Ensovibep, a novel trispecific DARPin candidate that protects against SARS-CoV-2 variants
et al., bioRxiv, doi:10.1101/2021.02.03.429164, Feb 2021
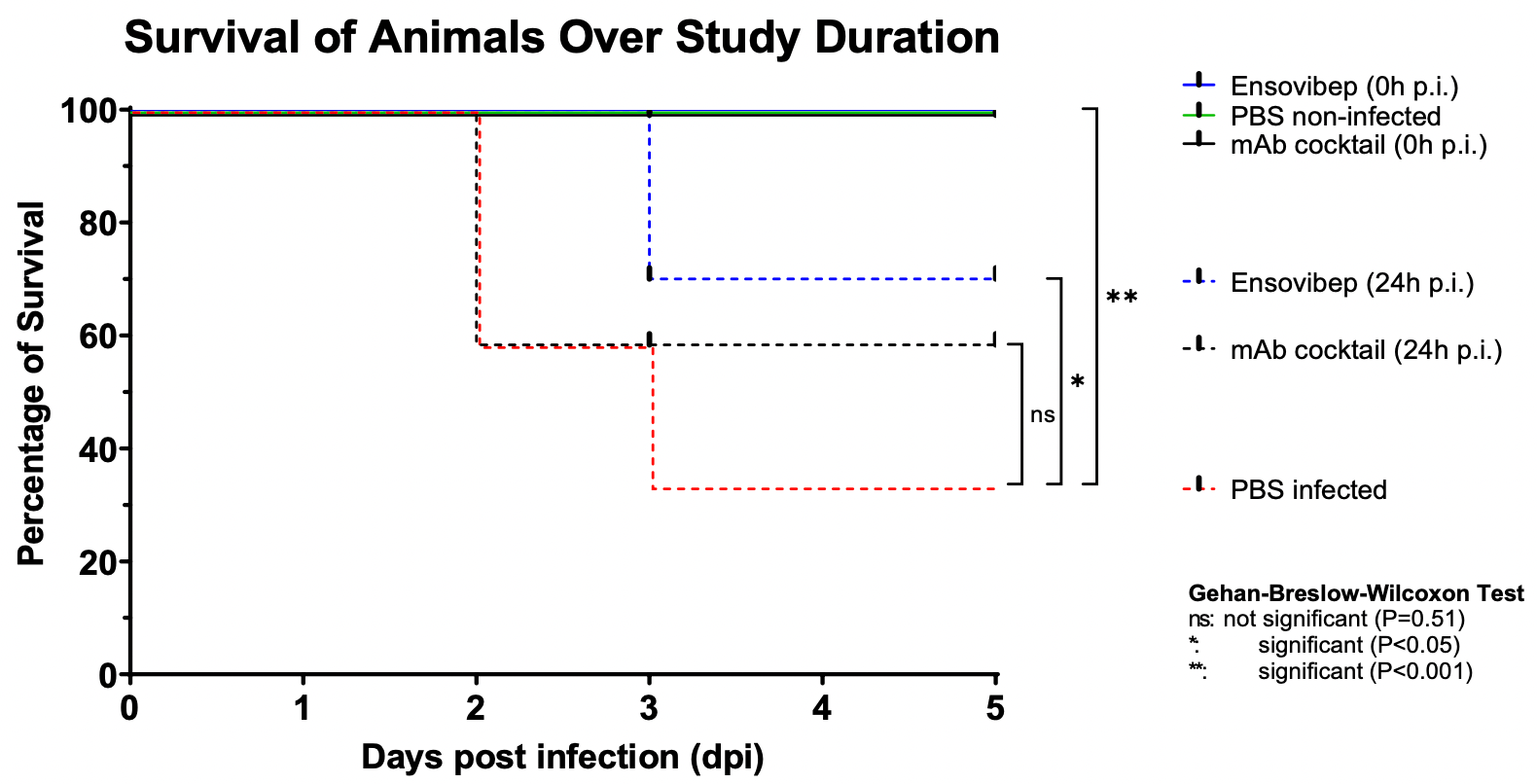
In silico, in vitro, and hamster study of ensovibep, a DARPin candidate that can engage all three units of the spike protein trimer to inhibit ACE2 interaction, showing efficacy for common variants, and efficacy comparable to casirivimab/imdevimab in hamsters.
Rothenberger et al., 3 Feb 2021, preprint, 65 authors.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
Ensovibep, a novel trispecific DARPin candidate that protects against SARS-CoV-2 variants
doi:10.1101/2021.02.03.429164
SARS-CoV-2 has infected millions of people globally and continues to undergo evolution. Emerging variants can be partially resistant to vaccine induced and therapeutic antibodies, emphasizing the urgent need for accessible, broad-spectrum therapeutics. Here, we report a comprehensive study of ensovibep, the first trispecific clinical DARPin candidate, that can simultaneously engage all three units of the spike protein trimer to potently inhibit ACE2 interaction, as revealed by structural analyses. The cooperative binding of the individual modules enables ensovibep to retain inhibitory potency against all frequent SARS-CoV-2 variants, including Omicron, as of December 2021. Moreover, viral passaging experiments show that ensovibep, when used as a single agent, can prevent development of escape mutations comparably to a cocktail of monoclonal antibodies (mAb). Finally, we demonstrate that the very high in vitro antiviral potency also translates into significant therapeutic protection and reduction of pathogenesis in Roborovski dwarf hamsters infected with either the SARS-CoV-2 wild-type or the Alpha variant. In this model, ensovibep prevents fatality and provides substantial protection equivalent to the standard of care mAb cocktail. These results support further clinical evaluation and indicate that ensovibep could be a valuable alternative to mAb cocktails and other treatments for COVID-19.
Supplementary Materials for Ensovibep Supplementary Figure 1: A-C
Supplementary Figure 4: Titration curves for ensovibep (MP0420) and its RBD-binding domains (i.e. R1, R2 and R3), REGN10933 and REGN10987 to determine IC50 neutralization potencies on multiple spike mutants or only for ensovibep (MP0420) on the variants, which are summarized in Figure 2. Reported is the mean +/− SEM (standard error of the mean).
Supplementary Figure 5: Overview of the experimental protocol for viral passaging: A patient SARS-CoV-2 isolate from early 2020 (1.5 ×10 6 pfu) was incubated in presence of increasing concentrations of DARPin candidate or antibody for 4 days on Vero E6 cells and virus-induced cytopathic effects (CPE) were determined by microscopy. For each DARPin and antibody condition, cultures showing significant cytopathic effect (≥20%) under the greatest selective pressure were selected and virus-containing supernatant collected to start a new culture passage on Vero E6 cells (bold circle), again under increasing concentrations of the corresponding DARPin candidate or antibody condition. Passaging of virus containing supernatant was continued in the same manner for a total of 4 passages.
References
Andreano, SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma, bioRxiv, doi:10.1101/2020.12.28.424451
Baum, Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies, Science, doi:10.1126/science.abd0831
Berger Rentsch, Zimmer, A vesicular stomatitis virus replicon-based bioassay for the rapid and sensitive determination of multi-species type I interferon, PLoS One, doi:10.1371/journal.pone.0025858
Binz, Design and characterization of MP0250, a tri-specific anti-HGF/anti-VEGF DARPin(R) drug candidate, MAbs, doi:10.1080/19420862.2017.1305529
Binz, High-affinity binders selected from designed ankyrin repeat protein libraries, Nat Biotechnol, doi:10.1038/nbt962
Bolger, Lohse, Usadel, Trimmomatic: a flexible trimmer for Illumina sequence data, Bioinformatics, doi:10.1093/bioinformatics/btu170
Cao, 1.529 escapes the majority of SARS-CoV-2 neutralizing antibodies of diverse epitopes, bioRxiv, doi:10.1101/2021.12.07.470392
Cao, De novo design of picomolar SARS-CoV-2 miniprotein inhibitors, Science, doi:10.1126/science.abd9909
Casalino, Beyond Shielding: The Roles of Glycans in the SARS-CoV-2 Spike Protein, ACS Cent Sci, doi:10.1021/acscentsci.0c01056
Cathcart, The dual function monoclonal antibodies VIR-7831 and VIR-7832 demonstrate potent in vitro and in vivo activity against SARS-CoV-2, bioRxiv, doi:10.1101/2021.03.09.434607v1
Cele, SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection, medRxiv, doi:10.1101/2021.12.08.21267417
Chaudhury, Benchmarking and analysis of protein docking performance in Rosetta v3.2, PLoS One, doi:10.1371/journal.pone.0022477
Choi, Proceedings of the ACM Conference on Bioinformatics, Computational Biology and Biomedicine
Choi, Sims, Murphy, Miller, Chan, Predicting the functional effect of amino acid substitutions and indels, PLoS One, doi:10.1371/journal.pone.0046688
Cianfrocco, Wong, Youn, Wagner, The Practice and Experience in Advanced Research Computing
Cingolani, A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3, Fly (Austin), doi:10.4161/fly.19695
Copin, In vitro and in vivo preclinical studies predict REGEN-COV protection against emergence of viral escape in humans, doi:10.1101/2021.03.10.434834
Copin, In vitro and in vivo preclinical studies predict REGEN-COV protection against emergence of viral escape in humans, bioRxiv, doi:10.1101/2021.03.10.434834v3
Copin, The monoclonal antibody combination REGEN-COV protects against SARS-CoV-2 mutational escape in preclinical and human studies, Cell, doi:10.1016/j.cell.2021.06.002
Corman, Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR, Euro Surveill, doi:10.2807/1560-7917.ES.2020.25.3.2000045
Corti, Purcell, Snell, Veesler, Tackling COVID-19 with neutralizing monoclonal antibodies, Cell, doi:10.1016/j.cell.2021.05.005
Danecek, Twelve years of SAMtools and BCFtools, GigaScience, doi:10.1093/gigascience/giab008
Duvaud, the Swiss Bioinformatics Resource Portal, as designed by its users, Nucleic Acids Res, doi:10.1093/nar/gkab225
Falsey, SARS-CoV-2 Neutralization with BNT162b2 Vaccine Dose 3, N Engl J Med, doi:10.1056/NEJMc2113468
Fiedler, MP0250, a VEGF and HGF neutralizing DARPin((R)) molecule shows high anti-tumor efficacy in mouse xenograft and patient-derived tumor models, Oncotarget, doi:10.18632/oncotarget.21738
Friedrich, Preclinical characterization of AMG 330, a CD3/CD33-bispecific T-cell-engaging antibody with potential for treatment of acute myelogenous leukemia, Mol Cancer Ther, doi:10.1158/1535-7163.MCT-13-0956
Garcia-Beltran, Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity, Cell, doi:10.1016/j.cell.2021.03.013
Gobeil, Effect of natural mutations of SARS-CoV-2 on spike structure, conformation, and antigenicity, Science, doi:10.1126/science.abi6226
Goddard, UCSF ChimeraX: Meeting modern challenges in visualization and analysis, Protein Sci, doi:10.1002/pro.3235
Greaney, Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition, Cell host & microbe, doi:10.1016/j.chom.2020.11.007
Gruber, Standardization of Reporting Criteria for Lung Pathology in SARS-CoV-2-infected Hamsters: What Matters?, Am J Respir Cell Mol Biol, doi:10.1165/rcmb.2020-0280LE
Gu, Eils, Schlesner, Complex heatmaps reveal patterns and correlations in multidimensional genomic data, Bioinformatics, doi:10.1093/bioinformatics/btw313
Hoffmann, SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor, Cell, doi:10.1016/j.cell.2020.02.052
Hoffmann, SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies, Cell, doi:10.1016/j.cell.2021.03.036
Huang, RosettaRemodel: a generalized framework for flexible backbone protein design, PLoS One, doi:10.1371/journal.pone.0024109
Hunt, Multivalent designed proteins protect against SARS-CoV-2 variants of concern, bioRxiv, doi:10.1101/2021.07.07.451375
Jones, LY-CoV555, a rapidly isolated potent neutralizing antibody, provides protection in a non-human primate model of SARS-CoV-2 infection, bioRxiv, doi:10.1101/2020.09.30.318972
Ku, Molecular determinants and mechanism for antibody cocktail preventing SARS-CoV-2 escape, Nat Commun, doi:10.1038/s41467-020-20789-7
Kumari, Neuroinvasion and Encephalitis Following Intranasal Inoculation of SARS-CoV-2 in K18-hACE2 Mice, Viruses, doi:10.3390/v13010132
Laffeber, De Koning, Kanaar, Lebbink, Experimental Evidence for Enhanced Receptor Binding by Rapidly Spreading SARS-CoV-2 Variants, Journal of Molecular Biology, doi:10.1016/j.jmb.2021.167058
Laskowski, Swindells, LigPlot+: multiple ligand-protein interaction diagrams for drug discovery, J Chem Inf Model, doi:10.1021/ci200227u
Leaver-Fay, ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules, Methods Enzymol, doi:10.1016/B978-0-12-381270-4.00019-6
Ledford, The race to make COVID antibody therapies cheaper and more potent, Nature, doi:10.1038/d41586-020-02965-3
Letko, Marzi, Munster, Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses, Nat Microbiol, doi:10.1038/s41564-020-0688-y
Li, Durbin, Fast and accurate short read alignment with Burrows-Wheeler transform, Bioinformatics, doi:10.1093/bioinformatics/btp324
Li, The Sequence Alignment/Map format and SAMtools, Bioinformatics, doi:10.1093/bioinformatics/btp352
Linsky, De novo design of potent and resilient hACE2 decoys to neutralize SARS-CoV-2, Science, doi:10.1126/science.abe0075
Liu, Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization, Cell host & microbe, doi:10.1016/j.chom.2021.01.014
Lu3, Peng1, Sterling4, Walsh, Structural impact on SARS SoV-2 spike protein by D614G substitution
Lusvarghi, Key substitutions in the spike protein of SARS-CoV-2 variants can predict resistance to monoclonal antibodies, but other substitutions can modify the effects, bioRxiv, doi:10.1101/2021.07.16.452748
Matsuyama, Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells, Proc Natl Acad Sci U S A, doi:10.1073/pnas.2002589117
Nao, Consensus and variations in cell line specificity among human metapneumovirus strains, PLoS One, doi:10.1371/journal.pone.0215822
Neerukonda, Establishment of a well-characterized SARS-CoV-2 lentiviral pseudovirus neutralization assay using 293T cells with stable expression of ACE2 and TMPRSS2, PLoS One, doi:10.1371/journal.pone.0248348
Nouailles, Temporal omics analysis in Syrian hamsters unravel cellular effector responses to moderate COVID-19, Nat Commun, doi:10.1038/s41467-021-25030-7
Osterrieder, Age-Dependent Progression of SARS-CoV-2 Infection in Syrian Hamsters, Viruses, doi:10.3390/v12070779
Pettersen, UCSF Chimera--a visualization system for exploratory research and analysis, J Comput Chem, doi:10.1002/jcc.20084
Pinto, Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody, Nature, doi:10.1038/s41586-020-2349-y
Planas, Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization, Nature, doi:10.1038/s41586-021-03777-9
Pulliam, Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa, medRxiv, doi:10.1101/2021.11.11.21266068
Robinson, Integrative genomics viewer, Nat Biotechnol, doi:10.1038/nbt.1754
Sanchez-Garcia, DeepEMhancer: a deep learning solution for cryo-EM volume post-processing, bioRxiv, doi:10.1101/2020.06.12.148296
Schoof, An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike, Science, doi:10.1126/science.abe3255
Shang, Structural basis of receptor recognition by SARS-CoV-2, Nature, doi:10.1038/s41586-020-2179-y
Shi, A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2, Nature, doi:10.1038/s41586-020-2381-y
Sim, SIFT web server: predicting effects of amino acid substitutions on proteins, Nucleic Acids Res, doi:10.1093/nar/gks539
Starr, Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding, Cell, doi:10.1016/j.cell.2020.08.012
Starr, Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding, Cell, doi:10.1016/j.cell.2020.08.012
Steiner, Half-life extension using serum albumin-binding DARPin(R) domains, Protein Eng Des Sel, doi:10.1093/protein/gzx022
Stumpp, Dawson, Binz, Beyond Antibodies: The DARPin((R)) Drug Platform, BioDrugs, doi:10.1007/s40259-020-00429-8
Tegally, Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa, medRxiv, doi:10.1101/2020.12.21.20248640
Thomson, Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity, Cell, doi:10.1016/j.cell.2021.01.037
Thomson, The circulating SARS-CoV-2 spike variant N439K maintains fitness while evading antibody-mediated immunity, bioRxiv, doi:10.1101/2020.11.04.355842
Torriani, Identification of Clotrimazole Derivatives as Specific Inhibitors of Arenavirus Fusion, J Virol, doi:10.1128/JVI.01744-18
Torriani, Macropinocytosis contributes to hantavirus entry into human airway epithelial cells, Virology, doi:10.1016/j.virol.2019.02.013
Tortorici, Veesler, Structural insights into coronavirus entry, Adv Virus Res, doi:10.1016/bs.aivir.2019.08.002
Trimpert, The Roborovski Dwarf Hamster Is A Highly Susceptible Model for a Rapid and Fatal Course of SARS-CoV-2 Infection, Cell reports, doi:10.1016/j.celrep.2020.108488
Voloch, Novel circulating lineage of SARS-CoV-2 in the state of Rio de Janeiro Brazil originated from B, medRxiv, doi:10.1101/2020.12.23.20248598
Walls, Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer, Nature, doi:10.1038/nature16988
Walls, Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein, Cell, doi:10.1016/j.cell.2020.02.058
Walls, Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion, Proc Natl Acad Sci U S A, doi:10.1073/pnas.1708727114
Walls, Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion, Cell, doi:10.1016/j.cell.2018.12.028
Walser, Highly potent anti-SARS-CoV-2 multivalent DARPin therapeutic candidates
Walter, Hutter, Garaeva, Scherer, Zimmermann, Highly potent bispecific sybodies neutralize SARS-CoV-2
Wang, Increased Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7 to Antibody Neutralization, bioRxiv, doi:10.1101/2021.01.25.428137
Wilm, LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cellpopulation heterogeneity from high-throughput sequencing datasets, Nucleic Acids Res, doi:10.1093/nar/gks918
Wollscheid, Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins, Nat Biotechnol, doi:10.1038/nbt.1532
Wrapp, Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation, Science, doi:10.1126/science.abb2507
Yi, Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies, Cell Mol Immunol, doi:10.1038/s41423-020-0458-z
Zahradnik, SARS-CoV-2 RBD in vitro evolution follows contagious mutation spread, yet generates an able infection inhibitor, bioRxiv, doi:10.1101/2021.01.06.425392v3
Zhang, Gctf: Real-time CTF determination and correction, J Struct Biol, doi:10.1016/j.jsb.2015.11.003
Zheng, MotionCor2: anisotropic correction of beam-induced motion for improved cryoelectron microscopy, Nat Methods, doi:10.1038/nmeth.4193
Zhou, A pneumonia outbreak associated with a new coronavirus of probable bat origin, Nature, doi:10.1038/s41586-020-2012-7
Zhou, Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera, Cell, doi:10.1016/j.cell.2021.02.037
Zivanov, New tools for automated high-resolution cryo-EM structure determination in RELION-3, Elife, doi:10.7554/eLife.42166
DOI record:
{
"DOI": "10.1101/2021.02.03.429164",
"URL": "http://dx.doi.org/10.1101/2021.02.03.429164",
"abstract": "<jats:title>Abstract</jats:title><jats:p>SARS-CoV-2 has infected millions of people globally and continues to undergo evolution. Emerging variants can be partially resistant to vaccine induced and therapeutic antibodies, emphasizing the urgent need for accessible, broad-spectrum therapeutics. Here, we report a comprehensive study of ensovibep, the first trispecific clinical DARPin candidate, that can simultaneously engage all three units of the spike protein trimer to potently inhibit ACE2 interaction, as revealed by structural analyses. The cooperative binding of the individual modules enables ensovibep to retain inhibitory potency against all frequent SARS-CoV-2 variants, including Omicron, as of December 2021. Moreover, viral passaging experiments show that ensovibep, when used as a single agent, can prevent development of escape mutations comparably to a cocktail of monoclonal antibodies (mAb). Finally, we demonstrate that the very high in vitro antiviral potency also translates into significant therapeutic protection and reduction of pathogenesis in Roborovski dwarf hamsters infected with either the SARS-CoV-2 wild-type or the Alpha variant. In this model, ensovibep prevents fatality and provides substantial protection equivalent to the standard of care mAb cocktail. These results support further clinical evaluation and indicate that ensovibep could be a valuable alternative to mAb cocktails and other treatments for COVID-19.</jats:p>",
"accepted": {
"date-parts": [
[
2021,
12,
17
]
]
},
"author": [
{
"affiliation": [],
"family": "Rothenberger",
"given": "Sylvia",
"sequence": "first"
},
{
"affiliation": [],
"family": "Hurdiss",
"given": "Daniel L.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Walser",
"given": "Marcel",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Malvezzi",
"given": "Francesca",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Mayor",
"given": "Jennifer",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Ryter",
"given": "Sarah",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Moreno",
"given": "Hector",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Liechti",
"given": "Nicole",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Bosshart",
"given": "Andreas",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Iss",
"given": "Chloe",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Calabro",
"given": "Valérie",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Cornelius",
"given": "Andreas",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Hospodarsch",
"given": "Tanja",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Neculcea",
"given": "Alexandra",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Looser",
"given": "Thamar",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Schlegel",
"given": "Anja",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Fontaine",
"given": "Simon",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Villemagne",
"given": "Denis",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Paladino",
"given": "Maria",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Kaufmann",
"given": "Yvonne",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Schaible",
"given": "Doris",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Schlegel",
"given": "Iris",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Schiegg",
"given": "Dieter",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Zitt",
"given": "Christof",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Sigrist",
"given": "Gabriel",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Straumann",
"given": "Marcel",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Wolter",
"given": "Julia",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Comby",
"given": "Marco",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Adler",
"given": "Julia M.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Eschke",
"given": "Kathrin",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Nascimento",
"given": "Mariana",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Abdelgawad",
"given": "Azza",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Gruber",
"given": "Achim D.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Bushe",
"given": "Judith",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Kershaw",
"given": "Olivia",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Lyoo",
"given": "Heyrhyoung",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Wang",
"given": "Chunyan",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Li",
"given": "Wentao",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Drulyte",
"given": "Ieva",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Du",
"given": "Wenjuan",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Kaspar Binz",
"given": "H.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Herrup",
"given": "Rachel",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Lusvarghi",
"given": "Sabrina",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Neerukonda",
"given": "Sabari Nath",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Vassell",
"given": "Russell",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Wang",
"given": "Wei",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Mangold",
"given": "Susanne",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Reichen",
"given": "Christian",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Radom",
"given": "Filip",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Knutson",
"given": "Charles G.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Balavenkatraman",
"given": "Kamal K.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Ramanathan",
"given": "Krishnan",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Lewis",
"given": "Seth",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Watson",
"given": "Randall",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Haeuptle",
"given": "Micha A.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Zürcher",
"given": "Alexander",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Dawson",
"given": "Keith M.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Steiner",
"given": "Daniel",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Weiss",
"given": "Carol D.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Amstutz",
"given": "Patrick",
"sequence": "additional"
},
{
"affiliation": [],
"family": "van Kuppeveld",
"given": "Frank J.M.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Stumpp",
"given": "Michael T.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Bosch",
"given": "Berend-Jan",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Engler",
"given": "Olivier",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Trimpert",
"given": "Jakob",
"sequence": "additional"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2021,
2,
4
]
],
"date-time": "2021-02-04T02:20:54Z",
"timestamp": 1612405254000
},
"deposited": {
"date-parts": [
[
2021,
12,
21
]
],
"date-time": "2021-12-21T09:55:26Z",
"timestamp": 1640080526000
},
"group-title": "Immunology",
"indexed": {
"date-parts": [
[
2021,
12,
22
]
],
"date-time": "2021-12-22T06:02:51Z",
"timestamp": 1640152971123
},
"institution": [
{
"name": "bioRxiv"
}
],
"is-referenced-by-count": 3,
"issued": {
"date-parts": [
[
2021,
2,
3
]
]
},
"link": [
{
"URL": "https://syndication.highwire.org/content/doi/10.1101/2021.02.03.429164",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "246",
"original-title": [],
"posted": {
"date-parts": [
[
2021,
2,
3
]
]
},
"prefix": "10.1101",
"published": {
"date-parts": [
[
2021,
2,
3
]
]
},
"publisher": "Cold Spring Harbor Laboratory",
"reference": [
{
"DOI": "10.1038/s41586-020-2012-7",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.1"
},
{
"DOI": "10.1038/s41586-020-2179-y",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.2"
},
{
"DOI": "10.1016/bs.aivir.2019.08.002",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.3"
},
{
"DOI": "10.1038/s41564-020-0688-y",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.4"
},
{
"DOI": "10.1016/j.cell.2020.02.058",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.5"
},
{
"DOI": "10.1038/nature16988",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.6"
},
{
"DOI": "10.1073/pnas.1708727114",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.7"
},
{
"DOI": "10.1016/j.cell.2020.02.052",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.8"
},
{
"key": "2021122101550814000_2021.02.03.429164v3.9",
"unstructured": "Jun Zhang 1, Yongfei Cai 1,2†, Tianshu Xiao 1,2, Jianming Lu 3, Hanqin Peng 1, Sarah M. Sterling 4,5, Richard M. Walsh Jr.4,5, Sophia Rits-Volloch 1, Haisun Zhu 6, Alec N. Woosley 6, Wei Yang 6, Piotr Sliz 1,2,5, Bing Chen 1,2*. Structural impact on SARS SoV-2 spike protein by D614G substitution. Science (2021)."
},
{
"DOI": "10.1016/j.cell.2021.03.013",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.10"
},
{
"DOI": "10.1016/j.chom.2020.11.007",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.11"
},
{
"DOI": "10.1101/2021.07.16.452748",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.12"
},
{
"DOI": "10.1016/j.cell.2020.08.012",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.13"
},
{
"DOI": "10.1016/j.cell.2021.01.037",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.14"
},
{
"DOI": "10.1101/2021.01.25.428137",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.15"
},
{
"DOI": "10.1038/s41423-020-0458-z",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.16"
},
{
"DOI": "10.1016/j.chom.2021.01.014",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.17"
},
{
"DOI": "10.1016/j.cell.2021.02.037",
"article-title": "Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera",
"doi-asserted-by": "crossref",
"first-page": "2348",
"journal-title": "Cell",
"key": "2021122101550814000_2021.02.03.429164v3.18",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1126/science.abi6226",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.19"
},
{
"DOI": "10.1101/2020.12.21.20248640",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.20"
},
{
"DOI": "10.1101/2020.12.23.20248598",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.21"
},
{
"DOI": "10.1101/2021.12.08.21267417",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.22"
},
{
"DOI": "10.1101/2020.11.04.355842",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.23"
},
{
"DOI": "10.1016/j.jmb.2021.167058",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.24"
},
{
"DOI": "10.1038/s41586-021-03777-9",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.25"
},
{
"DOI": "10.1038/d41586-020-02965-3",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.26"
},
{
"DOI": "10.1126/science.abd0831",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.27"
},
{
"DOI": "10.1016/j.cell.2021.06.002",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.28"
},
{
"DOI": "10.1038/s41467-020-20789-7",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.29"
},
{
"DOI": "10.1038/nbt962",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.30"
},
{
"DOI": "10.1101/2020.08.25.256339",
"doi-asserted-by": "crossref",
"key": "2021122101550814000_2021.02.03.429164v3.31",
"unstructured": "Walser, M. , et al. Highly potent anti-SARS-CoV-2 multivalent DARPin therapeutic candidates. bioRxiv (2020)."
},
{
"DOI": "10.1007/s40259-020-00429-8",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.32"
},
{
"DOI": "10.1080/19420862.2017.1305529",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.33"
},
{
"DOI": "10.18632/oncotarget.21738",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.34"
},
{
"DOI": "10.1093/protein/gzx022",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.35"
},
{
"DOI": "10.1101/2021.03.10.434834v3",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.36"
},
{
"DOI": "10.1101/2021.03.09.434607v1",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.37"
},
{
"DOI": "10.1016/j.celrep.2020.108488",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.38"
},
{
"DOI": "10.1016/j.cell.2018.12.028",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.39"
},
{
"DOI": "10.1016/j.cell.2021.05.005",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.40"
},
{
"DOI": "10.1056/NEJMc2113468",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.41"
},
{
"DOI": "10.1016/j.cell.2021.03.036",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.42"
},
{
"DOI": "10.1101/2021.11.11.21266068",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.43"
},
{
"DOI": "10.1021/acscentsci.0c01056",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.44"
},
{
"DOI": "10.1101/2020.12.28.424451",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.45"
},
{
"DOI": "10.3390/v12070779",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.46"
},
{
"DOI": "10.1038/s41467-021-25030-7",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.47"
},
{
"DOI": "10.3390/v13010132",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.48"
},
{
"DOI": "10.1126/science.abe3255",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.49"
},
{
"DOI": "10.1126/science.abd9909",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.50"
},
{
"DOI": "10.1126/science.abe0075",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.51"
},
{
"key": "2021122101550814000_2021.02.03.429164v3.52",
"unstructured": "Walter, J. D. , Hutter, C. A. J. , Garaeva, A. A. , Scherer, M. & Zimmermann, I . Highly potent bispecific sybodies neutralize SARS-CoV-2. bioRxiv (2020)."
},
{
"DOI": "10.1101/2021.07.07.451375",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.53"
},
{
"DOI": "10.1101/2020.09.30.318972",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.54"
},
{
"DOI": "10.1038/s41586-020-2349-y",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.55"
},
{
"DOI": "10.1038/s41586-020-2381-y",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.56"
},
{
"DOI": "10.1016/j.cell.2020.08.012",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.57"
},
{
"DOI": "10.1101/2021.01.06.425392v3",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.58"
},
{
"DOI": "10.1101/2021.12.07.470392",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.59"
},
{
"DOI": "10.7554/eLife.42166",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.60"
},
{
"DOI": "10.1038/nmeth.4193",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.61"
},
{
"DOI": "10.1016/j.jsb.2015.11.003",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.62"
},
{
"DOI": "10.1002/jcc.20084",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.63"
},
{
"DOI": "10.1126/science.abb2507",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.64"
},
{
"DOI": "10.1101/2020.06.12.148296",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.65"
},
{
"key": "2021122101550814000_2021.02.03.429164v3.66",
"unstructured": "Cianfrocco, M. A. , Wong, M. , Youn, C. & Wagner, R . in The Practice and Experience in Advanced Research Computing (New Orleans, LA 2017)."
},
{
"DOI": "10.1371/journal.pone.0022477",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.67"
},
{
"DOI": "10.1371/journal.pone.0024109",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.68"
},
{
"DOI": "10.1016/B978-0-12-381270-4.00019-6",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.69"
},
{
"DOI": "10.1021/ci200227u",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.70"
},
{
"DOI": "10.1002/pro.3235",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.71"
},
{
"DOI": "10.1371/journal.pone.0025858",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.72"
},
{
"DOI": "10.1016/j.virol.2019.02.013",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.73"
},
{
"DOI": "10.1128/JVI.01744-18",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.74"
},
{
"DOI": "10.1073/pnas.2002589117",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.75"
},
{
"DOI": "10.1371/journal.pone.0215822",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.76"
},
{
"DOI": "10.1371/journal.pone.0248348",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.77"
},
{
"DOI": "10.1038/nbt.1532",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.78"
},
{
"DOI": "10.1093/bioinformatics/btp352",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.79"
},
{
"DOI": "10.1093/bioinformatics/btu170",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.80"
},
{
"DOI": "10.1093/bioinformatics/btp324",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.81"
},
{
"DOI": "10.1093/nar/gks918",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.82"
},
{
"DOI": "10.4161/fly.19695",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.83"
},
{
"DOI": "10.1093/bioinformatics/btw313",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.84"
},
{
"DOI": "10.1101/2021.03.10.434834",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.85"
},
{
"DOI": "10.2807/1560-7917.ES.2020.25.3.2000045",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.86"
},
{
"DOI": "10.1165/rcmb.2020-0280LE",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.87"
},
{
"DOI": "10.1093/gigascience/giab008",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.88"
},
{
"DOI": "10.1038/nbt.1754",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.89"
},
{
"DOI": "10.1093/nar/gkab225",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.90"
},
{
"DOI": "10.1371/journal.pone.0046688",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.91"
},
{
"key": "2021122101550814000_2021.02.03.429164v3.92",
"unstructured": "Choi, Y . in Proceedings of the ACM Conference on Bioinformatics, Computational Biology and Biomedicine 414–417 (Association for Computing Machinery, Orlando, Florida, 2012)."
},
{
"DOI": "10.1093/nar/gks539",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.93"
},
{
"DOI": "10.1158/1535-7163.MCT-13-0956",
"doi-asserted-by": "publisher",
"key": "2021122101550814000_2021.02.03.429164v3.94"
}
],
"reference-count": 94,
"references-count": 94,
"relation": {},
"score": 1,
"short-container-title": [],
"short-title": [],
"source": "Crossref",
"subtitle": [],
"subtype": "preprint",
"title": [
"Ensovibep, a novel trispecific DARPin candidate that protects against SARS-CoV-2 variants"
],
"type": "posted-content"
}