An Allele of the MTHFR one-carbon metabolism gene predicts severity of COVID-19
et al., medRxiv, doi:10.1101/2025.02.28.25323089, Mar 2025
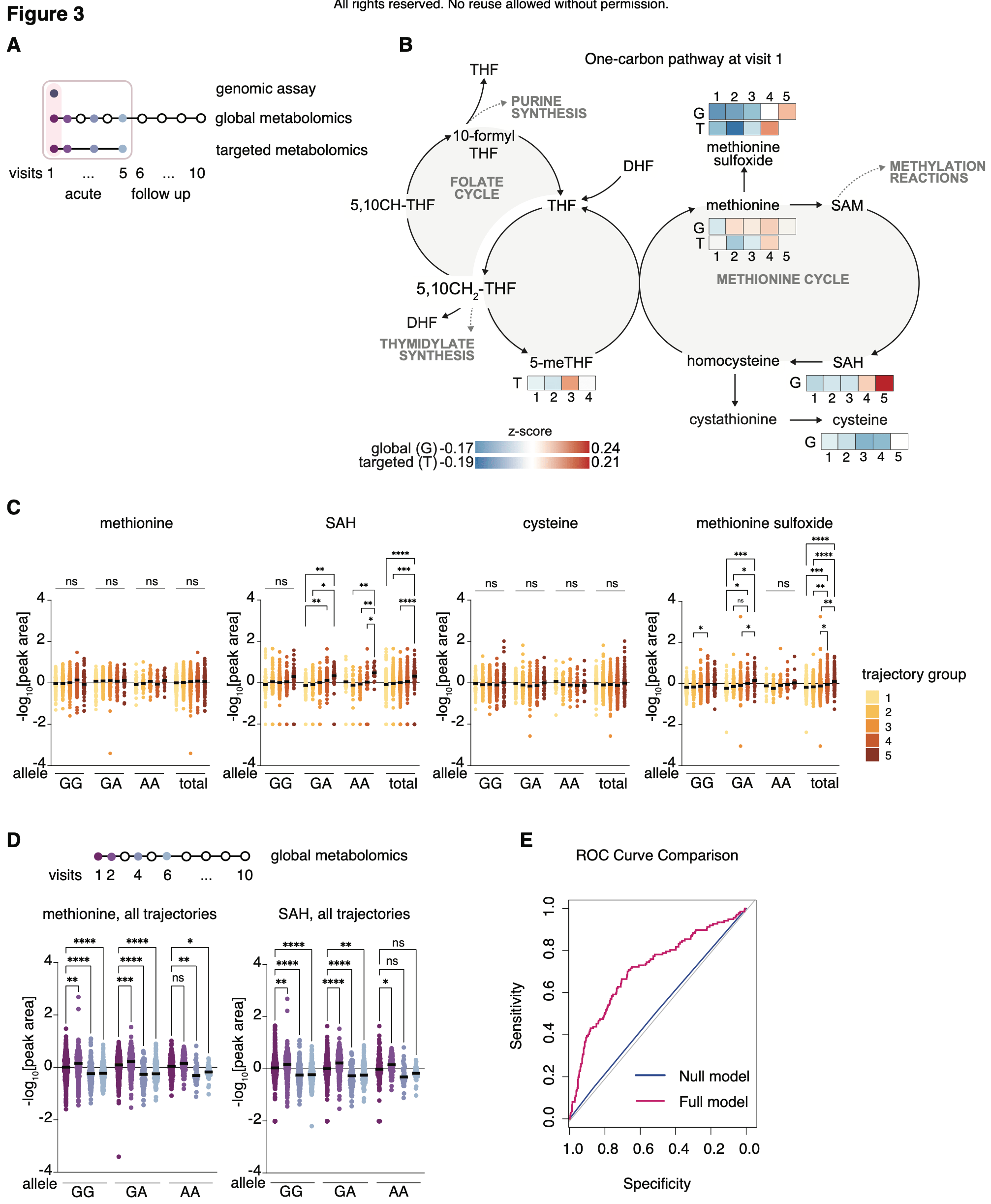
Analysis of plasma samples from 1,164 hospitalized COVID-19 patients revealing that the MTHFR C677T genetic variant, combined with alterations in one-carbon metabolism metabolites, predicts COVID-19 severity and risk of developing long COVID. A model combining MTHFR allele status with plasma levels of methionine, methionine-sulfoxide, and S-adenosylhomocysteine at hospital admission significantly improved prediction of COVID-19 severity and post-acute sequelae of SARS-CoV-2 compared to genetic information alone.
Petrova et al., 3 Mar 2025, retrospective, USA, preprint, 17 authors.
An Allele of the MTHFR one-carbon metabolism gene predicts severity of COVID-19
doi:10.1101/2025.02.28.25323089
While the public health burden of SARS-CoV-2 infection has lessened due to natural and vaccine-acquired immunity, the emergence of less virulent variants, and antiviral medications, COVID-19 continues to take a significant toll. There are > 10,000 new hospitalizations per week in the U.S., many of whom develop post-acute sequelae of SARS-CoV-2 (PASC), or "long COVID", with long-term health issues and compromised quality of life. Early identification of individuals at high risk of severe COVID-19 is key for monitoring and supporting respiratory status and improving outcomes. Therefore, precision tools for early detection of patients at high risk of severe disease can reduce morbidity and mortality. Here we report an untargeted and longitudinal metabolomic study of plasma derived from adult patients with COVID-19. Onecarbon metabolism, a pathway previously shown as critical for viral propagation and disease progression, and a potential target for COVID-19 treatment, scored strongly as differentially abundant in patients with severe COVID-19. A follow-up targeted metabolite profiling revealed that one arm of the one-carbon metabolism pathway, the methionine cycle, is a major driver of the metabolic profile associated with disease severity. The methionine cycle produces Sadenosylmethionine (SAM), the methyl group donor important for methylation of DNA, RNA, and proteins, and its high abundance was reported to correlate with disease severity. Further, genomic data from the profiled patients revealed a genetic contributor to methionine metabolism and identified the C677T allele of the MTHFR gene as a pre-existing predictor of disease trajectory -patients homozygous for the MTHFR C677T have higher incidence of experiencing NOTE: This preprint reports new research that has not been certified by peer review and should not be used to guide clinical practice. severe disease. Our results raise the possibility that screening for the common genetic MTHFR variant may be an actionable approach to stratify risk of COVID severity and may inform novel precision COVID-19 treatment strategies.
Yale School of Public Health, New Haven, CT 06510, USA: Denise Esserman, Leying Guan, Anderson Brito, Jessica Rothman, Nathan D. Grubaugh
IMPACC Network Competing Interests The Icahn School of Medicine at Mount Sinai has filed patent applications related to SARS-CoV-2 serological assays, NDV-based SARS-CoV-2 vaccines, influenza virus vaccines, and therapeutics, listing Florian Krammer and Viviana Simon as co-inventors. Mount Sinai has spun out Kantaro to market SARS-CoV-2 serological tests and Castlevax to develop SARS-CoV-2 vaccines, with Florian Krammer as co-founder and scientific advisory board member of Castlevax. Florian Krammer has consulted for Merck, Curevac, Seqirus, GSK, Pfizer, 3rd Rock Ventures, Sanofi, Gritstone, and Avimex. His laboratory collaborates with Dynavax on influenza vaccine development and with VIR on influenza virus therapeutics development. Ofer Levy is a named inventor on patents held by Boston Children's Hospital related to vaccine adjuvants and human in vitro platforms that model vaccine action. His laboratory has received research support from GlaxoSmithKline (GSK) and Pfizer and he is a co-founder and advisor to Ovax, Inc., which develops opioid vaccines. Charles Cairns consults for bioMérieux and receives grant funding from the Bill & Melinda Gates Foundation. James A. Overton is a consultant at Knocean Inc.
References
Admon, Iwashyna, Kamphuis, Gundel, Sahetya et al., Assessment of Symptom, Disability, and Financial Trajectories in Patients Hospitalized for COVID-19 at 6 Months, JAMA Netw Open, doi:10.1001/jamanetworkopen.2022.55795
Baden, Mendez, Lasky-Su, Tong, Rooks et al., None
Bailey, Gregory, Polymorphisms of methylenetetrahydrofolate reductase and other enzymes: metabolic significance, risks and impact on folate requirement, J Nutr, doi:10.1093/jn/129.5.919
Balnis, Madrid, Drake, Vancavage, Tiwari et al., Blood DNA methylation in post-acute sequelae of COVID-19 (PASC): a prospective cohort study, EBioMedicine, doi:10.1016/j.ebiom.2024.105251
Balnis, Madrid, Hogan, Drake, Adhikari et al., Persistent blood DNA methylation changes one year after SARS-CoV-2 infection, Clin Epigenetics, doi:10.1186/s13148-022-01313-8
Cai, Xie, Topol, Al-Aly, Three-year outcomes of post-acute sequelae of COVID-19, Nat Med, doi:10.1038/s41591-024-02987-8
Cairns, Haddad, Kutzler, Bernui, Cusimano et al., None
Celestino, Amarante, Vespero, Tavares, Yamauchi et al., Dermatological Manifestations in COVID-19: A Case Study of SARS-CoV-2 Infection in a Genetic Thrombophilic Patient with Mthfr Mutation, Pathogens, doi:10.3390/pathogens12030438
Chung, Fleisher, Churchwell for their support for the Precision Vaccines Program. Dr. Augustine's and Becker's co-authorship of this report does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases, the National Institutes of Health or any other agency of the United States Government
Danielle, Débora, Alessandra, Alexia, Débora et al., Correlating COVID-19 severity with biomarker profiles and patient prognosis, Sci Rep, doi:10.1038/s41598-024-71951-w
Diray-Arce, Conti, Petrova, Kanarek, Angelidou et al., Integrative Metabolomics to Identify Molecular Signatures of Responses to Vaccines and Infections, Metabolites, doi:10.3390/metabo10120492
Diray-Arce, Fourati, Jayavelu, Patel, Maguire et al., Multi-omic longitudinal study reveals immune correlates of clinical course among hospitalized COVID-19 patients, Cell Rep Med, doi:10.1016/j.xcrm.2023.101079
Ducker, Rabinowitz, One-Carbon Metabolism in Health and Disease, Cell Metab, doi:10.1016/j.cmet.2016.08.009
Erle, Calfee, Hendrickson, Kangelaris, Nguyen et al., None
Fernandez-Sesma, Simon, Krammer, Van Bakel, Kim-Schulze et al., None
Geng, receives research funding from Pfizer, Inc., through her institution. Catherine Hough receives research support
Grace, McComsey receives research grants from Redhill, Cognivue, Pfizer, and Genentech and consults for Gilead
Groff, Sun, Ssentongo, Ba, Parsons et al., Short-term and Long-term Rates of Postacute Sequelae of SARS-CoV-2 Infection: A Systematic Review, JAMA Netw Open, doi:10.1001/jamanetworkopen.2021.28568
Grubaugh, consults for Tempus Labs and the National Basketball Association
Gygi, Maguire, Patel, Shinde, Konstorum et al., Integrated longitudinal multiomics study identifies immune programs associated with acute COVID-19 severity and mortality, J Clin Invest, doi:10.1172/jci176640
Hafler, Montgomery, Shaw, Kleinstein, Gygi et al., Vicki Seyfer-Margolis is employed by MyOwnMed. Nadine Rouphael reports grants or contracts
Heller, Seshan, Moskowitz, Gönen, Inference for the difference in the area under the ROC curve derived from nested binary regression models, Biostatistics, doi:10.1093/biostatistics/kxw045
Hill, Mehta, Sharma, Mane, Singh et al., Risk factors associated with post-acute sequelae of SARS-CoV-2: an N3C and NIH RECOVER study, BMC Public Health, doi:10.1186/s12889-023-16916-w
Hu, Liu, Lei, Zeng, Li et al., Metabolic regulation of the immune system in health and diseases: mechanisms and interventions, Signal Transduct Target Ther, doi:10.1038/s41392-024-01954-6
Iwasaki, 4bio, Blue, Biologics, Biotherapeutics et al., Monika Kraft receives research funding from NIH, ALA, Sanofi, and AstraZeneca for asthma research. She consults for AstraZeneca, Sanofi, Chiesi, and GSK for severe asthma and is co-founder and CMO of RaeSedo, Inc., developing peptidomimetics for inflammatory lung disease. Esther Melamed receives research funding from Babson Diagnostics, honoraria from the Multiple Sclerosis Association of America, and has
James, Overton Precision Vaccines Program
Karakosta, Kontou, Xirou, Kabanarou, Acute Macular Neuroretinopathy Associated With COVID-19 Infection: Is Double Heterozygous Methylenetetrahydrofolate Reductase (MTHFR) Mutation an Underlying Risk Factor?, Cureus, doi:10.7759/cureus.34873
Karst, Hollenhorst, Achenbach, Life-threatening course in coronavirus disease 2019 (COVID-19): Is there a link to methylenetetrahydrofolic acid reductase (MTHFR) polymorphism and hyperhomocysteinemia?, Med Hypotheses, doi:10.1016/j.mehy.2020.110234
Kryukov, Ivanov, Karpov, Vasil'evich Aleksandrin, Dygai et al., Plasma S-Adenosylmethionine Is Associated with Lung Injury in COVID-19, Dis Markers, doi:10.1155/2021/7686374
Levy, Steen, Van Zalm, Fatou, Kinga et al., None
Lewis, Butler, Gilbert, A unified approach to model selection using the likelihood ratio test, Methods in Ecology and Evolution, doi:10.1111/j.2041-210X.2010.00063.x
Li, Lan, Zhang, Bassig, Holford et al., Role of one-carbon metabolizing pathway genes and genenutrient interaction in the risk of non-Hodgkin lymphoma, Cancer Causes Control, doi:10.1007/s10552-013-0264-3
Liew, Gupta, Methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism: epidemiology, metabolism and the associated diseases, Eur J Med Genet, doi:10.1016/j.ejmg.2014.10.004
Llamas, Perdomo, Magyar, Fulcher, Muenker et al., We thank the leadership of Boston Children's Hospital including Drs
Maecker, Pulendran, Nadeau, Rosenberg-Hasson, Leipold et al., School of Medicine at the University of California
Matthew, Altman, Jayavelu, Presnell, Kohr et al., None
Moll, Varga, Homocysteine and MTHFR Mutations, Circulation, doi:10.1161/circulationaha.114.013311
Moness, Mousa, Mousa, Adel, Ibrahim et al., Thrombophilia genetic mutations and their relation to disease severity among patients with COVID-19, PLoS One, doi:10.1371/journal.pone.0296668
Nefic, Mackic-Djurovic, Eminovic, The Frequency of the 677C>T and 1298A>C Polymorphisms in the Methylenetetrahydrofolate Reductase (MTHFR) Gene in the Population, Med Arch, doi:10.5455/medarh.2018.72.164-169
Ozonoff, Diray-Arce, Chen, Kho, Milliren et al., None
Ozonoff, Jayavelu, Liu, Melamed, Milliren et al., Features of acute COVID-19 associated with post-acute sequelae of SARS-CoV-2 phenotypes: results from the IMPACC study, Nat Commun, doi:10.1038/s41467-023-44090-5
Ozonoff, Schaenman, Jayavelu, Milliren, Calfee et al., Phenotypes of disease severity in a cohort of hospitalized COVID-19 patients: Results from the IMPACC study, EBioMedicine, doi:10.1016/j.ebiom.2022.104208
Perła ; Kaján, Jakubowski, COVID-19 and One-Carbon Metabolism, Int J Mol Sci, doi:10.3390/ijms23084181
Peters, Overton, Vita, Westendorf, Inc, None, Toronto, ON M
Petrova, Maynard, Wang, Kanarek, Regulatory mechanisms of one-carbon metabolism enzymes, J Biol Chem, doi:10.1016/j.jbc.2023.105457
Ponti, Pastorino, Manfredini, Ozben, Oliva et al., COVID-19 spreading across world correlates with C677T allele of the methylenetetrahydrofolate reductase (MTHFR) gene prevalence, J Clin Lab Anal, doi:10.1002/jcla.23798
Páez-Franco, Torres-Ruiz, Sosa-Hernández, Cervantes-Díaz, Romero-Ramírez et al., Metabolomics analysis reveals a modified amino acid metabolism that correlates with altered oxygen homeostasis in COVID-19 patients, Sci Rep, doi:10.1038/s41598-021-85788-0
Raghubeer, Matsha, Methylenetetrahydrofolate (MTHFR), the One-Carbon Cycle, and Cardiovascular Risks, Nutrients, doi:10.3390/nu13124562
Reed, Schaenman, Salehi-Rad, Rivera, Pickering et al., Jennifer Fulcher University of California
Rouphael, Bosinger, Boddapati, Tharp, Pellegrini et al., School of Medicine at Mount Sinai
Scott, Hutton, Michelotti, Wong, Prevention of Organ Failure
Scott, Tebbutt, Casey, Shannon Case Western Reserve University and
Sekaly, Fourati, Mccomsey, Harris, Sieg et al., None
Seyfert-Margolis, School, Medicine, None
Sherif, Gomez, Connors, Henrich, Reeves, Pathogenic mechanisms of post-acute sequelae of SARS-CoV-2 infection (PASC), Elife, doi:10.7554/eLife.86002
Stegmann, Dickmanns, Gerber, Nikolova, Klemke et al., The folate antagonist methotrexate diminishes replication of the coronavirus SARS-CoV-2 and enhances the antiviral efficacy of remdesivir in cell culture models, Virus Res, doi:10.1016/j.virusres.2021.198469
Su, Chen, Yuan, Lausted, Choi et al., Multi-Omics Resolves a Sharp Disease-State Shift between Mild and Moderate COVID-19, Cell, doi:10.1016/j.cell.2020.10.037
Vuong, Likelihood Ratio Tests for Model Selection and Non-Nested Hypotheses, Econometrica, doi:10.2307/1912557
Wang, Goodfellow, Walker, Blandford, Pfeffer et al., Trajectories of functional limitations, healthrelated quality of life and societal costs in individuals with long COVID: a population-based longitudinal cohort study, BMJ Open, doi:10.1136/bmjopen-2024-088538
Weisberg, Tran, Christensen, Sibani, Rozen, A second genetic polymorphism in methylenetetrahydrofolate reductase (MTHFR) associated with decreased enzyme activity, Mol Genet Metab, doi:10.1006/mgme.1998.2714
William, Messer, Hough, Siegel, Sullivan et al., None
Wilson, Danenberg, Johnston, Lenz, Ladner, Standing the test of time: targeting thymidylate biosynthesis in cancer therapy, Nat Rev Clin Oncol, doi:10.1038/nrclinonc.2014.51
Wulff Jam, A Comparison of Various Normalization Methods for LC/MS Metabolomics Data, Advances in Bioscience and Biotechnology, doi:10.4236/abb.2018.98022
Xiao, Nie, Pang, Wang, Hu et al., Integrated cytokine and metabolite analysis reveals immunometabolic reprogramming in COVID-19 patients with therapeutic implications, Nat Commun, doi:10.1038/s41467-021-21907-9
Zhang, Guo, Kim, Shah, Zhang et al., SARS-CoV-2 hijacks folate and one-carbon metabolism for viral replication, Nat Commun, doi:10.1038/s41467-021-21903-z
DOI record:
{
"DOI": "10.1101/2025.02.28.25323089",
"URL": "http://dx.doi.org/10.1101/2025.02.28.25323089",
"abstract": "<jats:title>Abstract</jats:title><jats:p>While the public health burden of SARS-CoV-2 infection has lessened due to natural and vaccine-acquired immunity, the emergence of less virulent variants, and antiviral medications, COVID-19 continues to take a significant toll. There are > 10,000 new hospitalizations per week in the U.S., many of whom develop post-acute sequelae of SARS-CoV-2 (PASC), or “long COVID”, with long-term health issues and compromised quality of life. Early identification of individuals at high risk of severe COVID-19 is key for monitoring and supporting respiratory status and improving outcomes. Therefore, precision tools for early detection of patients at high risk of severe disease can reduce morbidity and mortality. Here we report an untargeted and longitudinal metabolomic study of plasma derived from adult patients with COVID-19. One-carbon metabolism, a pathway previously shown as critical for viral propagation and disease progression, and a potential target for COVID-19 treatment, scored strongly as differentially abundant in patients with severe COVID-19. A follow-up targeted metabolite profiling revealed that one arm of the one-carbon metabolism pathway, the methionine cycle, is a major driver of the metabolic profile associated with disease severity. The methionine cycle produces S-adenosylmethionine (SAM), the methyl group donor important for methylation of DNA, RNA, and proteins, and its high abundance was reported to correlate with disease severity. Further, genomic data from the profiled patients revealed a genetic contributor to methionine metabolism and identified the C677T allele of the<jats:italic>MTHFR</jats:italic>gene as a pre-existing predictor of disease trajectory - patients homozygous for the<jats:italic>MTHFR</jats:italic>C677T have higher incidence of experiencing severe disease. Our results raise the possibility that screening for the common genetic<jats:italic>MTHFR</jats:italic>variant may be an actionable approach to stratify risk of COVID severity and may inform novel precision COVID-19 treatment strategies.</jats:p>",
"accepted": {
"date-parts": [
[
2025,
3,
3
]
]
},
"author": [
{
"ORCID": "https://orcid.org/0000-0001-9996-9353",
"affiliation": [],
"authenticated-orcid": false,
"family": "Petrova",
"given": "Boryana",
"sequence": "first"
},
{
"affiliation": [],
"family": "Syphurs",
"given": "Caitlin",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Culhane",
"given": "Andrew J",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Chen",
"given": "Jing",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-1573-7577",
"affiliation": [],
"authenticated-orcid": false,
"family": "Chen",
"given": "Ernie",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-7772-5910",
"affiliation": [],
"authenticated-orcid": false,
"family": "Cotsapas",
"given": "Chris",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Esserman",
"given": "Denise",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-8661-4454",
"affiliation": [],
"authenticated-orcid": false,
"family": "Montgomery",
"given": "Ruth",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-4957-1544",
"affiliation": [],
"authenticated-orcid": false,
"family": "Kleinstein",
"given": "Steven",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-8650-3693",
"affiliation": [],
"authenticated-orcid": false,
"family": "Smolen",
"given": "Kinga",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Mendez",
"given": "Kevin",
"sequence": "additional"
},
{
"affiliation": [],
"name": "IMPACC Network",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-6236-4705",
"affiliation": [],
"authenticated-orcid": false,
"family": "Lasky-Su",
"given": "Jessica",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-0179-6648",
"affiliation": [],
"authenticated-orcid": false,
"family": "Steen",
"given": "Hanno",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-5859-1945",
"affiliation": [],
"authenticated-orcid": false,
"family": "Levy",
"given": "Ofer",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0003-2183-4269",
"affiliation": [],
"authenticated-orcid": false,
"family": "Diray-Arce",
"given": "Joann",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-2068-3908",
"affiliation": [],
"authenticated-orcid": false,
"family": "Kanarek",
"given": "Naama",
"sequence": "additional"
}
],
"container-title": [],
"content-domain": {
"crossmark-restriction": false,
"domain": []
},
"created": {
"date-parts": [
[
2025,
3,
4
]
],
"date-time": "2025-03-04T00:10:13Z",
"timestamp": 1741047013000
},
"deposited": {
"date-parts": [
[
2025,
3,
5
]
],
"date-time": "2025-03-05T16:15:14Z",
"timestamp": 1741191314000
},
"group-title": "Infectious Diseases (except HIV/AIDS)",
"indexed": {
"date-parts": [
[
2025,
3,
5
]
],
"date-time": "2025-03-05T17:10:19Z",
"timestamp": 1741194619803,
"version": "3.38.0"
},
"institution": [
{
"name": "medRxiv"
}
],
"is-referenced-by-count": 0,
"issued": {
"date-parts": [
[
2025,
3,
3
]
]
},
"license": [
{
"URL": "https://www.medrxiv.org/about/FAQ#license",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
3,
3
]
],
"date-time": "2025-03-03T00:00:00Z",
"timestamp": 1740960000000
}
}
],
"link": [
{
"URL": "https://syndication.highwire.org/content/doi/10.1101/2025.02.28.25323089",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "246",
"original-title": [],
"posted": {
"date-parts": [
[
2025,
3,
3
]
]
},
"prefix": "10.1101",
"published": {
"date-parts": [
[
2025,
3,
3
]
]
},
"publisher": "Cold Spring Harbor Laboratory",
"reference": [
{
"DOI": "10.1136/bmjopen-2024-088538",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.1"
},
{
"key": "2025030508150628000_2025.02.28.25323089v1.2",
"unstructured": "CDC 2024. Available from: www.cdc.gov/covid/php/surveillance/burden-estimates.html."
},
{
"DOI": "10.1001/jamanetworkopen.2021.28568",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.3"
},
{
"DOI": "10.1001/jamanetworkopen.2022.55795",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.4"
},
{
"DOI": "10.1038/s41467-023-44090-5",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.5"
},
{
"DOI": "10.1186/s12889-023-16916-w",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.6"
},
{
"DOI": "10.1038/s41467-021-21903-z",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.7"
},
{
"DOI": "10.1016/j.cmet.2016.08.009",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.8"
},
{
"DOI": "10.1038/nrclinonc.2014.51",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.9"
},
{
"DOI": "10.3390/ijms23084181",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.10"
},
{
"DOI": "10.1016/j.virusres.2021.198469",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.11"
},
{
"DOI": "10.1006/mgme.1998.2714",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.12"
},
{
"DOI": "10.1016/j.ejmg.2014.10.004",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.13"
},
{
"DOI": "10.1016/j.mehy.2020.110234",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.14"
},
{
"DOI": "10.1002/jcla.23798",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.15"
},
{
"DOI": "10.1155/2021/7686374",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.16"
},
{
"DOI": "10.7759/cureus.34873",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.17"
},
{
"DOI": "10.3390/pathogens12030438",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.18"
},
{
"DOI": "10.1371/journal.pone.0296668",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.19"
},
{
"DOI": "10.1016/j.ebiom.2024.105251",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.20"
},
{
"DOI": "10.1186/s13148-022-01313-8",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.21"
},
{
"DOI": "10.1126/sciimmunol.abf3733",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.22"
},
{
"DOI": "10.1016/j.xcrm.2023.101079",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.23"
},
{
"DOI": "10.1172/jci176640",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.24"
},
{
"DOI": "10.4236/abb.2018.98022",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.25"
},
{
"DOI": "10.1016/j.cell.2020.10.037",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.26"
},
{
"DOI": "10.1038/s41467-021-21907-9",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.27"
},
{
"DOI": "10.1038/s41598-021-85788-0",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.28"
},
{
"DOI": "10.1016/j.jbc.2023.105457",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.29"
},
{
"DOI": "10.1093/jn/129.5.919",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.30"
},
{
"DOI": "10.1007/s10552-013-0264-3",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.31"
},
{
"DOI": "10.5455/medarh.2018.72.164-169",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.32"
},
{
"DOI": "10.1093/biostatistics/kxw045",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.33"
},
{
"DOI": "10.1111/j.2041-210X.2010.00063.x",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.34"
},
{
"DOI": "10.2307/1912557",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.35"
},
{
"DOI": "10.1038/s41591-024-02987-8",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.36"
},
{
"DOI": "10.7554/eLife.86002",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.37"
},
{
"DOI": "10.1038/s41598-024-71951-w",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.38"
},
{
"DOI": "10.3390/metabo10120492",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.39"
},
{
"DOI": "10.1038/s41392-024-01954-6",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.40"
},
{
"DOI": "10.3390/nu13124562",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.41"
},
{
"DOI": "10.1161/CIRCULATIONAHA.114.013311",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.42"
},
{
"DOI": "10.1016/j.ebiom.2022.104208",
"doi-asserted-by": "publisher",
"key": "2025030508150628000_2025.02.28.25323089v1.43"
}
],
"reference-count": 43,
"references-count": 43,
"relation": {},
"resource": {
"primary": {
"URL": "http://medrxiv.org/lookup/doi/10.1101/2025.02.28.25323089"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"subtype": "preprint",
"title": "An Allele of the MTHFR one-carbon metabolism gene predicts severity of COVID-19",
"type": "posted-content"
}