SARS-CoV-2 Helicase (NSP13) Interacts with Mammalian Polyamine and HSP Partners in Promoting Viral Replication
et al., Current Issues in Molecular Biology, doi:10.3390/cimb48010080, Jan 2026
In silico study showing that SARS-CoV-2 helicase NSP13 forms stable binding complexes with host heat shock proteins (HSP40, HSP70, HSP90) and ornithine decarboxylase (ODC). The findings provide computational evidence that targeting NSP13's recruitment of host heat shock proteins and ODC could disrupt viral replication processes.
Sitobo et al., 13 Jan 2026, South Africa, peer-reviewed, 8 authors.
Contact: makhoxh@unisa.ac.za (corresponding author).
In silico studies are an important part of preclinical research, however results may be very different in vivo.
SARS-CoV-2 Helicase (NSP13) Interacts with Mammalian Polyamine and HSP Partners in Promoting Viral Replication
Current Issues in Molecular Biology, doi:10.3390/cimb48010080
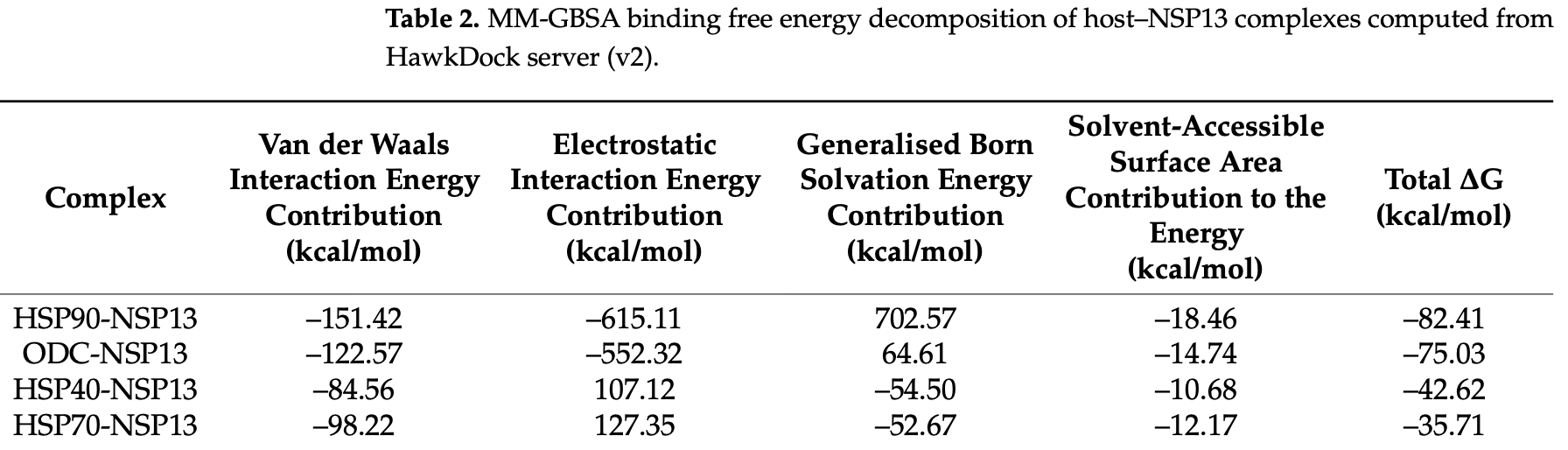
We present a computational study that precedes the potential interactions between SARS-CoV-2 helicase (NSP13) and selected host proteins implicated in chaperone-assisted folding and polyamine metabolism. Using structure-based modelling and protein-protein docking (BioLuminate v4.6), followed by all-atom molecular dynamics (MD) simulations (GROMACS v2018.6), and comparative MM-GBSA scoring (HawkDock v2), we evaluated the stability and interface properties of NSP13 complexes with cytosolic heat shock proteins; heat shock protein 40 (HSP40), heat shock protein 70 (HSP70), heat shock protein 90 (HSP90) and the polyamine biosynthesis enzyme ornithine decarboxylase (ODC). Docking, MD, and interface analyses indicate distinct complex behaviours: HSP70-NSP13 complexes sampled compact conformations, HSP90-NSP13 ensembles displayed greater conformational heterogeneity but more favourable comparative MM-GBSA estimates, and ODC-NSP13 interfaces were comparatively well packed. Per-residue contact mapping identified a small set of recurrent NSP13 residues, Lys22 and Asn51, as putative interaction hotspots. The reported findings herein generate testable hypotheses about NSP13 recruitment of host chaperones and modulation of polyamine metabolism that may inform downstream experimental studies.
Institutional Review Board Statement: Under U.S. federal regulations (45 CFR 46) , the database information used in this document does not involve any identifiable private information and therefore does not require IRB approval. Informed Consent Statement: Under U.S. federal regulations (45 CFR 46) , the database information used in this document does not involve any identifiable private information and therefore does not require patient informed consent.
Conflicts of Interest: The authors declare no conflicts of interest.
References
Adedeji, Marchand, Te, Velthuis, Snijder et al., Mechanism of nucleic acid unwinding by SARS-CoV helicase, PLoS ONE, doi:10.1371/journal.pone.0036521
Almuqdadi, Kifayat, Anwer, Alrehaili, Abid, Fragment-based virtual screening identifies novel leads against Plasmepsin IX (PlmIX) of Plasmodium falciparum: Homology modeling, molecular docking, and simulation approaches, Front. Pharmacol, doi:10.3389/fphar.2024.1387629
Ben Abdallah, Marino, Idorn, Reinert, Bregnhøj et al., The heat shock protein 90 inhibitor RGRN-305 attenuates SARS-CoV-2 spike protein-induced inflammation in vitro but lacks effectiveness as COVID-19 treatment in mice, PLoS ONE, doi:10.1371/journal.pone.0310915
Biebl, Delhommel, Faust, Zak, Agam et al., NudC guides client transfer between the Hsp40/70 and Hsp90 chaperone systems, Mol. Cell, doi:10.1016/j.molcel.2021.12.031
Brown, Doom, Lechuga-Peña, Watamura, Koppels, Stress and parenting during the global COVID-19 pandemic, Child Abus. Negl, doi:10.1016/j.chiabu.2020.104699
Calzadilla, Gazquez, Maiale, Rodriguez, Ruiz et al., Polyamines as indicators and modulators of the abiotic stress in plants
Chatterjee, Puri, Sharma, Pastor, Chaudhuri, Molecular chaperones: Structure-function relationship and their role in protein folding
Chen, Malone, Llewellyn, Grasso, Shelton et al., Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex, Cell, doi:10.1016/j.cell.2020.07.033
Coleman, Tay, Tan, Ong, Than et al., Viral load of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in respiratory aerosols emitted by patients with coronavirus disease 2019 (COVID-19) while breathing, talking, and singing, Clin. Infect. Dis, doi:10.1093/cid/ciab691
Daugherty, Guo, Heath, Dasmariñas, Jubilo et al., Risk of clinical sequelae after the acute phase of SARS-CoV-2 infection: Retrospective cohort study, BMJ, doi:10.1136/bmj.n1098
El Zowalaty, Young, Järhult, Environmental impact of the COVID-19 pandemic-a lesson for the future, Infect. Ecol. Epidemiol, doi:10.1080/20008686.2020.1768023
Firpo, Mastrodomenico, Hawkins, Prot, Levillayer et al., Targeting polyamines inhibits coronavirus infection by reducing cellular attachment and entry, ACS Infect. Dis, doi:10.1021/acsinfecdis.0c00491
Gerner, Meyskens, Jr, Polyamines and cancer: Old molecules, new understanding, Nat. Rev. Cancer, doi:10.1038/nrc1454
González-Esparragoza, Carrasco-Carballo, Rosas-Murrieta, Millán-Pérez Peña, Luna et al., In silico analysis of protein-protein interactions of putative endoplasmic reticulum metallopeptidase 1 in Schizosaccharomyces pombe, Curr. Issues Mol. Biol, doi:10.3390/cimb46050280
Grado Ń, Crime in the time of the plague: Fake news pandemic and the challenges to law-enforcement and intelligence community, Soc. Regist, doi:10.14746/sr.2020.4.2.10
Gralinski, Menachery, Return of the Coronavirus: 2019-nCoV, Viruses, doi:10.3390/v12020135
Grünberg, Leckner, Nilges, Complementarity of structure ensembles in protein-protein binding, Structure, doi:10.1016/j.str.2004.09.014
Heffner, Maio, Tip of the iceberg: A new wave of iron-sulfur cluster proteins found in viruses, Inorganics, doi:10.3390/inorganics12010034
Hou, Wang, Li, Wang, Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations, J. Chem. Inf. Model, doi:10.1021/ci100275a
Hoxie, Street, Hsp90 chaperones have an energetic hot-spot for binding inhibitors, Protein Sci
Hughes, Rees, Kalindjian, Philpott, Principles of early drug discovery, Br. J. Pharmacol, doi:10.1111/j.1476-5381.2010.01127.x
Ivanov, Ziebuhr, Human coronavirus 229E nonstructural protein 13: Characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5 ′ -triphosphatase activities, J. Virol, doi:10.1128/JVI.78.14.7833-7838.2004
Jia, Yan, Ren, Wu, Wang et al., Delicate structural coordination of the severe acute respiratory syndrome coronavirus Nsp13 upon ATP hydrolysis, Nucleic Acids Res, doi:10.1093/nar/gkz409
Jiang, Rossi, Kalodimos, Structural basis for client recognition and activity of Hsp40 chaperones, Science, doi:10.1126/science.aax1280
Kim, Hipp, Bracher, Hayer-Hartl, Ulrich Hartl, Molecular chaperone functions in protein folding and proteostasis, Annu. Rev. Biochem, doi:10.1146/annurev-biochem-060208-092442
Knox, Luke, Blatch, Pesce, Heat shock protein 40 (Hsp40) plays a key role in the virus life cycle, Virus Res, doi:10.1016/j.virusres.2011.06.013
Koes, Camacho, Pharmer, Efficient and exact pharmacophore search, J. Chem. Inf. Model
Laskowski, Macarthur, Moss, Thornton, Procheck, A program to check the stereochemical quality of protein structures, Appl. Crystallogr, doi:10.1107/S0021889892009944
Makhoba, Makumire, The capture of host cell's resources: The role of heat shock proteins and polyamines in SARS-CoV-2 (COVID-19) pathway to viral infection, Biomol. Concepts, doi:10.1515/bmc-2022-0008
Malinverni, Jost Lopez, De Los Rios, Hummer, Barducci, Modeling Hsp70/Hsp40 interaction by multi-scale molecular simulations and coevolutionary sequence analysis, eLife, doi:10.7554/eLife.23471
Mapa, Sikor, Kudryavtsev, Waegemann, Kalinin et al., The conformational dynamics of the mitochondrial Hsp70 chaperone, Mol. Cell, doi:10.1016/j.molcel.2010.03.010
Mayer, Hsp70 chaperone dynamics and molecular mechanism, Trends Biochem. Sci, doi:10.1016/j.tibs.2013.08.001
Mickolajczyk, Shelton, Grasso, Cao, Warrington et al., Force-dependent stimulation of RNA unwinding by SARS-CoV-2 nsp13 helicase, Biophys. J, doi:10.1016/j.bpj.2020.11.2276
Mounce, Olsen, Vignuzzi, Connor, Polyamines and their role in virus infection, Microbiol. Mol. Biol. Rev, doi:10.1128/MMBR.00029-17
Navhaya, Matsebatlela, Monama, Makhoba, In Silico Discovery and Evaluation of Inhibitors of the SARS-CoV-2 Spike Protein-HSPA8 Complex Towards Developing COVID-19 Therapeutic Drugs, Viruses, doi:10.3390/v16111726
Paladino, Vitale, Caruso Bavisotto, Conway De Macario, Cappello et al., The role of molecular chaperones in virus infection and implications for understanding and treating COVID-19, J. Clin. Med, doi:10.3390/jcm9113518
Pegg, Polyamine metabolism and its importance in neoplastic growth and as a target for chemotherapy, Cancer Res
Rehm, Bioinformatic tools for DNA/protein sequence analysis, functional assignment of genes, and protein classification, Appl. Microbiol. Biotechnol, doi:10.1007/s00253-001-0844-0
Richter, Buchner, Hsp90: Chaperoning signal transduction, J. Cell. Physiol, doi:10.1002/jcp.1131
Rudnev, Kulikova, Nikolsky, Malsagova, Kopylov et al., Current approaches in supersecondary structures investigation, Int. J. Mol. Sci, doi:10.3390/ijms222111879
Saponaro, Maione, Bonvin, Cantini, Understanding docking complexes of macromolecules using HADDOCK: The synergy between experimental data and computations, Bio-Protocol, doi:10.21769/BioProtoc.3793
Schoch, Ciufo, Domrachev, Hotton, Kannan et al., NCBI Taxonomy: A comprehensive update on curation, resources and tools, Database, doi:10.1093/database/baaa062
Sesethu, Nombalentle, Yamkela, Anelisa, Makumire et al., In silico evaluation of heat shock proteins reveals an interplay with polyamines as a survival strategy for the Plasmodium falciparum, INNOSC Theranostics Pharmacol. Sci, doi:10.36922/itps.1228
Tabor, Tabor, 1,4-Diaminobutane (putrescine), spermidine, and spermine, Annu. Rev. Biochem, doi:10.1146/annurev.bi.45.070176.001441
Tanner, Watt, Chai, Lu, Lin et al., The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5 ′ to 3 ′ viral helicases, J. Biol. Chem, doi:10.1074/jbc.C300328200
Vellingiri, Jayaramayya, Iyer, Narayanasamy, Govindasamy et al., COVID-19: A promising cure for the global panic, Sci. Total Environ, doi:10.1016/j.scitotenv.2020.138277
Wolber, Langer, LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters, J. Chem. Inf. Model, doi:10.1021/ci049885e
Wu, Chen, Cai, Xia, Zhou et al., Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China, JAMA Intern. Med, doi:10.1001/jamainternmed.2020.0994
Yamkela, Sitobo, Makhoba, In Silico Analysis of SARS-CoV-2 Non-Structural Proteins Reveals an Interaction with the Host's Heat Shock Proteins That May Contribute to Viral Replications and Development, Curr. Issues Mol. Biol, doi:10.3390/cimb45120638
Yazdani, Sirous, Brogi, Calderone, Structure-based high-throughput virtual screening and molecular dynamics simulation for the discovery of novel SARS-CoV-2 NSP3 Mac1 domain inhibitors, Viruses, doi:10.3390/v15122291
Zhou, Yang, Chi, Dong, Lv et al., Comorbidities and the risk of severe or fatal outcomes associated with coronavirus disease 2019: A systematic review and meta-analysis, Int. J. Infect. Dis
DOI record:
{
"DOI": "10.3390/cimb48010080",
"ISSN": [
"1467-3045"
],
"URL": "http://dx.doi.org/10.3390/cimb48010080",
"abstract": "<jats:p>We present a computational study that precedes the potential interactions between SARS-CoV-2 helicase (NSP13) and selected host proteins implicated in chaperone-assisted folding and polyamine metabolism. Using structure-based modelling and protein–protein docking (BioLuminate v4.6), followed by all-atom molecular dynamics (MD) simulations (GROMACS v2018.6), and comparative MM-GBSA scoring (HawkDock v2), we evaluated the stability and interface properties of NSP13 complexes with cytosolic heat shock proteins; heat shock protein 40 (HSP40), heat shock protein 70 (HSP70), heat shock protein 90 (HSP90) and the polyamine biosynthesis enzyme ornithine decarboxylase (ODC). Docking, MD, and interface analyses indicate distinct complex behaviours: HSP70-NSP13 complexes sampled compact conformations, HSP90-NSP13 ensembles displayed greater conformational heterogeneity but more favourable comparative MM-GBSA estimates, and ODC-NSP13 interfaces were comparatively well packed. Per-residue contact mapping identified a small set of recurrent NSP13 residues, Lys22 and Asn51, as putative interaction hotspots. The reported findings herein generate testable hypotheses about NSP13 recruitment of host chaperones and modulation of polyamine metabolism that may inform downstream experimental studies.</jats:p>",
"alternative-id": [
"cimb48010080"
],
"author": [
{
"affiliation": [
{
"name": "Department of Biochemistry and Microbiology, University of Fort Hare, Alice Campus, Alice 5700, South Africa"
}
],
"family": "Sitobo",
"given": "Zingisa",
"sequence": "first"
},
{
"ORCID": "https://orcid.org/0000-0003-1815-1272",
"affiliation": [
{
"name": "Department of Biochemistry, Microbiology and Biotechnology, University of Limpopo, Turfloop Campus, Sovenga 0727, South Africa"
}
],
"authenticated-orcid": false,
"family": "Navhaya",
"given": "Liberty T.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Biochemistry and Microbiology, University of Fort Hare, Alice Campus, Alice 5700, South Africa"
}
],
"family": "Nqumla",
"given": "Ntombekhaya",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Life and Consumer Sciences, College of Agriculture and Environmental Sciences, University of South Africa (UNISA), Florida Campus, Roodepoort 1709, South Africa"
}
],
"family": "Masenya",
"given": "Madipoane",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Department of Life and Consumer Sciences, College of Agriculture and Environmental Sciences, University of South Africa (UNISA), Florida Campus, Roodepoort 1709, South Africa"
}
],
"family": "Molapo",
"given": "Matsheliso",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-6939-1199",
"affiliation": [
{
"name": "Human Genetics Department, Computational Biology Division, Faculty of Health Sciences, University of Cape Town, Barnard Fuller Building, Anzio Rd, Observatory, Cape Town 7935, South Africa"
}
],
"authenticated-orcid": false,
"family": "Mthembu",
"given": "Yamkela",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-8076-3096",
"affiliation": [
{
"name": "Department of Life and Consumer Sciences, College of Agriculture and Environmental Sciences, University of South Africa (UNISA), Florida Campus, Roodepoort 1709, South Africa"
}
],
"authenticated-orcid": false,
"family": "Godlo",
"given": "Sesethu",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-7238-4619",
"affiliation": [
{
"name": "Department of Life and Consumer Sciences, College of Agriculture and Environmental Sciences, University of South Africa (UNISA), Florida Campus, Roodepoort 1709, South Africa"
}
],
"authenticated-orcid": false,
"family": "Makhoba",
"given": "Xolani H.",
"sequence": "additional"
}
],
"container-title": "Current Issues in Molecular Biology",
"container-title-short": "CIMB",
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2026,
1,
13
]
],
"date-time": "2026-01-13T18:40:09Z",
"timestamp": 1768329609000
},
"deposited": {
"date-parts": [
[
2026,
1,
13
]
],
"date-time": "2026-01-13T18:47:26Z",
"timestamp": 1768330046000
},
"funder": [
{
"DOI": "10.13039/501100001321",
"award": [
"RA22102665148"
],
"award-info": [
{
"award-number": [
"RA22102665148"
]
}
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100001321",
"id-type": "DOI"
}
],
"name": "National Research Foundation Incentive"
}
],
"indexed": {
"date-parts": [
[
2026,
1,
14
]
],
"date-time": "2026-01-14T02:19:09Z",
"timestamp": 1768357149813,
"version": "3.49.0"
},
"is-referenced-by-count": 0,
"issue": "1",
"issued": {
"date-parts": [
[
2026,
1,
13
]
]
},
"journal-issue": {
"issue": "1",
"published-online": {
"date-parts": [
[
2026,
1
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2026,
1,
13
]
],
"date-time": "2026-01-13T00:00:00Z",
"timestamp": 1768262400000
}
}
],
"link": [
{
"URL": "https://www.mdpi.com/1467-3045/48/1/80/pdf",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1968",
"original-title": [],
"page": "80",
"prefix": "10.3390",
"published": {
"date-parts": [
[
2026,
1,
13
]
]
},
"published-online": {
"date-parts": [
[
2026,
1,
13
]
]
},
"publisher": "MDPI AG",
"reference": [
{
"DOI": "10.1016/j.cell.2020.07.033",
"article-title": "Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex",
"author": "Chen",
"doi-asserted-by": "crossref",
"first-page": "1560",
"journal-title": "Cell",
"key": "ref_1",
"volume": "182",
"year": "2020"
},
{
"DOI": "10.1001/jamainternmed.2020.0994",
"article-title": "Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China",
"author": "Wu",
"doi-asserted-by": "crossref",
"first-page": "934",
"journal-title": "JAMA Intern. Med.",
"key": "ref_2",
"volume": "180",
"year": "2020"
},
{
"article-title": "Environmental impact of the COVID-19 pandemic–a lesson for the future",
"author": "Young",
"first-page": "1768023",
"journal-title": "Infect. Ecol. Epidemiol.",
"key": "ref_3",
"volume": "10",
"year": "2020"
},
{
"DOI": "10.1016/j.ijid.2020.07.029",
"article-title": "Comorbidities and the risk of severe or fatal outcomes associated with coronavirus disease 2019: A systematic review and meta-analysis",
"author": "Zhou",
"doi-asserted-by": "crossref",
"first-page": "47",
"journal-title": "Int. J. Infect. Dis.",
"key": "ref_4",
"volume": "99",
"year": "2020"
},
{
"DOI": "10.1074/jbc.C300328200",
"article-title": "The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5′ to 3′ viral helicases",
"author": "Tanner",
"doi-asserted-by": "crossref",
"first-page": "39578",
"journal-title": "J. Biol. Chem.",
"key": "ref_5",
"volume": "278",
"year": "2003"
},
{
"DOI": "10.1016/j.bpj.2020.11.2276",
"article-title": "Force-dependent stimulation of RNA unwinding by SARS-CoV-2 nsp13 helicase",
"author": "Mickolajczyk",
"doi-asserted-by": "crossref",
"first-page": "1020",
"journal-title": "Biophys. J.",
"key": "ref_6",
"volume": "120",
"year": "2021"
},
{
"DOI": "10.1371/journal.pone.0036521",
"doi-asserted-by": "crossref",
"key": "ref_7",
"unstructured": "Adedeji, A.O., Marchand, B., Te Velthuis, A.J., Snijder, E.J., Weiss, S., Eoff, R.L., Singh, K., and Sarafianos, S.G. (2012). Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLoS ONE, 7."
},
{
"DOI": "10.1093/nar/gkz409",
"article-title": "Delicate structural coordination of the severe acute respiratory syndrome coronavirus Nsp13 upon ATP hydrolysis",
"author": "Jia",
"doi-asserted-by": "crossref",
"first-page": "6538",
"journal-title": "Nucleic Acids Res.",
"key": "ref_8",
"volume": "47",
"year": "2019"
},
{
"DOI": "10.1126/science.aax1280",
"article-title": "Structural basis for client recognition and activity of Hsp40 chaperones",
"author": "Jiang",
"doi-asserted-by": "crossref",
"first-page": "1313",
"journal-title": "Science",
"key": "ref_9",
"volume": "365",
"year": "2019"
},
{
"DOI": "10.1146/annurev.bi.45.070176.001441",
"article-title": "1,4-Diaminobutane (putrescine), spermidine, and spermine",
"author": "Tabor",
"doi-asserted-by": "crossref",
"first-page": "285",
"journal-title": "Annu. Rev. Biochem.",
"key": "ref_10",
"volume": "45",
"year": "1976"
},
{
"article-title": "Polyamine metabolism and its importance in neoplastic growth and as a target for chemotherapy",
"author": "Pegg",
"first-page": "759",
"journal-title": "Cancer Res.",
"key": "ref_11",
"volume": "48",
"year": "1988"
},
{
"DOI": "10.1038/nrc1454",
"article-title": "Polyamines and cancer: Old molecules, new understanding",
"author": "Gerner",
"doi-asserted-by": "crossref",
"first-page": "781",
"journal-title": "Nat. Rev. Cancer",
"key": "ref_12",
"volume": "4",
"year": "2004"
},
{
"DOI": "10.1079/9781780642734.0109",
"doi-asserted-by": "crossref",
"key": "ref_13",
"unstructured": "Calzadilla, P.I., Gazquez, A., Maiale, S.J., Rodriguez, A.A., Ruiz, O.A., and Bernardina, M.A. (2014). Polyamines as indicators and modulators of the abiotic stress in plants. Plant Adaptation to Environmental Change: Significance of Amino Acids and Their Derivatives, CABI."
},
{
"DOI": "10.1146/annurev-biochem-060208-092442",
"article-title": "Molecular chaperone functions in protein folding and proteostasis",
"author": "Kim",
"doi-asserted-by": "crossref",
"first-page": "323",
"journal-title": "Annu. Rev. Biochem.",
"key": "ref_14",
"volume": "82",
"year": "2013"
},
{
"DOI": "10.1515/bmc-2022-0008",
"article-title": "The capture of host cell’s resources: The role of heat shock proteins and polyamines in SARS-CoV-2 (COVID-19) pathway to viral infection",
"author": "Makhoba",
"doi-asserted-by": "crossref",
"first-page": "220",
"journal-title": "Biomol. Concepts",
"key": "ref_15",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.1016/j.tibs.2013.08.001",
"article-title": "Hsp70 chaperone dynamics and molecular mechanism",
"author": "Mayer",
"doi-asserted-by": "crossref",
"first-page": "507",
"journal-title": "Trends Biochem. Sci.",
"key": "ref_16",
"volume": "38",
"year": "2013"
},
{
"DOI": "10.1007/978-3-319-74715-6_8",
"doi-asserted-by": "crossref",
"key": "ref_17",
"unstructured": "Chatterjee, B.K., Puri, S., Sharma, A., Pastor, A., and Chaudhuri, T.K. (2018). Molecular chaperones: Structure-function relationship and their role in protein folding. Regulation of Heat Shock Protein Responses, Springer International Publishing."
},
{
"DOI": "10.1002/jcp.1131",
"article-title": "Hsp90: Chaperoning signal transduction",
"author": "Richter",
"doi-asserted-by": "crossref",
"first-page": "281",
"journal-title": "J. Cell. Physiol.",
"key": "ref_18",
"volume": "188",
"year": "2001"
},
{
"DOI": "10.1136/bmj.n1098",
"article-title": "Risk of clinical sequelae after the acute phase of SARS-CoV-2 infection: Retrospective cohort study",
"author": "Daugherty",
"doi-asserted-by": "crossref",
"first-page": "n1098",
"journal-title": "BMJ",
"key": "ref_19",
"volume": "373",
"year": "2021"
},
{
"DOI": "10.3390/v16111726",
"doi-asserted-by": "crossref",
"key": "ref_20",
"unstructured": "Navhaya, L.T., Matsebatlela, T.M., Monama, M.Z., and Makhoba, X.H. (2024). In Silico Discovery and Evaluation of Inhibitors of the SARS-CoV-2 Spike Protein–HSPA8 Complex Towards Developing COVID-19 Therapeutic Drugs. Viruses, 16."
},
{
"DOI": "10.1093/database/baaa062",
"article-title": "NCBI Taxonomy: A comprehensive update on curation, resources and tools",
"author": "Schoch",
"doi-asserted-by": "crossref",
"first-page": "baaa062",
"journal-title": "Database",
"key": "ref_21",
"volume": "2020",
"year": "2020"
},
{
"DOI": "10.1007/s00253-001-0844-0",
"article-title": "Bioinformatic tools for DNA/protein sequence analysis, functional assignment of genes, and protein classification",
"author": "Rehm",
"doi-asserted-by": "crossref",
"first-page": "579",
"journal-title": "Appl. Microbiol. Biotechnol.",
"key": "ref_22",
"volume": "57",
"year": "2001"
},
{
"DOI": "10.3389/fphar.2024.1387629",
"doi-asserted-by": "crossref",
"key": "ref_23",
"unstructured": "Almuqdadi, H.T.A., Kifayat, S., Anwer, R., Alrehaili, J., and Abid, M. (2024). Fragment-based virtual screening identifies novel leads against Plasmepsin IX (PlmIX) of Plasmodium falciparum: Homology modeling, molecular docking, and simulation approaches. Front. Pharmacol., 15."
},
{
"DOI": "10.1107/S0021889892009944",
"article-title": "PROCHECK: A program to check the stereochemical quality of protein structures",
"author": "Laskowski",
"doi-asserted-by": "crossref",
"first-page": "283",
"journal-title": "Appl. Crystallogr.",
"key": "ref_24",
"volume": "26",
"year": "1993"
},
{
"DOI": "10.3390/cimb46050280",
"article-title": "In silico analysis of protein–protein interactions of putative endoplasmic reticulum metallopeptidase 1 in Schizosaccharomyces pombe.",
"author": "Luna",
"doi-asserted-by": "crossref",
"first-page": "4609",
"journal-title": "Curr. Issues Mol. Biol.",
"key": "ref_25",
"volume": "46",
"year": "2024"
},
{
"DOI": "10.1016/j.molcel.2021.12.031",
"article-title": "NudC guides client transfer between the Hsp40/70 and Hsp90 chaperone systems",
"author": "Biebl",
"doi-asserted-by": "crossref",
"first-page": "555",
"journal-title": "Mol. Cell",
"key": "ref_26",
"volume": "82",
"year": "2022"
},
{
"DOI": "10.21769/BioProtoc.3793",
"article-title": "Understanding docking complexes of macromolecules using HADDOCK: The synergy between experimental data and computations",
"author": "Saponaro",
"doi-asserted-by": "crossref",
"first-page": "e3793",
"journal-title": "Bio-Protocol",
"key": "ref_27",
"volume": "10",
"year": "2020"
},
{
"DOI": "10.1021/ci100275a",
"article-title": "Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations",
"author": "Hou",
"doi-asserted-by": "crossref",
"first-page": "69",
"journal-title": "J. Chem. Inf. Model.",
"key": "ref_28",
"volume": "51",
"year": "2011"
},
{
"DOI": "10.3390/ijms222111879",
"doi-asserted-by": "crossref",
"key": "ref_29",
"unstructured": "Rudnev, V.R., Kulikova, L.I., Nikolsky, K.S., Malsagova, K.A., Kopylov, A.T., and Kaysheva, A.L. (2021). Current approaches in supersecondary structures investigation. Int. J. Mol. Sci., 22."
},
{
"DOI": "10.1002/pro.3933",
"article-title": "Hsp90 chaperones have an energetic hot-spot for binding inhibitors",
"author": "Hoxie",
"doi-asserted-by": "crossref",
"first-page": "2101",
"journal-title": "Protein Sci.",
"key": "ref_30",
"volume": "29",
"year": "2020"
},
{
"DOI": "10.14746/sr.2020.4.2.10",
"article-title": "Crime in the time of the plague: Fake news pandemic and the challenges to law-enforcement and intelligence community",
"doi-asserted-by": "crossref",
"first-page": "133",
"journal-title": "Soc. Regist.",
"key": "ref_31",
"volume": "4",
"year": "2020"
},
{
"DOI": "10.1016/j.scitotenv.2020.138277",
"article-title": "COVID-19: A promising cure for the global panic",
"author": "Vellingiri",
"doi-asserted-by": "crossref",
"first-page": "138277",
"journal-title": "Sci. Total Environ.",
"key": "ref_32",
"volume": "725",
"year": "2020"
},
{
"DOI": "10.1111/j.1476-5381.2010.01127.x",
"article-title": "Principles of early drug discovery",
"author": "Hughes",
"doi-asserted-by": "crossref",
"first-page": "1239",
"journal-title": "Br. J. Pharmacol.",
"key": "ref_33",
"volume": "162",
"year": "2011"
},
{
"DOI": "10.1016/j.molcel.2010.03.010",
"article-title": "The conformational dynamics of the mitochondrial Hsp70 chaperone",
"author": "Mapa",
"doi-asserted-by": "crossref",
"first-page": "89",
"journal-title": "Mol. Cell",
"key": "ref_34",
"volume": "38",
"year": "2010"
},
{
"DOI": "10.1016/j.chiabu.2020.104699",
"article-title": "Stress and parenting during the global COVID-19 pandemic",
"author": "Brown",
"doi-asserted-by": "crossref",
"first-page": "104699",
"journal-title": "Child Abus. Negl.",
"key": "ref_35",
"volume": "110",
"year": "2020"
},
{
"DOI": "10.3390/v12020135",
"doi-asserted-by": "crossref",
"key": "ref_36",
"unstructured": "Gralinski, L.E., and Menachery, V.D. (2020). Return of the Coronavirus: 2019-nCoV. Viruses, 12."
},
{
"DOI": "10.36922/itps.1228",
"article-title": "In silico evaluation of heat shock proteins reveals an interplay with polyamines as a survival strategy for the Plasmodium falciparum",
"author": "Sesethu",
"doi-asserted-by": "crossref",
"first-page": "1228",
"journal-title": "INNOSC Theranostics Pharmacol. Sci.",
"key": "ref_37",
"volume": "7",
"year": "2023"
},
{
"DOI": "10.3390/cimb45120638",
"article-title": "In Silico Analysis of SARS-CoV-2 Non-Structural Proteins Reveals an Interaction with the Host’s Heat Shock Proteins That May Contribute to Viral Replications and Development",
"author": "Yamkela",
"doi-asserted-by": "crossref",
"first-page": "10225",
"journal-title": "Curr. Issues Mol. Biol.",
"key": "ref_38",
"volume": "45",
"year": "2023"
},
{
"DOI": "10.1093/cid/ciab691",
"article-title": "Viral load of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in respiratory aerosols emitted by patients with coronavirus disease 2019 (COVID-19) while breathing, talking, and singing",
"author": "Coleman",
"doi-asserted-by": "crossref",
"first-page": "1722",
"journal-title": "Clin. Infect. Dis.",
"key": "ref_39",
"volume": "74",
"year": "2022"
},
{
"DOI": "10.7554/eLife.23471",
"article-title": "Modeling Hsp70/Hsp40 interaction by multi-scale molecular simulations and coevolutionary sequence analysis",
"author": "Malinverni",
"doi-asserted-by": "crossref",
"first-page": "e23471",
"journal-title": "eLife",
"key": "ref_40",
"volume": "6",
"year": "2017"
},
{
"DOI": "10.1016/j.str.2004.09.014",
"article-title": "Complementarity of structure ensembles in protein–protein binding",
"author": "Leckner",
"doi-asserted-by": "crossref",
"first-page": "2125",
"journal-title": "Structure",
"key": "ref_41",
"volume": "12",
"year": "2004"
},
{
"DOI": "10.1128/JVI.78.14.7833-7838.2004",
"article-title": "Human coronavirus 229E nonstructural protein 13: Characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5′-triphosphatase activities",
"author": "Ivanov",
"doi-asserted-by": "crossref",
"first-page": "7833",
"journal-title": "J. Virol.",
"key": "ref_42",
"volume": "78",
"year": "2004"
},
{
"DOI": "10.3390/inorganics12010034",
"doi-asserted-by": "crossref",
"key": "ref_43",
"unstructured": "Heffner, A.L., and Maio, N. (2024). Tip of the iceberg: A new wave of iron–sulfur cluster proteins found in viruses. Inorganics, 12."
},
{
"DOI": "10.1371/journal.pone.0310915",
"doi-asserted-by": "crossref",
"key": "ref_44",
"unstructured": "Ben Abdallah, H., Marino, G., Idorn, M., Reinert, L.S., Bregnhøj, A., Paludan, S.R., and Johansen, C. (2024). The heat shock protein 90 inhibitor RGRN-305 attenuates SARS-CoV-2 spike protein-induced inflammation in vitro but lacks effectiveness as COVID-19 treatment in mice. PLoS ONE, 19."
},
{
"DOI": "10.1016/j.virusres.2011.06.013",
"article-title": "Heat shock protein 40 (Hsp40) plays a key role in the virus life cycle",
"author": "Knox",
"doi-asserted-by": "crossref",
"first-page": "15",
"journal-title": "Virus Res.",
"key": "ref_45",
"volume": "160",
"year": "2011"
},
{
"DOI": "10.3390/jcm9113518",
"doi-asserted-by": "crossref",
"key": "ref_46",
"unstructured": "Paladino, L., Vitale, A.M., Caruso Bavisotto, C., Conway de Macario, E., Cappello, F., Macario, A.J., and Marino Gammazza, A. (2020). The role of molecular chaperones in virus infection and implications for understanding and treating COVID-19. J. Clin. Med., 9."
},
{
"DOI": "10.1128/MMBR.00029-17",
"article-title": "Polyamines and their role in virus infection",
"author": "Mounce",
"doi-asserted-by": "crossref",
"first-page": "e00029-17",
"journal-title": "Microbiol. Mol. Biol. Rev.",
"key": "ref_47",
"volume": "81",
"year": "2017"
},
{
"DOI": "10.1021/acsinfecdis.0c00491",
"article-title": "Targeting polyamines inhibits coronavirus infection by reducing cellular attachment and entry",
"author": "Firpo",
"doi-asserted-by": "crossref",
"first-page": "1423",
"journal-title": "ACS Infect. Dis.",
"key": "ref_48",
"volume": "7",
"year": "2021"
},
{
"DOI": "10.3390/v15122291",
"doi-asserted-by": "crossref",
"key": "ref_49",
"unstructured": "Yazdani, B., Sirous, H., Brogi, S., and Calderone, V. (2023). Structure-based high-throughput virtual screening and molecular dynamics simulation for the discovery of novel SARS-CoV-2 NSP3 Mac1 domain inhibitors. Viruses, 15."
},
{
"DOI": "10.1021/ci049885e",
"article-title": "LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters",
"author": "Wolber",
"doi-asserted-by": "crossref",
"first-page": "160",
"journal-title": "J. Chem. Inf. Model.",
"key": "ref_50",
"volume": "45",
"year": "2005"
},
{
"DOI": "10.1021/ci200097m",
"article-title": "Pharmer: Efficient and exact pharmacophore search",
"author": "Koes",
"doi-asserted-by": "crossref",
"first-page": "1307",
"journal-title": "J. Chem. Inf. Model.",
"key": "ref_51",
"volume": "51",
"year": "2011"
}
],
"reference-count": 51,
"references-count": 51,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.mdpi.com/1467-3045/48/1/80"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "SARS-CoV-2 Helicase (NSP13) Interacts with Mammalian Polyamine and HSP Partners in Promoting Viral Replication",
"type": "journal-article",
"update-policy": "https://doi.org/10.3390/mdpi_crossmark_policy",
"volume": "48"
}