Analysis of phosphomotifs coupled to phosphoproteome and interactome unveils potential human kinase substrate proteins in SARS-CoV-2
et al., Frontiers in Cellular and Infection Microbiology, doi:10.3389/fcimb.2025.1554760, Jul 2025
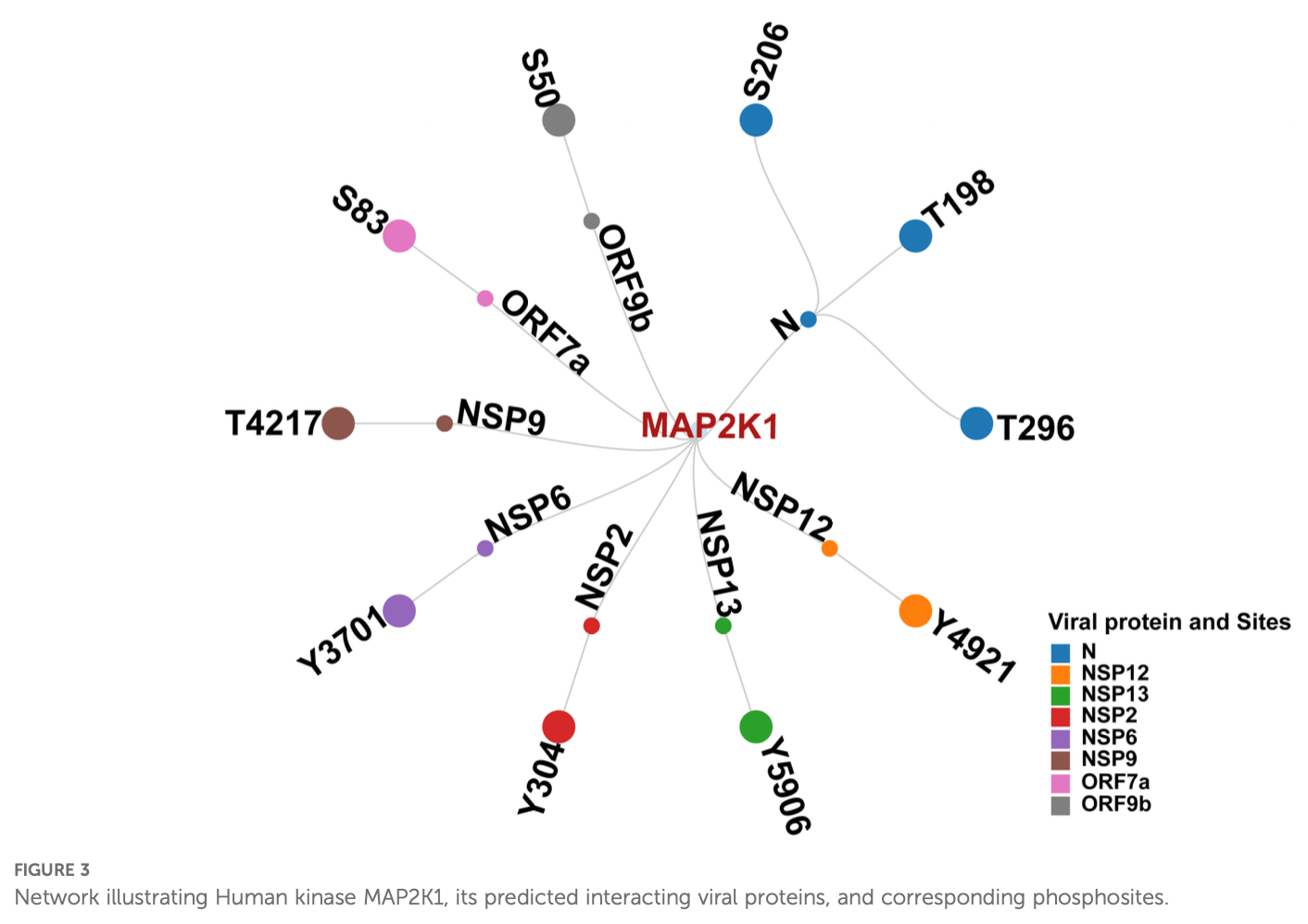
In silico study showing that MAP2K1 (MEK1) inhibition may reduce SARS-CoV-2 replication by disrupting viral protein phosphorylation.
Shaji et al., 9 Jul 2025, peer-reviewed, 9 authors.
Contact: rajrrnbt@gmail.com, rajeshraju@yenepoya.edu.in, ganesh@yenepoya.edu.in.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Analysis of phosphomotifs coupled to phosphoproteome and interactome unveils potential human kinase substrate proteins in SARS-CoV-2
Frontiers in Cellular and Infection Microbiology, doi:10.3389/fcimb.2025.1554760
Introduction: Viruses exploit host kinases to phosphorylate their proteins, enabling viral replication and interference with host-cell functions. Understanding phosphorylation in SARS-CoV-2 proteins necessitates identifying viral phosphoproteins, their phosphosites, and the host kinase-viral protein interactions critical for evading host antiviral responses. Methods: Employing the protein kinase substrate sequence-preference motifs derived by Poll B G. et. al., 2024 , we performed kinase-substrate phosphomotif pattern analysis on the SARS-CoV-2 proteome. We identified major host kinases by analyzing SARS-CoV-2 perturbed phosphoproteomes from various studies and cell systems. These kinases were subjected to interactome analysis and literature-based validation for the impact of kinase inhibitors on infection. Further, conservation of viral phosphosites across SARS CoV-2 variants were also assessed.
Results: The human kinome-substrate phosphomotif analysis predicted 49 kinases capable of phosphorylating 639 phosphosites across 33 SARS-CoV-2 proteins. From these, 24 kinases were also perturbed in SARS-CoV-2-infected phosphoproteomes. Literature review identified seven kinases, including MAP2K1, whose inhibition may reduce viral replication. MAP2K1 was found to target key viral phosphosites, including N protein (S206, T198) and ORF9b (S50), conserved across SARS-CoV-2 variants. Docking analysis showed MAP2K1 forms stronger, closer interactions with N protein compared to SRPK1, highlighting MAP2K1 as a potential host kinase for therapeutic targeting in SARS-CoV-2 infection.
Ethics statement Ethical approval was not required for the study involving humans in accordance with the local legislation and institutional requirements. Written informed consent to participate in this study was not required from the participants or the participants' legal guardians/next of kin in accordance with the national legislation and the institutional requirements.
Author contributions
Conflict of interest The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1554760/ full#supplementary-material
SUPPLEMENTARY FIGURE 1 Docking results of SRPK1 with experimentally validated phosphorylation sites on the N viral protein at T198 and S206.
SUPPLEMENTARY TABLE 1 List of predicted kinases, viral proteins, and their corresponding viral phosphosites.
SUPPLEMENTARY TABLE 2 The table highlights phosphosite-specific regulation (Upregulated or..
References
Al-Qaaneh, Alshammari, Aldahhan, Aldossary, Alkhalifah et al., Genome composition and genetic characterization of SARS-CoV-2, Saudi J. Biol. Sci, doi:10.1016/j.sjbs.2020.12.053
Arya, Kumari, Pandey, Mistry, Bihani et al., Structural insights into SARS-CoV-2 proteins, J. Mol. Biol, doi:10.1016/j.jmb.2020.11.024
Ayinde, Pinheiro, Ramos, Binding of SARS-CoV-2 protein ORF9b to mitochondrial translocase TOM70 prevents its interaction with chaperone HSP90, Biochimie, doi:10.1016/j.biochi.2022.05.016
Batson, Toop, Redondo, Babaei-Jadidi, Chaikuad et al., Development of potent, selective SRPK1 inhibitors as potential topical therapeutics for neovascular eye disease, ACS Chem. Biol, doi:10.1021/acschembio.6b01048
Berman, Westbrook, Feng, Gilliland, Bhat et al., The protein data bank, Nucleic Acids Res, doi:10.1093/nar/28.1.235
Blom, Gammeltoft, Brunak, Sequence and structure-based prediction of eukaryotic protein phosphorylation sites, J. Mol. Biol, doi:10.1006/jmbi.1999.3310
Bouhaddou, Memon, Meyer, White, Rezelj et al., The global phosphorylation landscape of SARS-coV-2 infection, Cell, doi:10.1016/j.cell.2020.06.034
Carabelli, Peacock, Thorne, Harvey, Hughes, SARS-CoV-2 variant biology: immune escape, transmission and fitness, Nat. Rev. Microbiol, doi:10.1038/s41579-022-00841-7
Chatterjee, Thakur, SARS-coV-2 infection triggers phosphorylation: potential target for anti-COVID-19 therapeutics, Front. Immunol, doi:10.3389/fimmu.2022.829474
Chen, Huang, Liao, Chang, Chu, GasPhos: protein phosphorylation site prediction using a new feature selection approach with a GA-aided ant colony system, Int. J. Mol. Sci, doi:10.3390/ijms21217891
Davidson, Williamson, Lewis, Shoemark, Carroll et al., Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein, Genome Med, doi:10.1186/s13073-020-00763-0
Durmus Tekir, Ulgen, Systems biology of pathogen-host interaction: networks of protein-protein interaction within pathogens and pathogenhuman interactions in the post-genomic era, Biotechnol. J, doi:10.1002/biot.201200110
Gopalakrishnan, Shivamurthy, Ahmed, Ummar, Ramesh et al., Positional distribution and conservation of major phosphorylated sites in the human kinome, Front. Mol. Biosci, doi:10.3389/fmolb.2025.1557835
Gordon, Jang, Bouhaddou, Xu, Obernier et al., A SARS-CoV-2 protein interaction map reveals targets for drug repurposing, Nature, doi:10.1038/s41586-020-2286-9
Granata, Pagani, Morenghi, Schiavone, Lezzi et al., Modulation of NBAS-related functions in the early response to SARS-coV-2 infection, Int. J. Mol. Sci, doi:10.3390/ijms24032634
Guirimand, Delmotte, Navratil, VirHostNet 2.0: surfing on the web of virus/host molecular interactions data, Nucleic Acids Res, doi:10.1093/nar/gku1121
Guo, Lei, Chang, Zhao, Wang et al., SARS-CoV-2 hijacks cellular kinase CDK2 to promote viral RNA synthesis, Signal Transduct Target Ther, doi:10.1038/s41392-022-01239-w
Gupta, Azumaya, Moritz, Pourmal, Diallo et al., CryoEM and AI reveal a structure of SARS-CoV-2 Nsp2, a multifunctional protein involved in key host processes, bioRxiv, doi:10.1101/2021.05.10.443524
Gustiananda, Sulistyo, Agustriawan, Andarini, Immunoinformatics analysis of SARS-coV-2 ORF1ab polyproteins to identify promiscuous and highly conserved T-cell epitopes to formulate vaccine for Indonesia and the world population, Vaccines (Basel), doi:10.3390/vaccines9121459
Haling, Sudhamsu, Yen, Sideris, Sandoval et al., Structure of the BRAF-MEK complex reveals a kinase activity independent role for BRAF in MAPK signaling, Cancer Cell, doi:10.1016/j.ccr.2014.07.007
Harrison, Lin, Wang, Mechanisms of SARS-coV-2 transmission and pathogenesis, Trends Immunol, doi:10.1016/j.it.2020.10.004
Hekman, Hume, Goel, Abo, Huang et al., Actionable cytopathogenic host responses of human alveolar type 2 cells to SARS-CoV-2, Mol. Cell, doi:10.1016/j.molcel.2020.12.028
Hekman, Hume, Goel, Abo, Huang et al., Actionable cytopathogenic host responses of human alveolar type 2 cells to SARS-coV-2, Mol. Cell, doi:10.1016/j.molcel.2020.11.028
Hornbeck, Kornhauser, Latham, Murray, Nandhikonda et al., 15 years of PhosphoSitePlus(R): integrating post-translationally modified sites, disease variants and isoforms, Nucleic Acids Res, doi:10.1093/nar/gky1159
Hornbeck, Zhang, Murray, Kornhauser, Latham et al., PhosphoSitePlus 2014: mutations, PTMs and recalibrations, Nucleic Acids Res, doi:10.1093/nar/gku1267
Huang, Yang, Xu, Xu, Liu, Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19, Acta Pharmacol. Sin, doi:10.1038/s41401-020-0485-4
Jacob, Van Den Broeke, Favoreel, Viral serine/threonine protein kinases, J. Virol, doi:10.1128/JVI.01369-10
Jakubiec, Jupin, Regulation of positive-strand RNA virus replication: the emerging role of phosphorylation, Virus Res, doi:10.1016/j.virusres.2007.07.012
Kakavandi, Zare, Vaezjalali, Dadashi, Azarian et al., Structural and non-structural proteins in SARS-CoV-2: potential aspects to COVID-19 treatment or prevention of progression of related diseases, Cell Commun. Signal, doi:10.1186/s12964-023-01104-5
Kerrien, Aranda, Breuza, Bridge, Broackes-Carter et al., The IntAct molecular interaction database in 2012, Nucleic Acids Res, doi:10.1093/nar/gkr1088
Kirchdoerfer, Ward, Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors, Nat. Commun, doi:10.1038/s41467-019-10280-3
Klann, Bojkova, Tascher, Ciesek, Munch et al., Growth factor receptor signaling inhibition prevents SARS-coV-2 replication, Mol. Cell, doi:10.1016/j.molcel.2020.08.006
Li, ClustalW-MPI: ClustalW analysis using distributed and parallel computing, Bioinformatics, doi:10.1093/bioinformatics/btg192
Li, Zhou, Li, Pan, Guo et al., Comprehensive characterization of human-virus protein-protein interactions reveals disease comorbidities and potential antiviral drugs, Comput. Struct. Biotechnol. J, doi:10.1016/j.csbj.2022.03.002
Liu, Huuskonen, Laitinen, Redchuk, Bogacheva et al., SARS-CoV-2-host proteome interactions for antiviral drug discovery, Mol. Syst. Biol, doi:10.15252/msb.202110396
Mahin, Soman, Modi, Raju, Keshava Prasad et al., Meta-analysis of the serum/plasma proteome identifies significant associations between COVID-19 with Alzheimer's/Parkinson's diseases, J. Neurovirol, doi:10.1007/s13365-023-01191-7
Mao, Liu, Phan, Renner, Sun et al., The TRAF3-DYRK1A-RAD54L2 complex maintains ACE2 expression to promote SARS-CoV-2 infection, J. Virol, doi:10.1128/jvi.00347-24
Michel, Mayer, Poch, Thompson, Characterization of accessory genes in coronavirus genomes, Virol. J, doi:10.1186/s12985-020-01402-1
Mizutani, Fukushi, Murakami, Hirano, Saijo et al., Tyrosine dephosphorylation of STAT3 in SARS coronavirus-infected Vero E6 cells, FEBS Lett, doi:10.1016/j.febslet.2004.10.005
Mondol, Hasib, Limon, Alam, Insights into Omicron's Low Fusogenicity through In Silico Molecular Studies on Spike-Furin Interactions, Bioinform. Biol. Insights, doi:10.1177/11779322231189371
Navhaya, Matsebatlela, Monama, Makhoba, In silico discovery and evaluation of inhibitors of the SARS-coV-2 spike protein-HSPA8 complex towards developing COVID-19 therapeutic drugs, Viruses, doi:10.3390/v16111726
Newman, Douangamath, Yadzani, Yosaatmadja, Aimon et al., Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase, Nat. Commun, doi:10.1038/s41467-021-25166-6
Noda, International agency for research on cancer, Jpn J. Clin. Oncol, doi:10.1093/jjco/29.11.592
Oughtred, Rust, Chang, Breitkreutz, Stark et al., The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions, Protein Sci, doi:10.1002/pro.3978
Pham, Phan, Seo, Kim, Song et al., Advancing the accuracy of SARS-CoV-2 phosphorylation site detection via meta-learning approach, Brief Bioinform, doi:10.1093/bib/bbad433
Poll, Leo, Deshpande, Jayatissa, Pisitkun et al., A resource database for protein kinase substrate sequence-preference motifs based on large-scale mass spectrometry data, Cell Commun. Signal, doi:10.1186/s12964-023-01436-2
Raj, Analysis of non-structural proteins, NSPs of SARS-CoV-2 as targets for computational drug designing, Biochem. Biophys. Rep, doi:10.1016/j.bbrep.2020.100847
Rex, Dagamajalu, Kandasamy, Raju, Prasad, SARS-CoV-2 signaling pathway map: A functional landscape of molecular mechanisms in COVID-19, J. Cell Commun. Signal, doi:10.1007/s12079-021-00632-4
Saethang, Hodge, Yang, Zhao, Kimkong et al., PTM-Logo: a program for generation of sequence logos based on positionspecific background amino-acid probabilities, Bioinformatics, doi:10.1093/bioinformatics/btz568
Sarkar, Chamucero-Millares, Rojas, Romulus and remus of inflammation: the conflicting roles of MAP2K1 and MAP2K2 in acute respiratory distress syndrome, Am. J. Respir. Cell Mol. Biol, doi:10.1165/rcmb.2022-0028ED
Saul, Karim, Ghita, Huang, Chiu et al., Anticancer pan-ErbB inhibitors reduce inflammation and tissue injury and exert broad-spectrum antiviral effects, J. Clin. Invest, doi:10.1172/JCI169510
Savastano, Ibanez De Opakua, Rankovic, Zweckstetter, Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerasecontaining condensates, Nat. Commun, doi:10.1038/s41467-020-19843-1
Schreiber, Viemann, Schoning, Schloer, Mecate Zambrano et al., The MEK1/2-inhibitor ATR-002 efficiently blocks SARS-CoV-2 propagation and alleviates pro-inflammatory cytokine/chemokine responses, Cell Mol. Life Sci, doi:10.1007/s00018-021-04085-1
Sommers, Loftus, Jones, Lee, Haren et al., Biochemical analysis of SARS-CoV-2 Nsp13 helicase implicated in COVID-19 and factors that regulate its catalytic functions, J. Biol. Chem, doi:10.1016/j.jbc.2023.102980
Soper, Yardumian, Chen, Yang, Ciervo et al., A repurposed drug interferes with nucleic acid to inhibit the dual activities of coronavirus nsp13, ACS Chem. Biol, doi:10.1021/acschembio.4c00244
Stewart, Lu, O'keefe, Valpadashi, Cruz-Zaragoza et al., The SARS-CoV-2 protein ORF3c is a mitochondrial modulator of innate immunity, iScience, doi:10.1016/j.isci.2023.108080
Strine, Cai, Wei, Alfajaro, Filler et al., DYRK1A promotes viral entry of highly pathogenic human coronaviruses in a kinase-independent manner, PloS Biol, doi:10.1371/journal.pbio.3002097
Stukalov, Girault, Grass, Karayel, Bergant et al., Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV, Nature, doi:10.1038/s41586-021-03493-4
Sugiyama, Cui, Redka, Karimzadeh, Rujas et al., Multiscale interactome analysis coupled with off-target drug predictions reveals drug repurposing candidates for human coronavirus disease, Sci. Rep, doi:10.1038/s41598-021-02432-7
Sugiyama, Imamura, Ishihama, Large-scale discovery of substrates of the human kinome, Sci. Rep, doi:10.1038/s41598-019-46385-4
Theobald, Simonis, Mudler, Gobel, Acton et al., Spleen tyrosine kinase mediates innate and adaptive immune crosstalk in SARS-CoV-2 mRNA vaccination, EMBO Mol. Med, doi:10.15252/emmm.202215888
Tutuncuoglu, Cakir, Batra, Bouhaddou, Eckhardt et al., The landscape of human cancer proteins targeted by SARS-coV-2, Nucleic Acids Res, doi:10.1093/nar/gkac1052
Weckbach, Schweizer, Kraechan, Bieber, Ishikawa-Ankerhold et al., Association of complement and MAPK activation with SARS-coV-2-associated myocardial inflammation, JAMA Cardiol, doi:10.1001/jamacardio.2021.5133
Wigerblad, Warner, Ramos-Benitez, Kardava, Tian et al., Spleen tyrosine kinase inhibition restores myeloid homeostasis in COVID-19, Sci. Adv, doi:10.1126/sciadv.ade8272
Wu, Cheng, Zhou, Sun, Zhang, The SARS-CoV-2 nucleocapsid protein: its role in the viral life cycle, structure and functions, and use as a potential target in the development of vaccines and diagnostics, Virol. J, doi:10.1186/s12985-023-01968-6
Wu, Shi, Pan, Wu, Hou et al., SARS-CoV-2 ORF9b inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO, Cell Rep, doi:10.1016/j.celrep.2021.108761
Wu, Zhao, Yu, Chen, Wang et al., Author Correction: A new coronavirus associated with human respiratory disease in China, Nature, doi:10.1038/s41586-020-2202-3
Xiang, Zou, Zhao, VPTMdb: a viral posttranslational modification database, Brief Bioinform, doi:10.1093/bib/bbaa251
Xie, Klemsz, Kacena, Sandusky, Zhang et al., Inhibition of MEK signaling prevents SARS-CoV2-induced lung damage and improves the survival of infected mice, J. Med. Virol, doi:10.1002/jmv.28094
Xu, Choi, Dai, Luo, Ladak et al., SARS-CoV-2 impairs interferon production via NSP2-induced repression of mRNA translation, Proc. Natl. Acad. Sci. U.S.A, doi:10.1073/pnas.2204539119
Yan, Zheng, Zeng, He, Cheng, Structural biology of SARS-CoV-2: open the door for novel therapies, Signal Transduct Target Ther, doi:10.1038/s41392-022-00884-5
Yang, Lian, Fu, Wuchty, Yang et al., HVIDB: a comprehensive database for human-virus protein-protein interactions, Brief Bioinform, doi:10.1093/bib/bbaa425
Yaron, Heaton, Levy, Johnson, Jordan et al., Host protein kinases required for SARS-CoV-2 nucleocapsid phosphorylation and viral replication, Sci. Signal, doi:10.1126/scisignal.abm0808
Yu, Guan, Miller, Lei, Liu, The role of SARS-CoV-2 nucleocapsid protein in antiviral immunity and vaccine development, Emerg. Microbes Infect, doi:10.1080/22221751.2022.2164219
Zhang, Li, Wang, Sun, Wei et al., Cardiovascular risk after SARS-coV-2 infection is mediated by IL18/IL18R1/HIF-1 signaling pathway axis, Front. Immunol, doi:10.3389/fimmu.2021.780804
Zhang, Xin, Liu, Jiang, Han et al., SARS-COV-2 protein NSP9 promotes cytokine production by targeting TBK1, Front. Immunol, doi:10.3389/fimmu.2023.1211816
Zhu, Jiang, Chen, Jouha, Wang et al., A postpandemic perspective: Evolution of SARS-CoV-2 early detection, Trac-Trends Analytical Chem, doi:10.1016/j.trac.2023.117458
Zumla, Chan, Azhar, Hui, Yuen, Coronaviruses -drug discovery and therapeutic options, Nat. Rev. Drug Discov, doi:10.1038/nrd.2015.37
DOI record:
{
"DOI": "10.3389/fcimb.2025.1554760",
"ISSN": [
"2235-2988"
],
"URL": "http://dx.doi.org/10.3389/fcimb.2025.1554760",
"abstract": "<jats:sec><jats:title>Introduction</jats:title><jats:p>Viruses exploit host kinases to phosphorylate their proteins, enabling viral replication and interference with host-cell functions. Understanding phosphorylation in SARS-CoV-2 proteins necessitates identifying viral phosphoproteins, their phosphosites, and the host kinase–viral protein interactions critical for evading host antiviral responses.</jats:p></jats:sec><jats:sec><jats:title>Methods</jats:title><jats:p>Employing the protein kinase substrate sequence-preference motifs derived by Poll B G. <jats:italic>et. al</jats:italic>., 2024, we performed kinase-substrate phosphomotif pattern analysis on the SARS-CoV-2 proteome. We identified major host kinases by analyzing SARS-CoV-2 perturbed phosphoproteomes from various studies and cell systems. These kinases were subjected to interactome analysis and literature-based validation for the impact of kinase inhibitors on infection. Further, conservation of viral phosphosites across SARS CoV-2 variants were also assessed.</jats:p></jats:sec><jats:sec><jats:title>Results</jats:title><jats:p>The human kinome–substrate phosphomotif analysis predicted 49 kinases capable of phosphorylating 639 phosphosites across 33 SARS-CoV-2 proteins. From these, 24 kinases were also perturbed in SARS-CoV-2-infected phosphoproteomes. Literature review identified seven kinases, including MAP2K1, whose inhibition may reduce viral replication. MAP2K1 was found to target key viral phosphosites, including N protein (S206, T198) and ORF9b (S50), conserved across SARS-CoV-2 variants. Docking analysis showed MAP2K1 forms stronger, closer interactions with N protein compared to SRPK1, highlighting MAP2K1 as a potential host kinase for therapeutic targeting in SARS-CoV-2 infection.</jats:p></jats:sec><jats:sec><jats:title>Discussion and Conclusions</jats:title><jats:p>This study presents a framework for predicting human kinases of specific SARS-CoV-2 protein phosphosites by integrating kinase specificity, virus–host interactions, and post-translational modifications. MAP2K1 was identified as a key host kinase, showing stronger interactions than SRPK1, and is proposed as an antiviral drug target for repurposing in SARS-CoV-2 infections.</jats:p></jats:sec>",
"alternative-id": [
"10.3389/fcimb.2025.1554760"
],
"article-number": "1554760",
"author": [
{
"affiliation": [],
"family": "Shaji",
"given": "Vineetha",
"sequence": "first"
},
{
"affiliation": [],
"family": "Rafi",
"given": "Ahmad",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Ahmed",
"given": "Mukhtar",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Gopalakrishnan",
"given": "Athira Perunelly",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Soman",
"given": "Sowmya",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Revikumar",
"given": "Amjesh",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Prasad",
"given": "Ganesh",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Jayanandan",
"given": "Abhithaj",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Raju",
"given": "Rajesh",
"sequence": "additional"
}
],
"container-title": "Frontiers in Cellular and Infection Microbiology",
"container-title-short": "Front. Cell. Infect. Microbiol.",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"frontiersin.org"
]
},
"created": {
"date-parts": [
[
2025,
7,
10
]
],
"date-time": "2025-07-10T05:47:22Z",
"timestamp": 1752126442000
},
"deposited": {
"date-parts": [
[
2025,
7,
10
]
],
"date-time": "2025-07-10T13:38:15Z",
"timestamp": 1752154695000
},
"funder": [
{
"DOI": "10.13039/501100002383",
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/501100002383",
"id-type": "DOI"
}
],
"name": "King Saud University"
}
],
"indexed": {
"date-parts": [
[
2025,
7,
10
]
],
"date-time": "2025-07-10T14:10:04Z",
"timestamp": 1752156604963,
"version": "3.41.2"
},
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2025,
7,
9
]
]
},
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
7,
9
]
],
"date-time": "2025-07-09T00:00:00Z",
"timestamp": 1752019200000
}
}
],
"link": [
{
"URL": "https://www.frontiersin.org/articles/10.3389/fcimb.2025.1554760/full",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "1965",
"original-title": [],
"prefix": "10.3389",
"published": {
"date-parts": [
[
2025,
7,
9
]
]
},
"published-online": {
"date-parts": [
[
2025,
7,
9
]
]
},
"publisher": "Frontiers Media SA",
"reference": [
{
"DOI": "10.1016/j.sjbs.2020.12.053",
"article-title": "Genome composition and genetic characterization of SARS-CoV-2",
"author": "Al-Qaaneh",
"doi-asserted-by": "publisher",
"first-page": "1978",
"journal-title": "Saudi J. Biol. Sci.",
"key": "B1",
"volume": "28",
"year": "2021"
},
{
"DOI": "10.1016/j.jmb.2020.11.024",
"article-title": "Structural insights into SARS-CoV-2 proteins",
"author": "Arya",
"doi-asserted-by": "publisher",
"first-page": "166725",
"journal-title": "J. Mol. Biol.",
"key": "B2",
"volume": "433",
"year": "2021"
},
{
"DOI": "10.1016/j.biochi.2022.05.016",
"article-title": "Binding of SARS-CoV-2 protein ORF9b to mitochondrial translocase TOM70 prevents its interaction with chaperone HSP90",
"author": "Ayinde",
"doi-asserted-by": "publisher",
"first-page": "99",
"journal-title": "Biochimie",
"key": "B3",
"volume": "200",
"year": "2022"
},
{
"DOI": "10.1021/acschembio.6b01048",
"article-title": "Development of potent, selective SRPK1 inhibitors as potential topical therapeutics for neovascular eye disease",
"author": "Batson",
"doi-asserted-by": "publisher",
"first-page": "825",
"journal-title": "ACS Chem. Biol.",
"key": "B4",
"volume": "12",
"year": "2017"
},
{
"DOI": "10.1093/nar/28.1.235",
"article-title": "The protein data bank",
"author": "Berman",
"doi-asserted-by": "publisher",
"first-page": "235",
"journal-title": "Nucleic Acids Res.",
"key": "B5",
"volume": "28",
"year": "2000"
},
{
"DOI": "10.1006/jmbi.1999.3310",
"article-title": "Sequence and structure-based prediction of eukaryotic protein phosphorylation sites",
"author": "Blom",
"doi-asserted-by": "publisher",
"first-page": "1351",
"journal-title": "J. Mol. Biol.",
"key": "B6",
"volume": "294",
"year": "1999"
},
{
"DOI": "10.1016/j.cell.2020.06.034",
"article-title": "The global phosphorylation landscape of SARS-coV-2 infection",
"author": "Bouhaddou",
"doi-asserted-by": "publisher",
"first-page": "685",
"journal-title": "Cell",
"key": "B7",
"volume": "182",
"year": "2020"
},
{
"DOI": "10.1038/s41579-022-00841-7",
"article-title": "SARS-CoV-2 variant biology: immune escape, transmission and fitness",
"author": "Carabelli",
"doi-asserted-by": "publisher",
"first-page": "162",
"journal-title": "Nat. Rev. Microbiol.",
"key": "B8",
"volume": "21",
"year": "2023"
},
{
"DOI": "10.3389/fimmu.2022.829474",
"article-title": "SARS-coV-2 infection triggers phosphorylation: potential target for anti-COVID-19 therapeutics",
"author": "Chatterjee",
"doi-asserted-by": "publisher",
"journal-title": "Front. Immunol.",
"key": "B9",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.3390/ijms21217891",
"article-title": "GasPhos: protein phosphorylation site prediction using a new feature selection approach with a GA-aided ant colony system",
"author": "Chen",
"doi-asserted-by": "publisher",
"journal-title": "Int. J. Mol. Sci.",
"key": "B10",
"volume": "21",
"year": "2020"
},
{
"DOI": "10.1186/s13073-020-00763-0",
"article-title": "Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein",
"author": "Davidson",
"doi-asserted-by": "publisher",
"first-page": "68",
"journal-title": "Genome Med.",
"key": "B11",
"volume": "12",
"year": "2020"
},
{
"DOI": "10.1002/biot.201200110",
"article-title": "Systems biology of pathogen-host interaction: networks of protein-protein interaction within pathogens and pathogen-human interactions in the post-genomic era",
"author": "Durmus Tekir",
"doi-asserted-by": "publisher",
"first-page": "85",
"journal-title": "Biotechnol. J.",
"key": "B12",
"volume": "8",
"year": "2013"
},
{
"DOI": "10.3389/fmolb.2025.1557835",
"article-title": "Positional distribution and conservation of major phosphorylated sites in the human kinome",
"author": "Gopalakrishnan",
"doi-asserted-by": "publisher",
"journal-title": "Front. Mol. Biosci.",
"key": "B13",
"volume": "12",
"year": "2025"
},
{
"DOI": "10.1038/s41586-020-2286-9",
"article-title": "A SARS-CoV-2 protein interaction map reveals targets for drug repurposing",
"author": "Gordon",
"doi-asserted-by": "publisher",
"first-page": "459",
"journal-title": "Nature",
"key": "B14",
"volume": "583",
"year": "2020"
},
{
"DOI": "10.3390/ijms24032634",
"article-title": "Modulation of NBAS-related functions in the early response to SARS-coV-2 infection",
"author": "Granata",
"doi-asserted-by": "publisher",
"journal-title": "Int. J. Mol. Sci.",
"key": "B15",
"volume": "24",
"year": "2023"
},
{
"DOI": "10.1093/nar/gku1121",
"article-title": "VirHostNet 2.0: surfing on the web of virus/host molecular interactions data",
"author": "Guirimand",
"doi-asserted-by": "publisher",
"first-page": "D583",
"journal-title": "Nucleic Acids Res.",
"key": "B16",
"volume": "43",
"year": "2015"
},
{
"DOI": "10.1038/s41392-022-01239-w",
"article-title": "SARS-CoV-2 hijacks cellular kinase CDK2 to promote viral RNA synthesis",
"author": "Guo",
"doi-asserted-by": "publisher",
"first-page": "400",
"journal-title": "Signal Transduct Target Ther.",
"key": "B17",
"volume": "7",
"year": "2022"
},
{
"DOI": "10.1101/2021.05.10.443524",
"article-title": "CryoEM and AI reveal a structure of SARS-CoV-2 Nsp2, a multifunctional protein involved in key host processes",
"author": "Gupta",
"doi-asserted-by": "publisher",
"first-page": "2021.05.10.443524",
"journal-title": "bioRxiv",
"key": "B18",
"year": "2021"
},
{
"DOI": "10.3390/vaccines9121459",
"article-title": "Immunoinformatics analysis of SARS-coV-2 ORF1ab polyproteins to identify promiscuous and highly conserved T-cell epitopes to formulate vaccine for Indonesia and the world population",
"author": "Gustiananda",
"doi-asserted-by": "publisher",
"first-page": "1459",
"journal-title": "Vaccines (Basel)",
"key": "B19",
"volume": "9",
"year": "2021"
},
{
"DOI": "10.1016/j.ccr.2014.07.007",
"article-title": "Structure of the BRAF-MEK complex reveals a kinase activity independent role for BRAF in MAPK signaling",
"author": "Haling",
"doi-asserted-by": "publisher",
"first-page": "402",
"journal-title": "Cancer Cell",
"key": "B20",
"volume": "26",
"year": "2014"
},
{
"DOI": "10.1016/j.it.2020.10.004",
"article-title": "Mechanisms of SARS-coV-2 transmission and pathogenesis",
"author": "Harrison",
"doi-asserted-by": "publisher",
"first-page": "1100",
"journal-title": "Trends Immunol.",
"key": "B21",
"volume": "41",
"year": "2020"
},
{
"DOI": "10.1016/j.molcel.2020.11.028",
"article-title": "Actionable cytopathogenic host responses of human alveolar type 2 cells to SARS-coV-2",
"author": "Hekman",
"doi-asserted-by": "publisher",
"first-page": "1104",
"journal-title": "Mol. Cell",
"key": "B22",
"volume": "80",
"year": "2020"
},
{
"DOI": "10.1016/j.molcel.2020.12.028",
"article-title": "Actionable cytopathogenic host responses of human alveolar type 2 cells to SARS-CoV-2",
"author": "Hekman",
"doi-asserted-by": "publisher",
"first-page": "2125",
"journal-title": "Mol. Cell",
"key": "B23",
"volume": "81",
"year": "2021"
},
{
"DOI": "10.1093/nar/gky1159",
"article-title": "15 years of PhosphoSitePlus(R): integrating post-translationally modified sites, disease variants and isoforms",
"author": "Hornbeck",
"doi-asserted-by": "publisher",
"first-page": "D433",
"journal-title": "Nucleic Acids Res.",
"key": "B24",
"volume": "47",
"year": "2019"
},
{
"DOI": "10.1093/nar/gku1267",
"article-title": "PhosphoSitePlus 2014: mutations, PTMs and recalibrations",
"author": "Hornbeck",
"doi-asserted-by": "publisher",
"first-page": "D512",
"journal-title": "Nucleic Acids Res.",
"key": "B25",
"volume": "43",
"year": "2015"
},
{
"DOI": "10.1038/s41401-020-0485-4",
"article-title": "Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19",
"author": "Huang",
"doi-asserted-by": "publisher",
"first-page": "1141",
"journal-title": "Acta Pharmacol. Sin.",
"key": "B26",
"volume": "41",
"year": "2020"
},
{
"DOI": "10.1128/JVI.01369-10",
"article-title": "Viral serine/threonine protein kinases",
"author": "Jacob",
"doi-asserted-by": "publisher",
"first-page": "1158",
"journal-title": "J. Virol.",
"key": "B27",
"volume": "85",
"year": "2011"
},
{
"DOI": "10.1016/j.virusres.2007.07.012",
"article-title": "Regulation of positive-strand RNA virus replication: the emerging role of phosphorylation",
"author": "Jakubiec",
"doi-asserted-by": "publisher",
"first-page": "73",
"journal-title": "Virus Res.",
"key": "B28",
"volume": "129",
"year": "2007"
},
{
"DOI": "10.1186/s12964-023-01104-5",
"article-title": "Structural and non-structural proteins in SARS-CoV-2: potential aspects to COVID-19 treatment or prevention of progression of related diseases",
"author": "Kakavandi",
"doi-asserted-by": "publisher",
"journal-title": "Cell Commun. Signal",
"key": "B29",
"volume": "21",
"year": "2023"
},
{
"DOI": "10.1093/nar/gkr1088",
"article-title": "The IntAct molecular interaction database in 2012",
"author": "Kerrien",
"doi-asserted-by": "publisher",
"first-page": "D841",
"journal-title": "Nucleic Acids Res.",
"key": "B30",
"volume": "40",
"year": "2012"
},
{
"DOI": "10.1038/s41467-019-10280-3",
"article-title": "Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors",
"author": "Kirchdoerfer",
"doi-asserted-by": "publisher",
"first-page": "2342",
"journal-title": "Nat. Commun.",
"key": "B31",
"volume": "10",
"year": "2019"
},
{
"DOI": "10.1016/j.molcel.2020.08.006",
"article-title": "Growth factor receptor signaling inhibition prevents SARS-coV-2 replication",
"author": "Klann",
"doi-asserted-by": "publisher",
"first-page": "164",
"journal-title": "Mol. Cell",
"key": "B32",
"volume": "80",
"year": "2020"
},
{
"DOI": "10.1093/bioinformatics/btg192",
"article-title": "ClustalW-MPI: ClustalW analysis using distributed and parallel computing",
"author": "Li",
"doi-asserted-by": "publisher",
"first-page": "1585",
"journal-title": "Bioinformatics",
"key": "B33",
"volume": "19",
"year": "2003"
},
{
"DOI": "10.1016/j.csbj.2022.03.002",
"article-title": "Comprehensive characterization of human-virus protein-protein interactions reveals disease comorbidities and potential antiviral drugs",
"author": "Li",
"doi-asserted-by": "publisher",
"first-page": "1244",
"journal-title": "Comput. Struct. Biotechnol. J.",
"key": "B34",
"volume": "20",
"year": "2022"
},
{
"DOI": "10.15252/msb.202110396",
"article-title": "SARS-CoV-2-host proteome interactions for antiviral drug discovery",
"author": "Liu",
"doi-asserted-by": "publisher",
"journal-title": "Mol. Syst. Biol.",
"key": "B35",
"volume": "17",
"year": "2021"
},
{
"DOI": "10.1007/s13365-023-01191-7",
"article-title": "Meta-analysis of the serum/plasma proteome identifies significant associations between COVID-19 with Alzheimer's/Parkinson's diseases",
"author": "Mahin",
"doi-asserted-by": "publisher",
"first-page": "57",
"journal-title": "J. Neurovirol",
"key": "B36",
"volume": "30",
"year": "2024"
},
{
"DOI": "10.1128/jvi.00347-24",
"article-title": "The TRAF3-DYRK1A-RAD54L2 complex maintains ACE2 expression to promote SARS-CoV-2 infection",
"author": "Mao",
"doi-asserted-by": "publisher",
"journal-title": "J. Virol.",
"key": "B37",
"volume": "98",
"year": "2024"
},
{
"DOI": "10.1186/s12985-020-01402-1",
"article-title": "Characterization of accessory genes in coronavirus genomes",
"author": "Michel",
"doi-asserted-by": "publisher",
"first-page": "131",
"journal-title": "Virol. J.",
"key": "B38",
"volume": "17",
"year": "2020"
},
{
"DOI": "10.1016/j.febslet.2004.10.005",
"article-title": "Tyrosine dephosphorylation of STAT3 in SARS coronavirus-infected Vero E6 cells",
"author": "Mizutani",
"doi-asserted-by": "publisher",
"first-page": "187",
"journal-title": "FEBS Lett.",
"key": "B39",
"volume": "577",
"year": "2004"
},
{
"DOI": "10.1177/11779322231189371",
"article-title": "Insights into Omicron's Low Fusogenicity through In Silico Molecular Studies on Spike-Furin Interactions",
"author": "Mondol",
"doi-asserted-by": "publisher",
"journal-title": "Bioinform. Biol. Insights",
"key": "B40",
"volume": "17",
"year": "2023"
},
{
"DOI": "10.3390/v16111726",
"article-title": "In silico discovery and evaluation of inhibitors of the SARS-coV-2 spike protein-HSPA8 complex towards developing COVID-19 therapeutic drugs",
"author": "Navhaya",
"doi-asserted-by": "publisher",
"journal-title": "Viruses",
"key": "B41",
"volume": "16",
"year": "2024"
},
{
"DOI": "10.1038/s41467-021-25166-6",
"article-title": "Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase",
"author": "Newman",
"doi-asserted-by": "publisher",
"first-page": "4848",
"journal-title": "Nat. Commun.",
"key": "B42",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1093/jjco/29.11.592",
"article-title": "International agency for research on cancer (http://www.iarc.fr/)",
"author": "Noda",
"doi-asserted-by": "publisher",
"journal-title": "Jpn J. Clin. Oncol.",
"key": "B43",
"volume": "29",
"year": "1999"
},
{
"DOI": "10.1002/pro.3978",
"article-title": "The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions",
"author": "Oughtred",
"doi-asserted-by": "publisher",
"first-page": "187",
"journal-title": "Protein Sci.",
"key": "B44",
"volume": "30",
"year": "2021"
},
{
"DOI": "10.1093/bib/bbad433",
"article-title": "Advancing the accuracy of SARS-CoV-2 phosphorylation site detection via meta-learning approach",
"author": "Pham",
"doi-asserted-by": "publisher",
"journal-title": "Brief Bioinform.",
"key": "B45",
"volume": "25",
"year": "2023"
},
{
"DOI": "10.1186/s12964-023-01436-2",
"article-title": "A resource database for protein kinase substrate sequence-preference motifs based on large-scale mass spectrometry data",
"author": "Poll",
"doi-asserted-by": "publisher",
"first-page": "137",
"journal-title": "Cell Commun. Signal",
"key": "B46",
"volume": "22",
"year": "2024"
},
{
"DOI": "10.1016/j.bbrep.2020.100847",
"article-title": "Analysis of non-structural proteins, NSPs of SARS-CoV-2 as targets for computational drug designing",
"author": "Raj",
"doi-asserted-by": "publisher",
"journal-title": "Biochem. Biophys. Rep.",
"key": "B47",
"volume": "25",
"year": "2021"
},
{
"DOI": "10.1007/s12079-021-00632-4",
"article-title": "SARS-CoV-2 signaling pathway map: A functional landscape of molecular mechanisms in COVID-19",
"author": "Rex",
"doi-asserted-by": "publisher",
"first-page": "601",
"journal-title": "J. Cell Commun. Signal",
"key": "B48",
"volume": "15",
"year": "2021"
},
{
"DOI": "10.1093/bioinformatics/btz568",
"article-title": "PTM-Logo: a program for generation of sequence logos based on position-specific background amino-acid probabilities",
"author": "Saethang",
"doi-asserted-by": "publisher",
"first-page": "5313",
"journal-title": "Bioinformatics",
"key": "B49",
"volume": "35",
"year": "2019"
},
{
"DOI": "10.1165/rcmb.2022-0028ED",
"article-title": "Romulus and remus of inflammation: the conflicting roles of MAP2K1 and MAP2K2 in acute respiratory distress syndrome",
"author": "Sarkar",
"doi-asserted-by": "publisher",
"first-page": "479",
"journal-title": "Am. J. Respir. Cell Mol. Biol.",
"key": "B50",
"volume": "66",
"year": "2022"
},
{
"DOI": "10.1172/JCI169510",
"article-title": "Anticancer pan-ErbB inhibitors reduce inflammation and tissue injury and exert broad-spectrum antiviral effects",
"author": "Saul",
"doi-asserted-by": "publisher",
"journal-title": "J. Clin. Invest.",
"key": "B51",
"volume": "133",
"year": "2023"
},
{
"DOI": "10.1038/s41467-020-19843-1",
"article-title": "Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates",
"author": "Savastano",
"doi-asserted-by": "publisher",
"first-page": "6041",
"journal-title": "Nat. Commun.",
"key": "B52",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1007/s00018-021-04085-1",
"article-title": "The MEK1/2-inhibitor ATR-002 efficiently blocks SARS-CoV-2 propagation and alleviates pro-inflammatory cytokine/chemokine responses",
"author": "Schreiber",
"doi-asserted-by": "publisher",
"first-page": "65",
"journal-title": "Cell Mol. Life Sci.",
"key": "B53",
"volume": "79",
"year": "2022"
},
{
"DOI": "10.1016/j.jbc.2023.102980",
"article-title": "Biochemical analysis of SARS-CoV-2 Nsp13 helicase implicated in COVID-19 and factors that regulate its catalytic functions",
"author": "Sommers",
"doi-asserted-by": "publisher",
"first-page": "1029805",
"journal-title": "J. Biol. Chem.",
"key": "B54",
"volume": "299",
"year": "2023"
},
{
"DOI": "10.1021/acschembio.4c00244",
"article-title": "A repurposed drug interferes with nucleic acid to inhibit the dual activities of coronavirus nsp13",
"author": "Soper",
"doi-asserted-by": "publisher",
"first-page": "1593",
"journal-title": "ACS Chem. Biol.",
"key": "B55",
"volume": "19",
"year": "2024"
},
{
"DOI": "10.1016/j.isci.2023.108080",
"article-title": "The SARS-CoV-2 protein ORF3c is a mitochondrial modulator of innate immunity",
"author": "Stewart",
"doi-asserted-by": "publisher",
"journal-title": "iScience",
"key": "B56",
"volume": "26",
"year": "2023"
},
{
"DOI": "10.1371/journal.pbio.3002097",
"article-title": "DYRK1A promotes viral entry of highly pathogenic human coronaviruses in a kinase-independent manner",
"author": "Strine",
"doi-asserted-by": "publisher",
"journal-title": "PloS Biol.",
"key": "B57",
"volume": "21",
"year": "2023"
},
{
"DOI": "10.1038/s41586-021-03493-4",
"article-title": "Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV",
"author": "Stukalov",
"doi-asserted-by": "publisher",
"first-page": "246",
"journal-title": "Nature",
"key": "B58",
"volume": "594",
"year": "2021"
},
{
"DOI": "10.1038/s41598-021-02432-7",
"article-title": "Multiscale interactome analysis coupled with off-target drug predictions reveals drug repurposing candidates for human coronavirus disease",
"author": "Sugiyama",
"doi-asserted-by": "publisher",
"first-page": "23315",
"journal-title": "Sci. Rep.",
"key": "B59",
"volume": "11",
"year": "2021"
},
{
"DOI": "10.1038/s41598-019-46385-4",
"article-title": "Large-scale discovery of substrates of the human kinome",
"author": "Sugiyama",
"doi-asserted-by": "publisher",
"first-page": "10503",
"journal-title": "Sci. Rep.",
"key": "B60",
"volume": "9",
"year": "2019"
},
{
"DOI": "10.15252/emmm.202215888",
"article-title": "Spleen tyrosine kinase mediates innate and adaptive immune crosstalk in SARS-CoV-2 mRNA vaccination",
"author": "Theobald",
"doi-asserted-by": "publisher",
"journal-title": "EMBO Mol. Med.",
"key": "B61",
"volume": "14",
"year": "2022"
},
{
"DOI": "10.1158/2159-8290.CD-20-0559",
"article-title": "The landscape of human cancer proteins targeted by SARS-coV-2",
"author": "Tutuncuoglu",
"doi-asserted-by": "publisher",
"first-page": "916",
"journal-title": "Cancer Discov.",
"key": "B62",
"volume": "10",
"year": "2020"
},
{
"DOI": "10.1093/nar/gkac1052",
"article-title": "UniProt: the universal protein knowledgebase in 2023",
"doi-asserted-by": "publisher",
"first-page": "D523",
"journal-title": "Nucleic Acids Res.",
"key": "B63",
"volume": "51",
"year": "2023"
},
{
"DOI": "10.1001/jamacardio.2021.5133",
"article-title": "Association of complement and MAPK activation with SARS-coV-2-associated myocardial inflammation",
"author": "Weckbach",
"doi-asserted-by": "publisher",
"first-page": "286",
"journal-title": "JAMA Cardiol.",
"key": "B64",
"volume": "7",
"year": "2022"
},
{
"DOI": "10.1126/sciadv.ade8272",
"article-title": "Spleen tyrosine kinase inhibition restores myeloid homeostasis in COVID-19",
"author": "Wigerblad",
"doi-asserted-by": "publisher",
"journal-title": "Sci. Adv.",
"key": "B65",
"volume": "9",
"year": "2023"
},
{
"DOI": "10.1186/s12985-023-01968-6",
"article-title": "The SARS-CoV-2 nucleocapsid protein: its role in the viral life cycle, structure and functions, and use as a potential target in the development of vaccines and diagnostics",
"author": "Wu",
"doi-asserted-by": "publisher",
"journal-title": "Virol. J.",
"key": "B66",
"volume": "20",
"year": "2023"
},
{
"DOI": "10.1016/j.celrep.2021.108761",
"article-title": "SARS-CoV-2 ORF9b inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO",
"author": "Wu",
"doi-asserted-by": "publisher",
"journal-title": "Cell Rep.",
"key": "B67",
"volume": "34",
"year": "2021"
},
{
"DOI": "10.1038/s41586-020-2202-3",
"article-title": "Author Correction: A new coronavirus associated with human respiratory disease in China",
"author": "Wu",
"doi-asserted-by": "publisher",
"journal-title": "Nature",
"key": "B68",
"volume": "580",
"year": "2020"
},
{
"DOI": "10.1093/bib/bbaa251",
"article-title": "VPTMdb: a viral posttranslational modification database",
"author": "Xiang",
"doi-asserted-by": "publisher",
"journal-title": "Brief Bioinform.",
"key": "B69",
"volume": "22",
"year": "2021"
},
{
"DOI": "10.1002/jmv.28094",
"article-title": "Inhibition of MEK signaling prevents SARS-CoV2-induced lung damage and improves the survival of infected mice",
"author": "Xie",
"doi-asserted-by": "publisher",
"first-page": "6097",
"journal-title": "J. Med. Virol.",
"key": "B70",
"volume": "94",
"year": "2022"
},
{
"DOI": "10.1073/pnas.2204539119",
"article-title": "SARS-CoV-2 impairs interferon production via NSP2-induced repression of mRNA translation",
"author": "Xu",
"doi-asserted-by": "publisher",
"journal-title": "Proc. Natl. Acad. Sci. U.S.A.",
"key": "B71",
"volume": "119",
"year": "2022"
},
{
"DOI": "10.1038/s41392-022-00884-5",
"article-title": "Structural biology of SARS-CoV-2: open the door for novel therapies",
"author": "Yan",
"doi-asserted-by": "publisher",
"first-page": "26",
"journal-title": "Signal Transduct Target Ther.",
"key": "B72",
"volume": "7",
"year": "2022"
},
{
"DOI": "10.1093/bib/bbaa425",
"article-title": "HVIDB: a comprehensive database for human-virus protein-protein interactions",
"author": "Yang",
"doi-asserted-by": "publisher",
"first-page": "832",
"journal-title": "Brief Bioinform.",
"key": "B73",
"volume": "22",
"year": "2021"
},
{
"DOI": "10.1126/scisignal.abm0808",
"article-title": "Host protein kinases required for SARS-CoV-2 nucleocapsid phosphorylation and viral replication",
"author": "Yaron",
"doi-asserted-by": "publisher",
"journal-title": "Sci. Signal",
"key": "B74",
"volume": "15",
"year": "2022"
},
{
"DOI": "10.1080/22221751.2022.2164219",
"article-title": "The role of SARS-CoV-2 nucleocapsid protein in antiviral immunity and vaccine development",
"author": "Yu",
"doi-asserted-by": "publisher",
"journal-title": "Emerg. Microbes Infect.",
"key": "B75",
"volume": "12",
"year": "2023"
},
{
"DOI": "10.3389/fimmu.2021.780804",
"article-title": "Cardiovascular risk after SARS-coV-2 infection is mediated by IL18/IL18R1/HIF-1 signaling pathway axis",
"author": "Zhang",
"doi-asserted-by": "publisher",
"journal-title": "Front. Immunol.",
"key": "B76",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.3389/fimmu.2023.1211816",
"article-title": "SARS-COV-2 protein NSP9 promotes cytokine production by targeting TBK1",
"author": "Zhang",
"doi-asserted-by": "publisher",
"journal-title": "Front. Immunol.",
"key": "B77",
"volume": "14",
"year": "2023"
},
{
"DOI": "10.1016/j.trac.2023.117458",
"article-title": "A post-pandemic perspective: Evolution of SARS-CoV-2 early detection",
"author": "Zhu",
"doi-asserted-by": "publisher",
"journal-title": "Trac-Trends Analytical Chem.",
"key": "B78",
"volume": "170",
"year": "2024"
},
{
"DOI": "10.1038/nrd.2015.37",
"article-title": "Coronaviruses - drug discovery and therapeutic options",
"author": "Zumla",
"doi-asserted-by": "publisher",
"first-page": "327",
"journal-title": "Nat. Rev. Drug Discov.",
"key": "B79",
"volume": "15",
"year": "2016"
}
],
"reference-count": 79,
"references-count": 79,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.frontiersin.org/articles/10.3389/fcimb.2025.1554760/full"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Analysis of phosphomotifs coupled to phosphoproteome and interactome unveils potential human kinase substrate proteins in SARS-CoV-2",
"type": "journal-article",
"update-policy": "https://doi.org/10.3389/crossmark-policy",
"volume": "15"
}