Pipelines, Parameters: Issues in 16S rRNA Gene Sequencing, mSphere, doi:10.1128/mSphere.01202-20
Al Kassaa, Hober, Hamze, Chihib, Drider, Antiviral potential of lactic acid bacteria and their bacteriocins, Probiotics Antimicrob. Proteins, doi:10.1007/s12602-014-9162-6
Albrich, Ghosh, Ahearn-Ford, Mikaeloff, Lunjani et al., A high-risk gut microbiota configuration associates with fatal hyperinflammatory immune and metabolic responses to SARS-CoV-2, Gut Microbes, doi:10.1080/19490976.2022.2073131
Asahara, Shimizu, Nomoto, Watanuki, Tanaka, Antibacterial effect of fermented milk containing Bifidobacterium breve, Bifidobacterium bifidum and Lactobacillus acidophilus against indigenous Escherichia coli infection in mice, Microb. Ecol. Health Dis, doi:10.1080/089106001750071663
Balvočiute, Huson, Silva, Rdp, NCBI and OTT-How do these taxonomies compare?, BMC Genom, doi:10.1186/s12864-017-3501-4
Bataineh, Henschel, Mousa, Daou, Waasia et al., Gut Microbiota Interplay With COVID-19 Reveals Links to Host Lipid Metabolism Among Middle Eastern Populations, Front. Microbiol, doi:10.3389/fmicb.2021.761067
Behrens, Gasparotto, Rampelotto, Escalona, Da Silva et al., Sodium propionate oral supplementation ameliorates depressive-like behavior through gut microbiome and histone 3 epigenetic regulation, J. Nutr. Biochem, doi:10.1016/j.jnutbio.2024.109660
Boge, Rémigy, Vaudaine, Tanguy, Bourdet-Sicard et al., A probiotic fermented dairy drink improves antibody response to influenza vaccination in the elderly in two randomised controlled trials, Vaccine, doi:10.1016/j.vaccine.2009.06.094
Breitwieser, Lu, Salzberg, A review of methods and databases for metagenomic classification and assembly, Brief. Bioinform, doi:10.1093/bib/bbx120
Brook, The role of anaerobic bacteria in bacteremia, Anaerobe, doi:10.1016/j.anaerobe.2009.12.001
Bucci, Ward, Bhattarai, Rojas-Correa, Purkayastha et al., The intestinal microbiota predicts COVID-19 severity and fatality regardless of hospital feeding method, mSystems, doi:10.1128/msystems.00310-23
Castillo, Perdigán, De Moreno De Leblanc, Oral administration of a probiotic Lactobacillus modulates cytokine production and TLR expression improving the immune response against Salmonella enterica serovar Typhimurium infection in mice, BMC Microbiol, doi:10.1186/1471-2180-11-177
Cheng, Luo, Wang, Zhang, Wang et al., Kidney disease is associated with in-hospital death of patients with COVID-19, Kidney Int, doi:10.1016/j.kint.2020.03.005
Cheng, Zhang, Li, Wu, Wu et al., Meta-analysis of 16S rRNA microbial data identified alterations of the gut microbiota in COVID-19 patients during the acute and recovery phases, BMC Microbiol, doi:10.1186/s12866-022-02686-9
Chollet, Heumel, Deruyter, Bouilloux, Delval et al., Faecalibacterium duncaniae as a novel next generation probiotic against influenza, Front. Immunol, doi:10.3389/fimmu.2024.1347676
Comin, Di Camillo, Pizzi, Vandin, Comparison of microbiome samples: Methods and computational challenges, Brief. Bioinform, doi:10.1093/bib/bbaa121
Costea, Zeller, Sunagawa, Pelletier, Alberti et al., Towards standards for human fecal sample processing in metagenomic studies, Nat. Biotechnol, doi:10.1038/nbt.3960
Crost, Coletto, Bell, Juge, Ruminococcus gnavus: Friend or foe for human health, FEMS Microbiol. Rev, doi:10.1093/femsre/fuad014
Dhar, Mohanty, Gut microbiota and COVID-19-possible link and implications, Virus Res, doi:10.1016/j.virusres.2020.198018
Everard, Belzer, Geurts, Ouwerkerk, Druart et al., Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity, Proc. Natl. Acad. Sci, doi:10.1073/pnas.1219451110
Fujimura, Slusher, Cabana, Lynch, V Role of the gut microbiota in defining human health, Expert Rev. Anti. Infect. Ther, doi:10.1586/eri.10.14
Gaibani, D'amico, Bartoletti, Lombardo, Rampelli et al., The gut microbiota of critically Ill patients with COVID-19, Front. Cell. Infect. Microbiol, doi:10.3389/fcimb.2021.670424
Galperine, Choi, Pagani, Kritikos, Papadimitriou-Olivgeris et al., Temporal changes in fecal microbiota of patients infected with COVID-19: A longitudinal cohort, BMC Infect. Dis, doi:10.1186/s12879-023-08511-6
Gophna, Konikoff, Nielsen, Oscillospira and related bacteria-From metagenomic species to metabolic features, Environ. Microbiol, doi:10.1111/1462-2920.13658
Gou, Fu, Yue, Chen, Cai et al., Gut microbiota, inflammation, and molecular signatures of host response to infection, J. Genet. Genom, doi:10.1016/j.jgg.2021.04.002
Gozalbo-Rovira, Rubio-Del-Campo, Santiso-Bellón, Vila-Vicent, Buesa et al., Interaction of intestinal bacteria with human rotavirus during infection in children, Int. J. Mol. Sci, doi:10.3390/ijms22031010
Gu, Chen, Wu, Chen, Gao et al., Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 Influenza, Clin. Infect. Dis, doi:10.1093/cid/ciaa709
Guan, Ni, Hu, Liang, Ou et al., Clinical Characteristics of Coronavirus Disease 2019 in China, N. Engl. J. Med, doi:10.1056/NEJMoa2002032
Gumenyuk, Golod, Silaeva, Sorokina, Ilyasov et al., Gut Microbiota Alterations and Their Relationship to the Disease Severity and Some Cytokine Profile Indicators in Patients with COVID-19, Bull. Russ. State Med. Univ, doi:10.24075/brsmu.2022.006
Hazan, Stollman, Bozkurt, Dave, Papoutsis et al., Lost microbes of COVID-19: Bifidobacterium, Faecalibacterium depletion and decreased microbiome diversity associated with SARS-CoV-2 infection severity, BMJ Open Gastroenterol, doi:10.1136/bmjgast-2022-000871
He, Zhao, Li, Faecalibacterium prausnitzii: A Next-Generation Probiotic in Gut Disease Improvement, Can. J. Infect. Dis. Med. Microbiol, doi:10.1155/2021/6666114
Heumel, De Rezende Rodovalho, Urien, Specque, Brito Rodrigues et al., Shotgun metagenomics and systemic targeted metabolomics highlight indole-3-propionic acid as a protective gut microbial metabolite against influenza infection, Gut Microbes, doi:10.1080/19490976.2024.2325067
Hu, Zhao, Yang, Gong, Sun et al., Akkermansia muciniphila Improves Host Defense Against Influenza Virus Infection, Front. Microbiol, doi:10.3389/fmicb.2020.586476
Islam, Foysal, Hoque, Mehedi, Rob et al., Dysbiosis of Oral and Gut Microbiomes in SARS-CoV-2 Infected Patients in Bangladesh: Elucidating the Role of Opportunistic Gut Microbes, Front. Med, doi:10.3389/fmed.2022.821777
Kandasamy, Letchumanan, Hong, Chua, Mutalib et al., The Role of Human Gut Microbe Ruminococcus gnavus in Inflammatory Diseases, Prog. Microbes Mol. Biol, doi:10.36877/pmmb.a0000396
Kawahara, Takahashi, Oishi, Tanaka, Masuda et al., Consecutive oral administration of Bifidobacterium longum MM-2 improves the defense system against influenza virus infection by enhancing natural killer cell activity in a murine model, Microbiol. Immunol, doi:10.1111/1348-0421.12210
Konikoff, Gophna, Oscillospira: A Central, Enigmatic Component of the Human Gut Microbiota, Trends Microbiol, doi:10.1016/j.tim.2016.02.015
Labarta-Bajo, Gramalla-Schmitz, Gerner, Kazane, Humphrey et al., CD8 T cells drive anorexia, dysbiosis, and blooms of a commensal with immunosuppressive potential after viral infection, Proc. Natl. Acad. Sci, doi:10.1073/pnas.2003656117
Lange, Rampelotto, Longo, De Freitas, Uribe-Cruz et al., Ornithine aspartate effects on bacterial composition and metabolic pathways in a rat model of steatotic liver disease, World J. Hepatol, doi:10.4254/wjh.v16.i5.832
Li, Ghosh, Mccann, Mallon, Hill et al., Robust cross-cohort gut microbiome associations with COVID-19 severity, Gut Microbes, doi:10.1080/19490976.2023.2242615
Li, Xu, Xu, Tang, Jiang et al., Faecalibacterium prausnitzii Attenuates CKD via Butyrate-Renal GPR43 Axis, Circ. Res, doi:10.1161/CIRCRESAHA.122.320184
Li, Yang, Zhou, Disoma, Dong et al., Microbiome Profiling Using Shotgun Metagenomic Sequencing Identified Unique Microorganisms in COVID-19 Patients with Altered Gut Microbiota, Front. Microbiol, doi:10.3389/fmicb.2021.712081
Ling, Linglong, Weixia, Hong, Protective effects of bifidobacterium on intestinal barrier function in LPS-induced enterocyte barrier injury of Caco-2 monolayers and in a rat NEC model, PLoS ONE, doi:10.1371/journal.pone.0161635
Machado, Oliveira, Lelis, De Paula, Guimarães et al., Oral Probiotic Bifidobacterium longum Supplementation Improves Metabolic Parameters and Alters the Expression of the Renin-Angiotensin System in Obese Mice Liver, Biol. Res. Nurs, doi:10.1177/1099800420942942
Maddah, Goodarzi, Asadi-Yousefabad, Abbasluo, Shariati et al., Evaluation of the gut microbiome associated with COVID-19, Inform. Med. Unlocked, doi:10.1016/j.imu.2023.101239
Maeda, Motooka, Kawasaki, Oki, Noda et al., Longitudinal alterations of the gut mycobiota and microbiota on COVID-19 severity, BMC Infect. Dis, doi:10.1186/s12879-022-07358-7
Mattar, Teitelbaum, Drongowski, Yongyi, Harmon et al., Probiotics up-regulate MUC-2 mucin gene expression in a Caco-2 cell-culture model, Pediatr. Surg. Int, doi:10.1007/s00383-002-0855-7
Maubert, Michon, Chain, Marquant, Miquel et al., Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn's disease, Gut
Mazmanian, Round, Kasper, A microbial symbiosis factor prevents intestinal inflammatory disease, Nature, doi:10.1038/nature07008
Mena Canata, Benfato, Pereira, Ramos Pereira, Hackenhaar et al., Comparative Analysis of the Gut Microbiota of Bat Species with Different Feeding Habits, Biology, doi:10.3390/biology13060363
Niu, Cui, Yang, Li, Yao et al., Microbiota-derived acetate enhances host antiviral response via NLRP3, Nat. Commun, doi:10.1038/s41467-023-36323-4
Ottman, Reunanen, Meijerink, Pietila, Kainulainen et al., Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function, PLoS ONE, doi:10.1371/journal.pone.0173004
Pezzini, Rampelotto, Dall'agnol, Guerreiro, Longo et al., Changes in the gut microbiota of rats after exposure to the fungicide Mancozeb, Toxicol. Appl. Pharmacol, doi:10.1016/j.taap.2023.116480
Qin, Li, Raes, Arumugam, Burgdorf et al., A human gut microbial gene catalogue established by metagenomic sequencing, Nature, doi:10.1038/nature08821
Quast, Pruesse, Yilmaz, Gerken, Schweer et al., The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools, Nucleic Acids Res, doi:10.1093/nar/gks1219
Ramakrishna, Kujawski, Chu, Li, Mazmanian et al., Bacteroides fragilis polysaccharide A induces IL-10 secreting B and T cells that prevent viral encephalitis, Nat. Commun, doi:10.1038/s41467-019-09884-6
Reau, Suen, The Ruminococci: Key symbionts of the gut ecosystem, J. Microbiol, doi:10.1007/s12275-018-8024-4
Reinold, Farahpour, Fehring, Dolff, Konik et al., A Pro-Inflammatory Gut Microbiome Characterizes SARS-CoV-2 Infected Patients and a Reduction in the Connectivity of an Anti-Inflammatory Bacterial Network Associates with Severe COVID-19, Front. Cell. Infect. Microbiol, doi:10.3389/fcimb.2021.747816
Reuben, Beugnon, Jurburg, COVID-19 alters human microbiomes: A meta-analysis, Front. Cell. Infect. Microbiol, doi:10.3389/fcimb.2023.1211348
Rocchi, Giovanetti, Benedetti, Borsetti, Ceccarelli et al., Gut Microbiota and COVID-19: Potential Implications for Disease Severity, Pathogens, doi:10.3390/pathogens11091050
Rodríguez-Díaz, García-Mantrana, Vila-Vicent, Gozalbo-Rovira, Buesa et al., Relevance of secretor status genotype and microbiota composition in susceptibility to rotavirus and norovirus infections in humans, Sci. Rep, doi:10.1038/srep45559
Rognes, Flouri, Nichols, Quince, Mahé et al., A versatile open source tool for metagenomics, PeerJ, doi:10.7717/peerj.2584
Rooks, Garrett, Gut microbiota, metabolites and host immunity, Nat. Rev. Immunol, doi:10.1038/nri.2016.42
Rueca, Fontana, Bartolini, Piselli, Mazzarelli et al., Investigation of Nasal/Oropharyngeal Microbial Community of COVID-19 Patients by 16S rDNA Sequencing, Int. J. Environ. Res. Public Health, doi:10.3390/ijerph18042174
Schloss, Westcott, Ryabin, Hall, Hartmann et al., Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities, Appl. Environ. Microbiol, doi:10.1128/AEM.01541-09
Segata, Izard, Waldron, Gevers, Miropolsky et al., Metagenomic biomarker discovery and explanation, Genome Biol, doi:10.1186/gb-2011-12-6-r60
Shane, Mody, Crump, Tarr, Steiner et al., Infectious Diseases Society of America Clinical Practice Guidelines for the Diagnosis and Management of Infectious Diarrhea, Clin. Infect. Dis, doi:10.1093/cid/cix669
Shin, Lee, Lee, Kim, Whon et al., An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice, Gut, doi:10.1136/gutjnl-2012-303839
Sinha, Abu-Ali, Vogtmann, Fodor, Ren et al., Assessment of variation in microbial community amplicon sequencing by the Microbiome Quality Control (MBQC) project consortium, Nat. Biotechnol, doi:10.1038/nbt.3981
Stefan, Kim, Iwasaki, Kasper, Commensal Microbiota Modulation of Natural Resistance to Virus Infection, Cell, doi:10.1016/j.cell.2020.10.047
Szajewska, Ruszczy Ński, Radzikowski, Probiotics in the prevention of antibiotic-associated diarrhea in children: A meta-analysis of randomized controlled trials, J. Pediatr, doi:10.1016/j.jpeds.2006.04.053
Talukdar, Bandopadhyay, Ray, Paul, Sarif et al., Association of gut microbial dysbiosis with disease severity, response to therapy and disease outcomes in Indian patients with COVID-19, Gut Pathog, doi:10.1186/s13099-023-00546-z
Turroni, Taverniti, Ruas-Madiedo, Duranti, Guglielmetti et al., Bifidobacterium bifidum PRL2010 Modulates the Host Innate Immune Response, Appl. Environ. Microbiol, doi:10.1128/AEM.03313-13
Wang, Lkhagva, Kim, Zhai, Islam et al., The gut microbe pair of Oribacterium sp. GMB0313 and Ruminococcus sp. GMB0270 confers complete protection against SARS-CoV-2 infection by activating CD8+ T cell-mediated immunity, Gut Microbes, doi:10.1080/19490976.2024.2342497
Wexler, Bacteroides: The good, the bad, and the nitty-gritty, Clin. Microbiol. Rev, doi:10.1128/CMR.00008-07
Wu, Cheng, Jiang, Tang, Ming et al., Altered oral and gut microbiota and its association with SARS-CoV-2 viral load in COVID-19 patients during hospitalization, NPJ Biofilms Microbiomes, doi:10.1038/s41522-021-00232-5
Xia, Gu, Guo, Feng, Chen et al., Gut Microbiota Mediates the Preventive Effects of Dietary Capsaicin Against Depression-Like Behavior Induced by Lipopolysaccharide in Mice, Front. Cell. Infect. Microbiol, doi:10.3389/fcimb.2021.627608
Xu, Chen, Tang, Guidelines for the diagnosis and treatment of coronavirus disease 2019 (COVID-19) in China, Glob. Health Med, doi:10.35772/ghm.2020.01015
Xu, Hao, Zai, Song, Huang et al., A new perspective on gut-lung axis affected through resident microbiome and their implications on immune response in respiratory diseases, Arch. Microbiol, doi:10.1007/s00203-024-03843-6
Xu, Zhang, Guo, Xiao, Fu et al., Integrated analysis of gut microbiome and host immune responses in COVID-19, Front. Med, doi:10.1007/s11684-022-0921-6
Yeoh, Zuo, Lui, Zhang, Liu et al., Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19, Gut, doi:10.1136/gutjnl-2020-323020
Yoda, Miyazawa, Hosoda, Hiramatsu, Yan et al., Lactobacillus GG-fermented milk prevents DSS-induced colitis and regulates intestinal epithelial homeostasis through activation of epidermal growth factor receptor, Eur. J. Nutr, doi:10.1007/s00394-013-0506-x
Zhai, Feng, Arjan, Chen, A next generation probiotic, Akkermansia muciniphila, Crit. Rev. Food Sci. Nutr, doi:10.1080/10408398.2018.1517725
Zhang, Hu, Feng, Hu, Wang et al., Influenza infection elicits an expansion of gut population of endogenous Bifidobacterium animalis which protects mice against infection, Genome Biol, doi:10.1186/s13059-020-02007-1
Zhang, Zhu, Xu, Liu, Qiu et al., Bacteroides fragilis protects against antibiotic-associated Diarrhea in rats by modulating intestinal defenses, Front. Immunol, doi:10.3389/fimmu.2018.01040
Zheng, Wittouck, Salvetti, Franz, Harris et al., A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae, Int. J. Syst. Evol. Microbiol, doi:10.1099/ijsem.0.004107
Zuo, Zhang, Lui, Yeoh, Li et al., Alterations in gut microbiota of patients with COVID-19 during time of hospitalization, Gastroenterology, doi:10.1053/j.gastro.2020.05.048
DOI record:
{
"DOI": "10.3390/microorganisms12112353",
"ISSN": [
"2076-2607"
],
"URL": "http://dx.doi.org/10.3390/microorganisms12112353",
"abstract": "<jats:p>The gut microbiota plays a crucial role in modulating the immune response during COVID-19, with several studies reporting significant alterations in specific bacterial genera, including Akkermansia, Bacteroides, Bifidobacterium, Faecalibacterium, Lactobacillus, Oscillospira, and Ruminococcus. These genera are symbionts of the gut microbiota and contribute to host health. However, comparing results across studies is challenging due to differences in analysis methods and reference databases. We screened 16S rRNA raw datasets available in public databases on COVID-19, focusing on the V3–V4 region of the bacterial genome. In total, seven studies were included. All samples underwent the same bioinformatics pipeline, evaluating the differential abundance of these seven bacterial genera at each level of severity. The reanalysis identified significant changes in differential abundance. Bifidobacterium emerged as a potential biomarker of disease severity and a therapeutic target. Bacteroides presented a complex pattern, possibly related to disease-associated inflammation or opportunistic pathogen growth. Lactobacillus showed significant changes in abundance across the COVID-19 stages. On the other hand, Akkermansia and Faecalibacterium did not show significant differences, while Oscillospira and Ruminococcus produced statistically significant results but with limited relevance to COVID-19 severity. Our findings reveal new insights into the differential abundance of key bacterial genera in COVID-19, particularly Bifidobacterium and Bacteroides.</jats:p>",
"alternative-id": [
"microorganisms12112353"
],
"author": [
{
"affiliation": [
{
"name": "Graduate Program in Genetics and Molecular Biology, Universidade Federal do Rio Grande do Sul, Porto Alegre 91501-970, Brazil"
}
],
"family": "Taufer",
"given": "Clarissa Reginato",
"sequence": "first"
},
{
"ORCID": "http://orcid.org/0000-0002-1089-6766",
"affiliation": [
{
"name": "Graduate Program in Genetics and Molecular Biology, Universidade Federal do Rio Grande do Sul, Porto Alegre 91501-970, Brazil"
},
{
"name": "Graduate Program in Health and Human Development, Universidade La Salle, Canoas 92010-000, Brazil"
}
],
"authenticated-orcid": false,
"family": "da Silva",
"given": "Juliana",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0002-8992-9697",
"affiliation": [
{
"name": "Bioinformatics and Biostatistics Core Facility, Instituto de Ciências Básicas da Saúde, Universidade Federal do Rio Grande do Sul, Porto Alegre 91501-970, Brazil"
}
],
"authenticated-orcid": false,
"family": "Rampelotto",
"given": "Pabulo Henrique",
"sequence": "additional"
}
],
"container-title": "Microorganisms",
"container-title-short": "Microorganisms",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2024,
11,
18
]
],
"date-time": "2024-11-18T12:45:27Z",
"timestamp": 1731933927000
},
"deposited": {
"date-parts": [
[
2024,
11,
18
]
],
"date-time": "2024-11-18T13:26:12Z",
"timestamp": 1731936372000
},
"funder": [
{
"DOI": "10.13039/501100002322",
"award": [
"88887.513461/2020-00",
"88887.798411/2022-00"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100002322",
"id-type": "DOI"
}
],
"name": "CAPES"
}
],
"indexed": {
"date-parts": [
[
2024,
11,
19
]
],
"date-time": "2024-11-19T05:24:21Z",
"timestamp": 1731993861385,
"version": "3.28.0"
},
"is-referenced-by-count": 0,
"issue": "11",
"issued": {
"date-parts": [
[
2024,
11,
18
]
]
},
"journal-issue": {
"issue": "11",
"published-online": {
"date-parts": [
[
2024,
11
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2024,
11,
18
]
],
"date-time": "2024-11-18T00:00:00Z",
"timestamp": 1731888000000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/2076-2607/12/11/2353/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "2353",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2024,
11,
18
]
]
},
"published-online": {
"date-parts": [
[
2024,
11,
18
]
]
},
"publisher": "MDPI AG",
"reference": [
{
"key": "ref_1",
"unstructured": "World Health Organization (2023). Clinical Management of COVID-19: Living Guideline."
},
{
"DOI": "10.1056/NEJMoa2002032",
"article-title": "Clinical Characteristics of Coronavirus Disease 2019 in China",
"author": "Guan",
"doi-asserted-by": "crossref",
"first-page": "1708",
"journal-title": "N. Engl. J. Med.",
"key": "ref_2",
"volume": "382",
"year": "2020"
},
{
"DOI": "10.1016/j.kint.2020.03.005",
"article-title": "Kidney disease is associated with in-hospital death of patients with COVID-19",
"author": "Cheng",
"doi-asserted-by": "crossref",
"first-page": "829",
"journal-title": "Kidney Int.",
"key": "ref_3",
"volume": "97",
"year": "2020"
},
{
"DOI": "10.1053/j.gastro.2020.05.048",
"article-title": "Alterations in gut microbiota of patients with COVID-19 during time of hospitalization",
"author": "Zuo",
"doi-asserted-by": "crossref",
"first-page": "944",
"journal-title": "Gastroenterology",
"key": "ref_4",
"volume": "159",
"year": "2020"
},
{
"DOI": "10.1016/j.jgg.2021.04.002",
"article-title": "Gut microbiota, inflammation, and molecular signatures of host response to infection",
"author": "Gou",
"doi-asserted-by": "crossref",
"first-page": "792",
"journal-title": "J. Genet. Genom.",
"key": "ref_5",
"volume": "48",
"year": "2021"
},
{
"DOI": "10.1136/gutjnl-2020-323020",
"article-title": "Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19",
"author": "Yeoh",
"doi-asserted-by": "crossref",
"first-page": "698",
"journal-title": "Gut",
"key": "ref_6",
"volume": "70",
"year": "2021"
},
{
"DOI": "10.1093/cid/ciaa709",
"article-title": "Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 Influenza",
"author": "Gu",
"doi-asserted-by": "crossref",
"first-page": "2669",
"journal-title": "Clin. Infect. Dis.",
"key": "ref_7",
"volume": "71",
"year": "2020"
},
{
"DOI": "10.3389/fmicb.2021.761067",
"doi-asserted-by": "crossref",
"key": "ref_8",
"unstructured": "Al Bataineh, M.T., Henschel, A., Mousa, M., Daou, M., Waasia, F., Kannout, H., Khalili, M., Kayasseh, M.A., Alkhajeh, A., and Uddin, M. (2021). Gut Microbiota Interplay With COVID-19 Reveals Links to Host Lipid Metabolism Among Middle Eastern Populations. Front. Microbiol., 12."
},
{
"DOI": "10.1128/msystems.00310-23",
"article-title": "The intestinal microbiota predicts COVID-19 severity and fatality regardless of hospital feeding method",
"author": "Bucci",
"doi-asserted-by": "crossref",
"first-page": "e00310-23",
"journal-title": "mSystems",
"key": "ref_9",
"volume": "8",
"year": "2023"
},
{
"DOI": "10.3390/ijerph18042174",
"doi-asserted-by": "crossref",
"key": "ref_10",
"unstructured": "Rueca, M., Fontana, A., Bartolini, B., Piselli, P., Mazzarelli, A., Copetti, M., Binda, E., Perri, F., Gruber, C.E.M., and Nicastri, E. (2021). Investigation of Nasal/Oropharyngeal Microbial Community of COVID-19 Patients by 16S rDNA Sequencing. Int. J. Environ. Res. Public Health, 18."
},
{
"DOI": "10.1136/bmjgast-2022-000871",
"article-title": "Lost microbes of COVID-19: Bifidobacterium, Faecalibacterium depletion and decreased microbiome diversity associated with SARS-CoV-2 infection severity",
"author": "Hazan",
"doi-asserted-by": "crossref",
"first-page": "e000871",
"journal-title": "BMJ Open Gastroenterol.",
"key": "ref_11",
"volume": "9",
"year": "2022"
},
{
"DOI": "10.1186/s12879-022-07358-7",
"doi-asserted-by": "crossref",
"key": "ref_12",
"unstructured": "Maeda, Y., Motooka, D., Kawasaki, T., Oki, H., Noda, Y., Adachi, Y., Niitsu, T., Okamoto, S., Tanaka, K., and Fukushima, K. (2022). Longitudinal alterations of the gut mycobiota and microbiota on COVID-19 severity. BMC Infect. Dis., 22."
},
{
"DOI": "10.1080/10408398.2018.1517725",
"article-title": "A next generation probiotic, Akkermansia muciniphila",
"author": "Zhai",
"doi-asserted-by": "crossref",
"first-page": "3227",
"journal-title": "Crit. Rev. Food Sci. Nutr.",
"key": "ref_13",
"volume": "59",
"year": "2019"
},
{
"DOI": "10.1371/journal.pone.0173004",
"doi-asserted-by": "crossref",
"key": "ref_14",
"unstructured": "Ottman, N., Reunanen, J., Meijerink, M., Pietila, T.E., Kainulainen, V., Klievink, J., Huuskonen, L., Aalvink, S., Skurnik, M., and Boeren, S. (2017). Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS ONE, 12."
},
{
"DOI": "10.1073/pnas.1219451110",
"article-title": "Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity",
"author": "Everard",
"doi-asserted-by": "crossref",
"first-page": "9066",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_15",
"volume": "110",
"year": "2013"
},
{
"DOI": "10.1136/gutjnl-2012-303839",
"article-title": "An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice",
"author": "Shin",
"doi-asserted-by": "crossref",
"first-page": "727",
"journal-title": "Gut",
"key": "ref_16",
"volume": "63",
"year": "2014"
},
{
"DOI": "10.3389/fmicb.2020.586476",
"doi-asserted-by": "crossref",
"key": "ref_17",
"unstructured": "Hu, X., Zhao, Y., Yang, Y., Gong, W., Sun, X., Yang, L., Zhang, Q., and Jin, M. (2021). Akkermansia muciniphila Improves Host Defense Against Influenza Virus Infection. Front. Microbiol., 11."
},
{
"DOI": "10.1038/s41467-023-36323-4",
"article-title": "Microbiota-derived acetate enhances host antiviral response via NLRP3",
"author": "Niu",
"doi-asserted-by": "crossref",
"first-page": "642",
"journal-title": "Nat. Commun.",
"key": "ref_18",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.3389/fimmu.2018.01040",
"doi-asserted-by": "crossref",
"key": "ref_19",
"unstructured": "Zhang, W., Zhu, B., Xu, J., Liu, Y., Qiu, E., Li, Z., Li, Z., He, Y., Zhou, H., and Bai, Y. (2018). Bacteroides fragilis protects against antibiotic-associated Diarrhea in rats by modulating intestinal defenses. Front. Immunol., 9."
},
{
"DOI": "10.1016/j.cell.2020.10.047",
"article-title": "Commensal Microbiota Modulation of Natural Resistance to Virus Infection",
"author": "Stefan",
"doi-asserted-by": "crossref",
"first-page": "1312",
"journal-title": "Cell",
"key": "ref_20",
"volume": "183",
"year": "2020"
},
{
"DOI": "10.1038/nature07008",
"article-title": "A microbial symbiosis factor prevents intestinal inflammatory disease",
"author": "Mazmanian",
"doi-asserted-by": "crossref",
"first-page": "620",
"journal-title": "Nature",
"key": "ref_21",
"volume": "453",
"year": "2008"
},
{
"DOI": "10.1038/s41467-019-09884-6",
"article-title": "Bacteroides fragilis polysaccharide A induces IL-10 secreting B and T cells that prevent viral encephalitis",
"author": "Ramakrishna",
"doi-asserted-by": "crossref",
"first-page": "2153",
"journal-title": "Nat. Commun.",
"key": "ref_22",
"volume": "10",
"year": "2019"
},
{
"DOI": "10.1038/nri.2016.42",
"article-title": "Gut microbiota, metabolites and host immunity",
"author": "Rooks",
"doi-asserted-by": "crossref",
"first-page": "341",
"journal-title": "Nat. Rev. Immunol.",
"key": "ref_23",
"volume": "16",
"year": "2016"
},
{
"DOI": "10.1586/eri.10.14",
"article-title": "V Role of the gut microbiota in defining human health",
"author": "Fujimura",
"doi-asserted-by": "crossref",
"first-page": "435",
"journal-title": "Expert Rev. Anti. Infect. Ther.",
"key": "ref_24",
"volume": "8",
"year": "2010"
},
{
"DOI": "10.1111/1348-0421.12210",
"article-title": "Consecutive oral administration of Bifidobacterium longum MM-2 improves the defense system against influenza virus infection by enhancing natural killer cell activity in a murine model",
"author": "Kawahara",
"doi-asserted-by": "crossref",
"first-page": "1",
"journal-title": "Microbiol. Immunol.",
"key": "ref_25",
"volume": "59",
"year": "2015"
},
{
"article-title": "Antibacterial effect of fermented milk containing Bifidobacterium breve, Bifidobacterium bifidum and Lactobacillus acidophilus against indigenous Escherichia coli infection in mice",
"author": "Asahara",
"first-page": "16",
"journal-title": "Microb. Ecol. Health Dis.",
"key": "ref_26",
"volume": "13",
"year": "2001"
},
{
"DOI": "10.3389/fimmu.2024.1347676",
"doi-asserted-by": "crossref",
"key": "ref_27",
"unstructured": "Chollet, L., Heumel, S., Deruyter, L., Bouilloux, F., Delval, L., Robert, V., Gevaert, M.H., Pichavant, M., Sencio, V., and Robil, C. (2024). Faecalibacterium duncaniae as a novel next generation probiotic against influenza. Front. Immunol., 15."
},
{
"DOI": "10.1016/j.jpeds.2006.04.053",
"article-title": "Probiotics in the prevention of antibiotic-associated diarrhea in children: A meta-analysis of randomized controlled trials",
"author": "Szajewska",
"doi-asserted-by": "crossref",
"first-page": "E38",
"journal-title": "J. Pediatr.",
"key": "ref_28",
"volume": "149",
"year": "2006"
},
{
"DOI": "10.1093/cid/cix669",
"article-title": "2017 Infectious Diseases Society of America Clinical Practice Guidelines for the Diagnosis and Management of Infectious Diarrhea",
"author": "Shane",
"doi-asserted-by": "crossref",
"first-page": "e45",
"journal-title": "Clin. Infect. Dis.",
"key": "ref_29",
"volume": "65",
"year": "2017"
},
{
"DOI": "10.1007/s00394-013-0506-x",
"article-title": "Lactobacillus GG-fermented milk prevents DSS-induced colitis and regulates intestinal epithelial homeostasis through activation of epidermal growth factor receptor",
"author": "Yoda",
"doi-asserted-by": "crossref",
"first-page": "105",
"journal-title": "Eur. J. Nutr.",
"key": "ref_30",
"volume": "53",
"year": "2014"
},
{
"DOI": "10.1016/j.vaccine.2009.06.094",
"article-title": "A probiotic fermented dairy drink improves antibody response to influenza vaccination in the elderly in two randomised controlled trials",
"author": "Boge",
"doi-asserted-by": "crossref",
"first-page": "5677",
"journal-title": "Vaccine",
"key": "ref_31",
"volume": "27",
"year": "2009"
},
{
"DOI": "10.1111/1462-2920.13658",
"article-title": "Oscillospira and related bacteria—From metagenomic species to metabolic features",
"author": "Gophna",
"doi-asserted-by": "crossref",
"first-page": "835",
"journal-title": "Environ. Microbiol.",
"key": "ref_32",
"volume": "19",
"year": "2017"
},
{
"DOI": "10.1016/j.tim.2016.02.015",
"article-title": "Oscillospira: A Central, Enigmatic Component of the Human Gut Microbiota",
"author": "Konikoff",
"doi-asserted-by": "crossref",
"first-page": "523",
"journal-title": "Trends Microbiol.",
"key": "ref_33",
"volume": "24",
"year": "2016"
},
{
"DOI": "10.3389/fcimb.2021.627608",
"doi-asserted-by": "crossref",
"key": "ref_34",
"unstructured": "Xia, J., Gu, L., Guo, Y., Feng, H., Chen, S., Jurat, J., Fu, W., and Zhang, D. (2021). Gut Microbiota Mediates the Preventive Effects of Dietary Capsaicin Against Depression-Like Behavior Induced by Lipopolysaccharide in Mice. Front. Cell. Infect. Microbiol., 11."
},
{
"DOI": "10.1038/nature08821",
"article-title": "A human gut microbial gene catalogue established by metagenomic sequencing",
"author": "Qin",
"doi-asserted-by": "crossref",
"first-page": "59",
"journal-title": "Nature",
"key": "ref_35",
"volume": "464",
"year": "2010"
},
{
"DOI": "10.1007/s12275-018-8024-4",
"article-title": "The Ruminococci: Key symbionts of the gut ecosystem",
"author": "Suen",
"doi-asserted-by": "crossref",
"first-page": "199",
"journal-title": "J. Microbiol.",
"key": "ref_36",
"volume": "56",
"year": "2018"
},
{
"DOI": "10.3390/ijms22031010",
"doi-asserted-by": "crossref",
"key": "ref_37",
"unstructured": "Gozalbo-Rovira, R., Rubio-Del-campo, A., Santiso-Bellón, C., Vila-Vicent, S., Buesa, J., Delgado, S., Molinero, N., Margolles, A., Yebra, M.J., and Collado, M.C. (2021). Interaction of intestinal bacteria with human rotavirus during infection in children. Int. J. Mol. Sci., 22."
},
{
"DOI": "10.1038/srep45559",
"doi-asserted-by": "crossref",
"key": "ref_38",
"unstructured": "Rodríguez-Díaz, J., García-Mantrana, I., Vila-Vicent, S., Gozalbo-Rovira, R., Buesa, J., Monedero, V., and Collado, M.C. (2017). Relevance of secretor status genotype and microbiota composition in susceptibility to rotavirus and norovirus infections in humans. Sci. Rep., 7."
},
{
"DOI": "10.1080/19490976.2024.2342497",
"article-title": "The gut microbe pair of Oribacterium sp. GMB0313 and Ruminococcus sp. GMB0270 confers complete protection against SARS-CoV-2 infection by activating CD8+ T cell-mediated immunity",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "2342497",
"journal-title": "Gut Microbes",
"key": "ref_39",
"volume": "16",
"year": "2024"
},
{
"DOI": "10.1016/j.virusres.2020.198018",
"article-title": "Gut microbiota and COVID-19-possible link and implications",
"author": "Dhar",
"doi-asserted-by": "crossref",
"first-page": "198018",
"journal-title": "Virus Res.",
"key": "ref_40",
"volume": "285",
"year": "2020"
},
{
"DOI": "10.3390/pathogens11091050",
"doi-asserted-by": "crossref",
"key": "ref_41",
"unstructured": "Rocchi, G., Giovanetti, M., Benedetti, F., Borsetti, A., Ceccarelli, G., Zella, D., Altomare, A., Ciccozzi, M., and Guarino, M.P.L. (2022). Gut Microbiota and COVID-19: Potential Implications for Disease Severity. Pathogens, 11."
},
{
"DOI": "10.1371/journal.pone.0161635",
"doi-asserted-by": "crossref",
"key": "ref_42",
"unstructured": "Ling, X., Linglong, P., Weixia, D., and Hong, W. (2016). Protective effects of bifidobacterium on intestinal barrier function in LPS-induced enterocyte barrier injury of Caco-2 monolayers and in a rat NEC model. PLoS ONE, 11."
},
{
"article-title": "Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease",
"author": "Maubert",
"first-page": "415",
"journal-title": "Gut",
"key": "ref_43",
"volume": "65",
"year": "2017"
},
{
"DOI": "10.1161/CIRCRESAHA.122.320184",
"article-title": "Faecalibacterium prausnitzii Attenuates CKD via Butyrate-Renal GPR43 Axis",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "E120",
"journal-title": "Circ. Res.",
"key": "ref_44",
"volume": "131",
"year": "2022"
},
{
"DOI": "10.1007/s12602-014-9162-6",
"article-title": "Antiviral potential of lactic acid bacteria and their bacteriocins",
"author": "Hober",
"doi-asserted-by": "crossref",
"first-page": "177",
"journal-title": "Probiotics Antimicrob. Proteins",
"key": "ref_45",
"volume": "6",
"year": "2014"
},
{
"DOI": "10.1007/s00203-024-03843-6",
"doi-asserted-by": "crossref",
"key": "ref_46",
"unstructured": "Xu, C., Hao, M., Zai, X., Song, J., Huang, Y., Gui, S., and Chen, J. (2024). A new perspective on gut-lung axis affected through resident microbiome and their implications on immune response in respiratory diseases. Arch. Microbiol., 206."
},
{
"DOI": "10.1007/s11684-022-0921-6",
"article-title": "Integrated analysis of gut microbiome and host immune responses in COVID-19",
"author": "Xu",
"doi-asserted-by": "crossref",
"first-page": "263",
"journal-title": "Front. Med.",
"key": "ref_47",
"volume": "16",
"year": "2022"
},
{
"DOI": "10.1038/nbt.3960",
"article-title": "Towards standards for human fecal sample processing in metagenomic studies",
"author": "Costea",
"doi-asserted-by": "crossref",
"first-page": "1069",
"journal-title": "Nat. Biotechnol.",
"key": "ref_48",
"volume": "35",
"year": "2017"
},
{
"DOI": "10.1038/nbt.3981",
"article-title": "Assessment of variation in microbial community amplicon sequencing by the Microbiome Quality Control (MBQC) project consortium",
"author": "Sinha",
"doi-asserted-by": "crossref",
"first-page": "1077",
"journal-title": "Nat. Biotechnol.",
"key": "ref_49",
"volume": "35",
"year": "2017"
},
{
"DOI": "10.3389/fcimb.2023.1211348",
"doi-asserted-by": "crossref",
"key": "ref_50",
"unstructured": "Reuben, R.C., Beugnon, R., and Jurburg, S.D. (2023). COVID-19 alters human microbiomes: A meta-analysis. Front. Cell. Infect. Microbiol., 13."
},
{
"DOI": "10.1186/s12866-022-02686-9",
"doi-asserted-by": "crossref",
"key": "ref_51",
"unstructured": "Cheng, X., Zhang, Y., Li, Y., Wu, Q., Wu, J., Park, S.K., Guo, C., and Lu, J. (2022). Meta-analysis of 16S rRNA microbial data identified alterations of the gut microbiota in COVID-19 patients during the acute and recovery phases. BMC Microbiol., 22."
},
{
"DOI": "10.1080/19490976.2023.2242615",
"article-title": "Robust cross-cohort gut microbiome associations with COVID-19 severity",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "2242615",
"journal-title": "Gut Microbes",
"key": "ref_52",
"volume": "15",
"year": "2023"
},
{
"DOI": "10.1128/AEM.01541-09",
"article-title": "Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities",
"author": "Schloss",
"doi-asserted-by": "crossref",
"first-page": "7537",
"journal-title": "Appl. Environ. Microbiol.",
"key": "ref_53",
"volume": "75",
"year": "2009"
},
{
"DOI": "10.4254/wjh.v16.i5.832",
"article-title": "Ornithine aspartate effects on bacterial composition and metabolic pathways in a rat model of steatotic liver disease",
"author": "Lange",
"doi-asserted-by": "crossref",
"first-page": "832",
"journal-title": "World J. Hepatol.",
"key": "ref_54",
"volume": "16",
"year": "2024"
},
{
"DOI": "10.1016/j.jnutbio.2024.109660",
"doi-asserted-by": "crossref",
"key": "ref_55",
"unstructured": "Behrens, L.M.P., Gasparotto, J., Rampelotto, P.H., Escalona, M.A.R., da Silva, L.d.S., Carazza-Kessler, F.G., Barbosa, C.P., Campos, M.S., Dorn, M., and Gelain, D.P. (2024). Sodium propionate oral supplementation ameliorates depressive-like behavior through gut microbiome and histone 3 epigenetic regulation. J. Nutr. Biochem., 130."
},
{
"DOI": "10.3390/biology13060363",
"doi-asserted-by": "crossref",
"key": "ref_56",
"unstructured": "Mena Canata, D.A., Benfato, M.S., Pereira, F.D., Ramos Pereira, M.J., Hackenhaar, F.S., Mann, M.B., Frazzon, A.P.G., and Rampelotto, P.H. (2024). Comparative Analysis of the Gut Microbiota of Bat Species with Different Feeding Habits. Biology, 13."
},
{
"DOI": "10.7717/peerj.2584",
"article-title": "VSEARCH: A versatile open source tool for metagenomics",
"author": "Rognes",
"doi-asserted-by": "crossref",
"first-page": "e2584",
"journal-title": "PeerJ",
"key": "ref_57",
"volume": "2016",
"year": "2016"
},
{
"DOI": "10.1016/j.taap.2023.116480",
"article-title": "Changes in the gut microbiota of rats after exposure to the fungicide Mancozeb",
"author": "Pezzini",
"doi-asserted-by": "crossref",
"first-page": "116480",
"journal-title": "Toxicol. Appl. Pharmacol.",
"key": "ref_58",
"volume": "466",
"year": "2023"
},
{
"DOI": "10.1093/nar/gks1219",
"article-title": "The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools",
"author": "Quast",
"doi-asserted-by": "crossref",
"first-page": "590",
"journal-title": "Nucleic Acids Res.",
"key": "ref_59",
"volume": "41",
"year": "2012"
},
{
"DOI": "10.1186/gb-2011-12-6-r60",
"doi-asserted-by": "crossref",
"key": "ref_60",
"unstructured": "Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W.S., and Huttenhower, C. (2011). Metagenomic biomarker discovery and explanation. Genome Biol., 12."
},
{
"DOI": "10.35772/ghm.2020.01015",
"article-title": "Guidelines for the diagnosis and treatment of coronavirus disease 2019 (COVID-19) in China",
"author": "Xu",
"doi-asserted-by": "crossref",
"first-page": "66",
"journal-title": "Glob. Health Med.",
"key": "ref_61",
"volume": "2",
"year": "2020"
},
{
"DOI": "10.1080/19490976.2022.2073131",
"article-title": "A high-risk gut microbiota configuration associates with fatal hyperinflammatory immune and metabolic responses to SARS-CoV-2",
"author": "Albrich",
"doi-asserted-by": "crossref",
"first-page": "2073131",
"journal-title": "Gut Microbes",
"key": "ref_62",
"volume": "14",
"year": "2022"
},
{
"DOI": "10.3389/fcimb.2021.670424",
"doi-asserted-by": "crossref",
"key": "ref_63",
"unstructured": "Gaibani, P., D’Amico, F., Bartoletti, M., Lombardo, D., Rampelli, S., Fornaro, G., Coladonato, S., Siniscalchi, A., Re, M.C., and Viale, P. (2021). The gut microbiota of critically Ill patients with COVID-19. Front. Cell. Infect. Microbiol., 11."
},
{
"DOI": "10.1186/s12879-023-08511-6",
"doi-asserted-by": "crossref",
"key": "ref_64",
"unstructured": "Galperine, T., Choi, Y., Pagani, J.L., Kritikos, A., Papadimitriou-Olivgeris, M., Méan, M., Scherz, V., Opota, O., Greub, G., and Guery, B. (2023). Temporal changes in fecal microbiota of patients infected with COVID-19: A longitudinal cohort. BMC Infect. Dis., 23."
},
{
"DOI": "10.3389/fmed.2022.821777",
"doi-asserted-by": "crossref",
"key": "ref_65",
"unstructured": "Rafiqul Islam, S.M., Foysal, M.J., Hoque, M.N., Mehedi, H.M.H., Rob, M.A., Salauddin, A., Tanzina, A.Y., Biswas, S., Noyon, S.H., and Siddiki, A.M.A.M.Z. (2022). Dysbiosis of Oral and Gut Microbiomes in SARS-CoV-2 Infected Patients in Bangladesh: Elucidating the Role of Opportunistic Gut Microbes. Front. Med., 9."
},
{
"DOI": "10.3389/fcimb.2021.747816",
"doi-asserted-by": "crossref",
"key": "ref_66",
"unstructured": "Reinold, J., Farahpour, F., Fehring, C., Dolff, S., Konik, M., Korth, J., van Baal, L., Hoffmann, D., Buer, J., and Witzke, O. (2021). A Pro-Inflammatory Gut Microbiome Characterizes SARS-CoV-2 Infected Patients and a Reduction in the Connectivity of an Anti-Inflammatory Bacterial Network Associates with Severe COVID-19. Front. Cell. Infect. Microbiol., 11."
},
{
"DOI": "10.1186/s13099-023-00546-z",
"article-title": "Association of gut microbial dysbiosis with disease severity, response to therapy and disease outcomes in Indian patients with COVID-19",
"author": "Talukdar",
"doi-asserted-by": "crossref",
"first-page": "22",
"journal-title": "Gut Pathog.",
"key": "ref_67",
"volume": "15",
"year": "2023"
},
{
"DOI": "10.1038/s41522-021-00232-5",
"doi-asserted-by": "crossref",
"key": "ref_68",
"unstructured": "Wu, Y., Cheng, X., Jiang, G., Tang, H., Ming, S., Tang, L., Lu, J., Guo, C., Shan, H., and Huang, X. (2021). Altered oral and gut microbiota and its association with SARS-CoV-2 viral load in COVID-19 patients during hospitalization. NPJ Biofilms Microbiomes, 7."
},
{
"DOI": "10.1177/1099800420942942",
"article-title": "Oral Probiotic Bifidobacterium longum Supplementation Improves Metabolic Parameters and Alters the Expression of the Renin-Angiotensin System in Obese Mice Liver",
"author": "Machado",
"doi-asserted-by": "crossref",
"first-page": "100",
"journal-title": "Biol. Res. Nurs.",
"key": "ref_69",
"volume": "23",
"year": "2021"
},
{
"article-title": "Primer, Pipelines, Parameters: Issues in 16S rRNA Gene Sequencing",
"author": "Matchado",
"first-page": "10",
"journal-title": "mSphere",
"key": "ref_70",
"volume": "6",
"year": "2021"
},
{
"DOI": "10.1093/bib/bbx120",
"article-title": "A review of methods and databases for metagenomic classification and assembly",
"author": "Breitwieser",
"doi-asserted-by": "crossref",
"first-page": "1125",
"journal-title": "Brief. Bioinform.",
"key": "ref_71",
"volume": "20",
"year": "2018"
},
{
"DOI": "10.1186/s12864-017-3501-4",
"doi-asserted-by": "crossref",
"key": "ref_72",
"unstructured": "Balvočiute, M., and Huson, D.H. (2017). SILVA, RDP, Greengenes, NCBI and OTT—How do these taxonomies compare?. BMC Genom., 18."
},
{
"DOI": "10.1093/bib/bbaa121",
"article-title": "Comparison of microbiome samples: Methods and computational challenges",
"author": "Comin",
"doi-asserted-by": "crossref",
"first-page": "88",
"journal-title": "Brief. Bioinform.",
"key": "ref_73",
"volume": "22",
"year": "2021"
},
{
"DOI": "10.1080/19490976.2024.2325067",
"article-title": "Shotgun metagenomics and systemic targeted metabolomics highlight indole-3-propionic acid as a protective gut microbial metabolite against influenza infection",
"author": "Heumel",
"doi-asserted-by": "crossref",
"first-page": "2325067",
"journal-title": "Gut Microbes",
"key": "ref_74",
"volume": "16",
"year": "2024"
},
{
"DOI": "10.1073/pnas.2003656117",
"article-title": "CD8 T cells drive anorexia, dysbiosis, and blooms of a commensal with immunosuppressive potential after viral infection",
"author": "Gerner",
"doi-asserted-by": "crossref",
"first-page": "24998",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "ref_75",
"volume": "117",
"year": "2020"
},
{
"DOI": "10.1007/s00383-002-0855-7",
"article-title": "Probiotics up-regulate MUC-2 mucin gene expression in a Caco-2 cell-culture model",
"author": "Mattar",
"doi-asserted-by": "crossref",
"first-page": "586",
"journal-title": "Pediatr. Surg. Int.",
"key": "ref_76",
"volume": "18",
"year": "2002"
},
{
"DOI": "10.1186/1471-2180-11-177",
"doi-asserted-by": "crossref",
"key": "ref_77",
"unstructured": "Castillo, N.A., Perdigán, G., and De Moreno De Leblanc, A. (2011). Oral administration of a probiotic Lactobacillus modulates cytokine production and TLR expression improving the immune response against Salmonella enterica serovar Typhimurium infection in mice. BMC Microbiol., 11."
},
{
"DOI": "10.1099/ijsem.0.004107",
"article-title": "A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae",
"author": "Zheng",
"doi-asserted-by": "crossref",
"first-page": "2782",
"journal-title": "Int. J. Syst. Evol. Microbiol.",
"key": "ref_78",
"volume": "70",
"year": "2020"
},
{
"DOI": "10.1016/j.anaerobe.2009.12.001",
"article-title": "The role of anaerobic bacteria in bacteremia",
"author": "Brook",
"doi-asserted-by": "crossref",
"first-page": "183",
"journal-title": "Anaerobe",
"key": "ref_79",
"volume": "16",
"year": "2010"
},
{
"DOI": "10.1128/CMR.00008-07",
"article-title": "Bacteroides: The good, the bad, and the nitty-gritty",
"author": "Wexler",
"doi-asserted-by": "crossref",
"first-page": "593",
"journal-title": "Clin. Microbiol. Rev.",
"key": "ref_80",
"volume": "20",
"year": "2007"
},
{
"DOI": "10.3389/fmicb.2021.712081",
"doi-asserted-by": "crossref",
"key": "ref_81",
"unstructured": "Li, S., Yang, S., Zhou, Y., Disoma, C., Dong, Z., Du, A., Zhang, Y., Chen, Y., Huang, W., and Chen, J. (2021). Microbiome Profiling Using Shotgun Metagenomic Sequencing Identified Unique Microorganisms in COVID-19 Patients with Altered Gut Microbiota. Front. Microbiol., 12."
},
{
"DOI": "10.1128/AEM.03313-13",
"article-title": "Bifidobacterium bifidum PRL2010 Modulates the Host Innate Immune Response",
"author": "Turroni",
"doi-asserted-by": "crossref",
"first-page": "730",
"journal-title": "Appl. Environ. Microbiol.",
"key": "ref_82",
"volume": "80",
"year": "2014"
},
{
"DOI": "10.1186/s13059-020-02007-1",
"doi-asserted-by": "crossref",
"key": "ref_83",
"unstructured": "Zhang, Q., Hu, J., Feng, J., Hu, X., Wang, T., Gong, W., Huang, K., Guo, Y., Zou, Z., and Lin, X. (2020). Influenza infection elicits an expansion of gut population of endogenous Bifidobacterium animalis which protects mice against infection. Genome Biol., 21."
},
{
"DOI": "10.1016/j.imu.2023.101239",
"article-title": "Evaluation of the gut microbiome associated with COVID-19",
"author": "Maddah",
"doi-asserted-by": "crossref",
"first-page": "101239",
"journal-title": "Inform. Med. Unlocked",
"key": "ref_84",
"volume": "38",
"year": "2023"
},
{
"DOI": "10.24075/brsmu.2022.006",
"doi-asserted-by": "crossref",
"key": "ref_85",
"unstructured": "Gumenyuk, L.N., Golod, M.V., Silaeva, N.V., Sorokina, L.E., Ilyasov, S.S., Androschyuk, N.A., Krivoshapko, O.R., Velilyaev, A.M., and Asanova, L.N. (2022). Gut Microbiota Alterations and Their Relationship to the Disease Severity and Some Cytokine Profile Indicators in Patients with COVID-19. Bull. Russ. State Med. Univ., 22–29."
},
{
"DOI": "10.1155/2021/6666114",
"doi-asserted-by": "crossref",
"key": "ref_86",
"unstructured": "He, X., Zhao, S., and Li, Y. (2021). Faecalibacterium prausnitzii: A Next-Generation Probiotic in Gut Disease Improvement. Can. J. Infect. Dis. Med. Microbiol., 2021."
},
{
"DOI": "10.1093/femsre/fuad014",
"doi-asserted-by": "crossref",
"key": "ref_87",
"unstructured": "Crost, E.H., Coletto, E., Bell, A., and Juge, N. (2023). Ruminococcus gnavus: Friend or foe for human health. FEMS Microbiol. Rev., 47."
},
{
"DOI": "10.36877/pmmb.a0000396",
"article-title": "The Role of Human Gut Microbe Ruminococcus gnavus in Inflammatory Diseases",
"author": "Kandasamy",
"doi-asserted-by": "crossref",
"first-page": "1",
"journal-title": "Prog. Microbes Mol. Biol.",
"key": "ref_88",
"volume": "6",
"year": "2023"
}
],
"reference-count": 88,
"references-count": 88,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.mdpi.com/2076-2607/12/11/2353"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "In Silico Analysis of Probiotic Bacteria Changes Across COVID-19 Severity Stages",
"type": "journal-article",
"volume": "12"
}