Targeting G9a-m6A translational mechanism of SARS-CoV-2 pathogenesis for multifaceted therapeutics of COVID-19 and its sequalae
et al., iScience, doi:10.1016/j.isci.2025.112632, May 2025
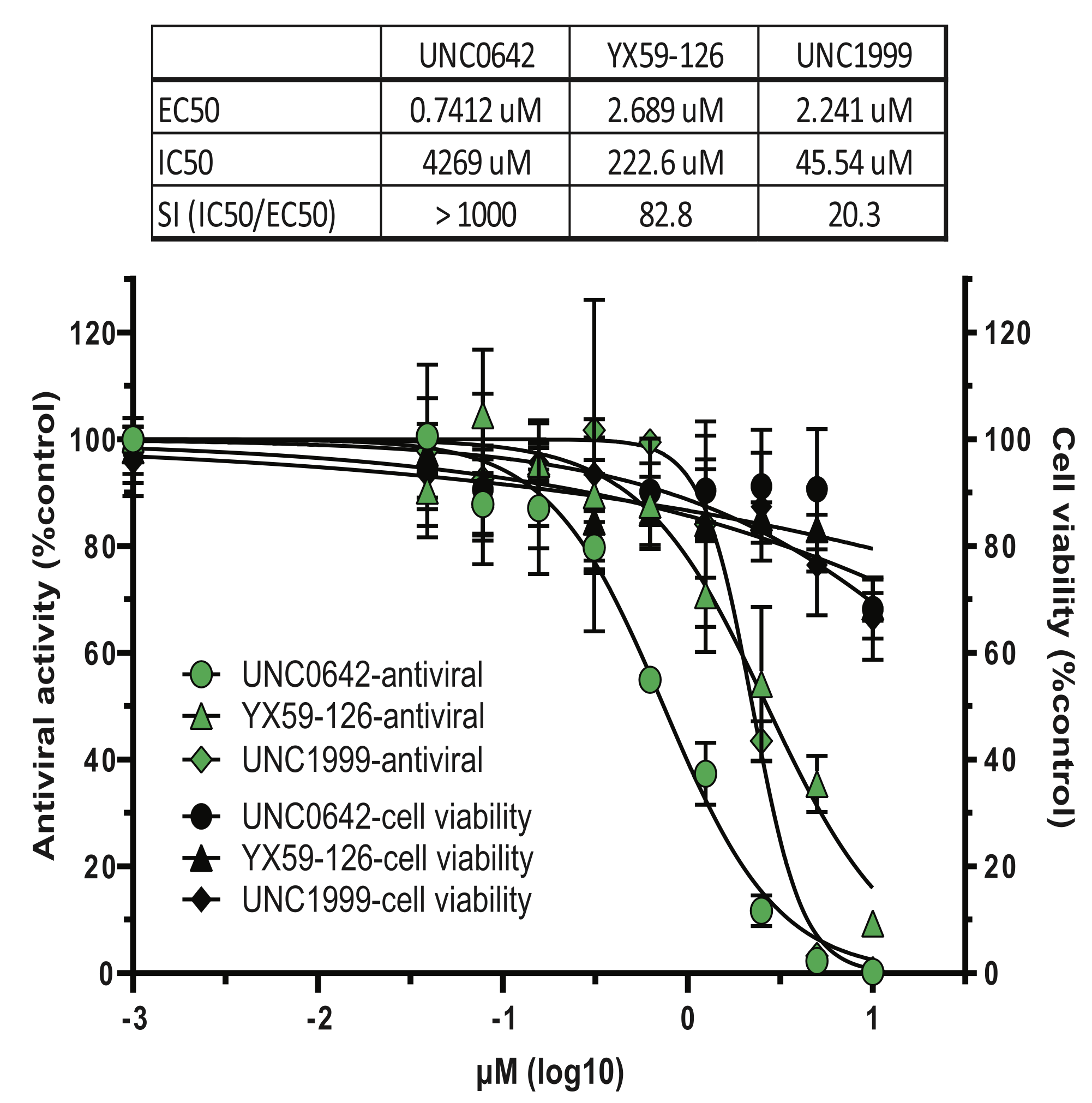
In vitro and mouse study showing that G9a and EZH2 methyltransferase inhibitors (UNC0642, MS1262, UNC1999) block SARS-CoV-2 replication in A549-hACE2 cells with good selectivity indices.
Muneer et al., 11 May 2025, USA, peer-reviewed, 16 authors.
Contact: xianc@email.unc.edu (corresponding author), xianc@email.unc.edu (corresponding author).
Targeting G9a-m6A translational mechanism of SARS-CoV-2 pathogenesis for multifaceted therapeutics of COVID-19 and its sequalae
iScience, doi:10.1016/j.isci.2025.112632
N6-methyladenosine (m6A) modification pathway is hijacked by several RNA viruses, including SARS-CoV-2, making it an attractive host-directed target for development of broad-spectrum antivirals. Here, we show that histone methyltransferase G9a, through its interaction with METTL3, regulates SARS-CoV-2-mediated rewiring of host m6A methylome to ultimately promote turnover, abundance, secretion and/or phosphorylation of various viral receptors and proteases, transcription factors, cytokines/chemokines, coagulation and angiogenesis associated proteins, and fibrosis markers. More importantly, drugs targeting G9a and its associated protein EZH2 are potent inhibitors of SARS-CoV-2 replication and reverse multi-omic effects of coronavirus infection in human alveolar epithelial cells (A549-hACE2) and COVID-19 patient peripheral blood mononuclear cells (PBMCs)-with similar changes seen in multiorgan autopsy samples from COVID-19 patients. Altogether, we extend G9a function(s) beyond transcription to translational regulation during COVID-19 pathogenesis and show that targeting this master regulatory complex represents a new strategy (drug-class) that can be leveraged to combat emerging anti-viral resistance and infections.
AUTHOR CONTRIBUTIONS X.C. conceived, designed, supervised the project, and wrote the manuscript. A.M. analyzed and interpreted data and wrote the manuscript. L.X. performed inhibitor treatment, sample preparation and processing for MS/MS experimental analysis and analyzed data. F.Z., H.S., and J.S. performed RNA-seq, MeRIP-seq, and analyzed the data. X.X., P.-Y.S., and P.W. performed inhibitor treatment and collected antiviral and cytotoxicity data. J.A.W. analyzed RNAseq and MeRIP-seq data. Y.X., X.Y., and J.J. provided UNC0965, UNC0642, UNC1999, and MS1262 targeting G9a/Ezh2.
DECLARATION OF INTERESTS X.C. is the founder of TransChromix, LLC. J.J. is a cofounder and equity shareholder in Cullgen, Inc., a scientific cofounder and scientific advisory board member of Onsero Therapeutics, Inc., and a consultant for Cullgen, Inc., EpiCypher, Inc., Accent Therapeutics, Inc, and Tavotek Biotherapeutics, Inc. The Jin laboratory received research funds from Celgene Corporation, Levo Therapeutics Inc., Cullgen, Inc. and Cullinan Oncology, Inc. The indication of using clinically trialed G9a/Ezh2 inhibitors for COVID-19 therapy is protected by US provisional patent application #63/113,211 as ''Use of Method''.
STAR★METHODS Detailed methods are provided in the online version of this paper and include the following:
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS Cell lines and primary cultures of human origin Human alveolar epithelial cells that overexpress hACE2 receptor..
References
Abavisani, Rahimian, Mahdavi, Tokhanbigli, Mollapour Siasakht et al., Mutations in SARS-CoV-2 structural proteins: a global analysis, Virol. J
Alcantara, Higuchi, Kirita, Matoba, Hoshino, Deep Mutational Scanning to Predict Escape from Bebtelovimab in SARS-CoV-2 Omicron Subvariants, Vaccines
Amicone, Borges, Alves, Isidro, Ze et al., Mutation rate of SARS-CoV-2 and emergence of mutators during experimental evolution, Evol. Med. Public Health
An, Xie, Liao, Jiang, Dong et al., Systematic analysis of clinical relevance and molecular characterization of m6A in COVID-19 patients, Genes Dis
Ankney, Muneer, Chen, Relative and absolute quantitation in mass spectrometry-based proteomics, Annu. Rev. Anal. Chem
Arunachalam, Wimmers, Mok, Perera, Scott et al., Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans, Science
Ayaz, Crea, Targeting SARS-CoV-2 using polycomb inhibitors as antiviral agents, Future Medicine
Batista-Roche, Go ´mez-Gil, Lund, Berlanga-Robles, Garcı ´a-Gasca et al., Global m6A RNA Methylation in SARS-CoV-2 Positive Nasopharyngeal Samples in a Mexican Population: A First Approximation Study, J. Alzheimers Dis. Rep
Bergant, Yamada, Grass, Tsukamoto, Lavacca et al., Attenuation of SARS-CoV-2 replication and associated inflammation by concomitant targeting of viral and host cap 2'-O-ribose methyltransferases, EMBO J
Bojkova, Klann, Koch, Widera, Krause et al., Proteomics of SARS-CoV-2-infected host cells reveals therapy targets, Nature, doi:10.1038/s41586-020-2332-7
Bolger, Lohse, Usadel, Trimmomatic: a flexible trimmer for Illumina sequence data, Bioinformatics
Bouhaddou, Memon, Meyer, White, Rezelj et al., The Global Phosphorylation Landscape of SARS-CoV-2 Infection, Cell, doi:10.1016/j.cell.2020.06.034
Boutajangout, Frontera, Debure, Vedvyas, Faustin et al., Plasma biomarkers of neurodegeneration and neuroinflammation in hospitalized COVID-19 patients with and without new neurological symptoms, Alzheimer's Dement
Burgess, Depledge, Thompson, Srinivas, Grande et al., Targeting the m(6)A RNA modification pathway blocks SARS-CoV-2 and HCoV-OC43 replication, Genes Dev, doi:10.1101/gad.348320.121
Butler, Mozsary, Meydan, Foox, Rosiene et al., Shotgun transcriptome, spatial omics, and isothermal profiling of SARS-CoV-2 infection reveals unique host responses, viral diversification, and drug interactions, Nat. Commun
Carvelli, Demaria, Ve ´ly, Batista, Benmansour et al., Association of COVID-19 inflammation with activation of the C5a-C5aR1 axis, Nature
Cox, Peacock, Harvey, Hughes, Wright et al., COVID-19 Genomics UK COG-UK Consortium (2023). SARS-CoV-2 variant evasion of monoclonal antibodies based on in vitro studies, Nat. Rev. Microbiol
Daniloski, Jordan, Wessels, Hoagland, Kasela et al., Identification of required host factors for SARS-CoV-2 infection in human cells, Cell
Davidson, Williamson, Lewis, Shoemark, Carroll et al., Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein, Genome Med
Diao, Wang, Tan, Chen, Liu et al., Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19), Front. Immunol, doi:10.3389/fimmu.2020.00827
Dobin, Davis, Schlesinger, Drenkow, Zaleski et al., STAR: ultrafast universal RNA-seq aligner, Bioinformatics
Dong, Du, Gardner, An interactive web-based dashboard to track COVID-19 in real time, Lancet Infect. Dis
Gandhi, Klein, Robertson, Pen ˜a-Herna ´ndez, Lin et al., De novo emergence of a remdesivir resistance mutation during treatment of persistent SARS-CoV-2 infection in an immunocompromised patient: a case report, Nat. Commun
Gavriilaki, Kokoris, COVID-19 sequelae: can long-term effects be predicted?, Lancet Infect. Dis, doi:10.1016/S1473-3099(22)00529-1
Ge, Li, Yang, Chmura, Zhu et al., Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor, Nature
Gordon, Hiatt, Bouhaddou, Rezelj, Ulferts et al., Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms, Science
Gordon, Jang, Bouhaddou, Xu, Obernier et al., A SARS-CoV-2 protein interaction map reveals targets for drug repurposing, Nature
Gu, Wang, Zhang, Li, Guo et al., Probing long COVID through a proteomic lens: a comprehensive two-year longitudinal cohort study of hospitalised survivors, EBioMedicine
Gutlapalli, Ganipineni, Danda, Fabian, Okorie et al., Exploring the Potential of Broadly Neutralizing Antibodies for Treating SARS-CoV-2 Variants of Global Concern in 2023: A Comprehensive Clinical Review, Cureus
Han, Yang, Liu, Liu, Wu et al., Prominent changes in blood coagulation of patients with SARS-CoV-2 infection, Clin. Chem. Lab. Med
Hao, Hao, Chen, Chen, Zhang et al., N 6-methyladenosine modification and METTL3 modulate enterovirus 71 replication, Nucleic Acids Res
Heilmann, Costacurta, Moghadasi, Ye, Pavan et al., SARS-CoV-2 3CLpro mutations selected in a VSV-based system confer resistance to nirmatrelvir, ensitrelvir, and GC376, Sci. Transl. Med
Hu, Lewandowski, Tan, Zhang, Morgan et al., Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir, doi:10.1101/2022.06.28.497978
Hu, Song, Zhou, Jiao, Li, METTL3 Accelerates Breast Cancer Progression via Regulating EZH2 m6A Modification, J. Healthc. Eng
Huttlin, Ting, Bruckner, Gebreab, Gygi et al., The BioPlex Network: A Systematic Exploration of the Human Interactome, Cell, doi:10.1016/j.cell.2015.06.043
Ishizaki, Bi, Nguyen, Maeno, Hara et al., Neutralizing-antibody response to SARS-CoV-2 for 12 months after the COVID-19 workplace outbreaks in Japan, PLoS One
Izadpanah, Rappaport, Datta, Epitranscriptomics of SARS-CoV-2 infection, Front. Cell Dev. Biol
Kaneko, Esmail, Voss, Martin, Slessarev et al., System-wide hematopoietic and immune signaling aberrations in COVID-19 revealed by deep proteome and phosphoproteome analysis, doi:10.1101/2021.03.19.21253675
Kee, Thudium, Renner, Glastad, Palozola et al., SARS-CoV-2 disrupts host epigenetic regulation via histone mimicry, Nature
Kim, Siddiqui, Hepatitis B virus X protein recruits methyltransferases to affect cotranscriptional N6-methyladenosine modification of viral/host RNAs, Proc. Natl. Acad. Sci
Klann, Bojkova, Tascher, Ciesek, Mu ¨nch et al., Growth factor receptor signaling inhibition prevents SARS-CoV-2 replication, Mol. Cell
Konze, Ma, Li, Barsyte-Lovejoy, Parton et al., An orally bioavailable chemical probe of the Lysine Methyltransferases EZH2 and EZH1, ACS Chem. Biol, doi:10.1021/cb400133j
Konze, Pattenden, Liu, Barsyte-Lovejoy, Li et al., A chemical tool for in vitro and in vivo precipitation of lysine methyltransferase G9a, ChemMed-Chem
Krause, Fleming, Longini, Peto, Briand et al., SARS-CoV-2 variants and vaccines, N. Engl. J. Med
Kumar, Khandelwal, Chander, Nagori, Verma et al., S-adenosylmethionine-dependent methyltransferase inhibitor DZNep blocks transcription and translation of SARS-CoV-2 genome with a low tendency to select for drug-resistant viral variants, Antiviral Res
Laurent, Sofianatos, Komarova, Gimeno, Tehrani et al., Global BioID-based SARS-CoV-2 proteins proximal interactome unveils novel ties between viral polypeptides and host factors involved in multiple COVID19-associated mechanisms, doi:10.1101/2020.08.28.272955
Li, Guo, Tian, Wang, Yang et al., Virus-Host Interactome and Proteomic Survey Reveal Potential Virulence Factors Influencing SARS-CoV-2 Pathogenesis, Med
Li, Handsaker, Wysoker, Fennell, Ruan et al., The sequence alignment/map format and SAMtools, Bioinformatics
Li, Hui, Bray, Gonzalez, Zeller et al., METTL3 regulates viral m6A RNA modification and host cell innate immune responses during SARS-CoV-2 infection, Cell Rep
Liao, Liu, Yuan, Wen, Xu et al., Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19, Nat. Med, doi:10.1038/s41591-020-0901-9
Lin, Choe, Du, Triboulet, Gregory, The m6A methyltransferase METTL3 promotes translation in human cancer cells, Mol. Cell
Lineburg, Smith, The Persistence of SARS-CoV-2 and Its Role in Long Covid, NEJM Evid, doi:10.1056/EVIDe2300165
Liu, Barsyte-Lovejoy, Li, Xiong, Korboukh et al., Discovery of an in vivo chemical probe of the lysine methyltransferases G9a and GLP, J. Med. Chem
Liu, Hu, Luo, Xiong, Chen et al., USP7-Based Deubiquitinase-Targeting Chimeras Stabilize AMPK, J. Am. Chem. Soc
Liu, Li, Liu, Yao, Wang et al., SARS-CoV-2 cell tropism and multiorgan infection, Cell Discov
Liu, Wang, Zhang, Cui, Shen et al., Viral sepsis is a complication in patients with Novel Corona Virus Disease (COVID-19), Med. Drug Discov
Liu, Xu, Li, Ye, Zhou et al., The m(6)A methylome of SARS-CoV-2 in host cells, Cell Res, doi:10.1038/s41422-020-00465-7
Liu, Zhuo, Wang, Zhang, Li et al., METTL3 plays multiple functions in biological processes, Am. J. Cancer Res
Lu, Li, Ao, Huang, Liu et al., The risk of COVID-19 can be predicted by a nomogram based on m6A-related genes, Infect. Genet. Evol
Malbec, Celerier, Bizet, Calonne, Hofmann-Winkler et al., The RNA demethylase FTO controls m6A marking on SARS-CoV-2 and classifies COVID-19 severity in patients, doi:10.1101/2022.06.27.497749
Malovannaya, Lanz, Jung, Bulynko, Le et al., Analysis of the human endogenous coregulator complexome, Cell, doi:10.1016/j.cell.2011.05.006
Meganck, Baric, Developing therapeutic approaches for twenty-first-century emerging infectious viral diseases, Nat. Med
Menachery, Eisfeld, Scha ¨fer, Josset, Sims et al., Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses, mBio
Meng, Zhang, Wang, Zhang, Yang et al., RBM15-mediated N6-methyladenosine modification affects COVID-19 severity by regulating the expression of multitarget genes, Cell Death Dis
Menon, Otto, Sealfon, Nair, Wong et al., SARS-CoV-2 receptor networks in diabetic kidney disease, BK-Virus nephropathy and COVID-19 associated acute kidney injury, doi:10.1101/2020.05.09.20096511
Merad, Martin, Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages, Nat. Rev. Immunol, doi:10.1038/s41577-020-0331-4
Muneer, Non-Canonical Translation Regulatory Function of G9a in Chronic Inflammation Associated Diseases (The University of North Carolina at Chapel Hill)
Muneer, Wang, Xie, Zhang, Wu et al., Non-canonical function of histone methyltransferase G9a in the translational regulation of chronic inflammation, Cell Chem. Biol, doi:10.1016/j.chembiol.2023.09.012
Nie, Qian, Sun, Huang, Dong et al., Multi-organ proteomic landscape of COVID-19 autopsies, Cell, doi:10.1016/j.cell.2021.01.004
Paranjpe, Russak, De Freitas, Lala, Miotto et al., Clinical Characteristics of Hospitalized Covid-19 Patients in New York City, doi:10.1101/2020.04.19.20062117
Park, Foox, Hether, Beheshti, Saravia-Butler et al., Systemic tissue and cellular disruption from SARS-CoV-2 infection revealed in COVID-19 autopsies and spatial omics tissue maps, Preprint at bioRxiv, doi:10.1101/2021.03.08.434433
Park, Kim, Kim, Kim, Lee et al., In-depth blood proteome profiling analysis revealed distinct functional characteristics of plasma proteins between severe and non-severe COVID-19 patients, Sci. Rep
Picelli, Faridani, Bjo ¨rklund, Winberg, Sagasser et al., Full-length RNA-seq from single cells using Smartseq2, Nat. Protoc
Qing, Chen, Wang, m6A regulator-mediated methylation modification patterns and characteristics in COVID-19 patients, Front. Public Health
Qiu, Hua, Li, Zhou, Chen, m6A Regulator-Mediated Methylation Modification Patterns and Characteristics of Immunity in Blood Leukocytes of COVID-19 Patients, Front. Immunol
Ramlall, Thangaraj, Meydan, Foox, Butler et al., Immune complement and coagulation dysfunction in adverse outcomes of SARS-CoV-2 infection, Nat. Med, doi:10.1038/s41591-020-1021-2
Roundtree, Evans, Pan, He, Dynamic RNA Modifications in Gene Expression Regulation, Cell, doi:10.1016/j.cell.2017.05.045
Sacco, Bland, Horner, WTAP targets the METTL3 m6A-methyltransferase complex to cytoplasmic hepatitis C virus RNA to regulate infection, J. Virol
Sakai, Masuda, Tarumoto, Aihara, Tsunoda et al., Genomescale CRISPR-Cas9 screen identifies novel host factors as potential therapeutic targets for SARS-CoV-2 infection, doi:10.1101/2023.03.06.531431
Samavarchi-Tehrani, Abdouni, Knight, Astori, Samson et al., A SARS-CoV-2-host proximity interactome, doi:10.1101/2020.09.03.282103
Schneider, Luna, Hoffmann, Sa ´nchez-Rivera, Leal et al., Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks, Cell
Shen, Yi, Sun, Bi, Du et al., Proteomic and Metabolomic Characterization of COVID-19 Patient Sera, Cell, doi:10.1016/j.cell.2020.05.032
Straining, Eighmy, Tazemetostat: EZH2 Inhibitor, J. Adv. Pract. Oncol, doi:10.6004/jadpro.2022.13.2.7
Stukalov, Girault, Grass, Bergant, Karayel et al., Multi-level proteomics reveals host-perturbation strategies of SARS-CoV-2 and SARS-CoV, doi:10.1101/2020.06.17.156455
Su, Chen, Yuan, Lausted, Choi et al., Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19, Cell
Su, Yuan, Chen, Ng, Wang et al., Multiple early factors anticipate postacute COVID-19 sequelae, Cell
Sugiyama, Suzuki, Aoyama, Toyota, Nakagawa et al., Long-term observation of antibody titers against SARS-CoV-2 following vaccination, Public Health Pract
Sun, Li, Ju, Rao, Huang et al., In vivo structural characterization of the SARS-CoV-2 RNA genome identifies host proteins vulnerable to repurposed drugs, Cell
Swenson, Swenson, Pathophysiology of acute respiratory distress syndrome and COVID-19 lung injury, Crit. Care Clin
Tay, Poh, Re ´nia, Macary, Ng, The trinity of COVID-19: immunity, inflammation and intervention, Nat. Rev. Immunol
Wan, Xu, Wei, Zhang, Han et al., Phosphorylation of EZH2 by AMPK suppresses PRC2 methyltransferase activity and oncogenic function, Mol. Cell
Wang, Lee, Kim, Xiong, Hasani et al., SARS-CoV-2 restructures host chromatin architecture, Nat. Microbiol
Wang, Li, Yang, Huang, Zhang et al., Serological evidence of bat SARS-related coronavirus infection in humans, China, Virol. Sin
Wang, Muneer, Xie, Zhang, Wu et al., Novel gene-specific translation mechanism of dysregulated, chronic inflammation reveals promising, multifaceted COVID-19 therapeutics, doi:10.1101/2020.11.14.382416
Wang, Simoneau, Kulsuptrakul, Bouhaddou, Travisano et al., Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses, Cell
Wei, Alfajaro, Deweirdt, Hanna, Lu-Culligan et al., Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection, Cell
Weng, Wang, An, Cassin, Vissers et al., Epitranscriptomic m6A regula-tion of axon regeneration in the adult mammalian nervous system, Neuron
Wessel, Flu ¨gge, A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids, Anal. Biochem
Wrobel, Xie, Wang, Liu, Rashid et al., Multi-omic Dissection of Oncogenically Active Epiproteomes Identifies Drivers of Proliferative and Invasive Breast Tumors, iScience, doi:10.1016/j.isci.2019.07.001
Xie, Muruato, Zhang, Lokugamage, Fontes-Garfias et al., A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19, Nat. Commun
Xie, Sheehy, Xiong, Muneer, Wrobel et al., Novel brain-penetrant inhibitor of G9a methylase blocks Alzheimer disease proteopathology for precision medication, doi:10.1101/2023.10.25.23297491
Yaron, Heaton, Levy, Johnson, Jordan et al., The FDA-approved drug Alectinib compromises SARS-CoV-2 nucleocapsid phosphorylation and inhibits viral infection in vitro, Preprint at bioRxiv, doi:10.1101/2020.08.14.251207
Yi, Li, Meng, Li, Li et al., A PRC2-independent function for EZH2 in regulating rRNA 2'-O methylation and IRES-dependent translation, Nat. Cell Biol, doi:10.1038/s41556-021-00653-6
Zannella, Rinaldi, Boccia, Chianese, Sasso et al., Regulation of m6A methylation as a new therapeutic option against COVID-19, Pharmaceuticals
Zhang, Hao, Ma, Zhang, Hu et al., Methyltransferase-like 3 modulates severe acute respiratory syndrome coronavirus-2 RNA N6-methyladenosine modification and replication, mBio
Zhao, Hu, Ohtsuki, Jin, Wu et al., Hyperactivity of the CD155 immune checkpoint suppresses anti-viral immunity in patients with coronary artery disease, Nat. Cardiovasc. Res
Zhao, Roundtree, He, Post-transcriptional gene regulation by mRNA modifications, Nat. Rev. Mol. Cell Biol, doi:10.1038/nrm.2016.132
Zhou, Ji, Chen, Bi, Li et al., Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses, Cell
Zhou, Liu, Gupta, Paramo, Hou et al., A comprehensive SARS-CoV-2-human protein-protein interactome reveals COVID-19 pathobiology and potential host therapeutic targets, Nat. Biotechnol, doi:10.1038/s41587-022-01474-0
Zhou, Zhou, Pache, Chang, Khodabakhshi et al., Metascape provides a biologist-oriented resource for the analysis of systems-level datasets, Nat. Commun
Zhu, Feng, Hu, Wang, Yu et al., A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry, Nat. Commun
Zhu, Jiang, Zhan, Wang, Xia et al., Durability of neutralization against Omicron subvariants after vaccination and breakthrough infection, Cell Rep, doi:10.1016/j.celrep.2023.112075
DOI record:
{
"DOI": "10.1016/j.isci.2025.112632",
"ISSN": [
"2589-0042"
],
"URL": "http://dx.doi.org/10.1016/j.isci.2025.112632",
"alternative-id": [
"S2589004225008934"
],
"article-number": "112632",
"assertion": [
{
"label": "This article is maintained by",
"name": "publisher",
"value": "Elsevier"
},
{
"label": "Article Title",
"name": "articletitle",
"value": "Targeting G9a-m6A translational mechanism of SARS-CoV-2 pathogenesis for multifaceted therapeutics of COVID-19 and its sequalae"
},
{
"label": "Journal Title",
"name": "journaltitle",
"value": "iScience"
},
{
"label": "CrossRef DOI link to publisher maintained version",
"name": "articlelink",
"value": "https://doi.org/10.1016/j.isci.2025.112632"
},
{
"label": "Content Type",
"name": "content_type",
"value": "article"
},
{
"label": "Copyright",
"name": "copyright",
"value": "© 2025 The Author(s). Published by Elsevier Inc."
}
],
"author": [
{
"affiliation": [],
"family": "Muneer",
"given": "Adil",
"sequence": "first"
},
{
"affiliation": [],
"family": "Xie",
"given": "Ling",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Xie",
"given": "Xuping",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Zhang",
"given": "Feng",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Wrobel",
"given": "John A.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Xiong",
"given": "Yan",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Yu",
"given": "Xufen",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Wang",
"given": "Charles",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Gheorghe",
"given": "Ciprian",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Wu",
"given": "Ping",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Song",
"given": "Juan",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Ming",
"given": "Guo-Li",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Jin",
"given": "Jian",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Song",
"given": "Hongjun",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Shi",
"given": "Pei-Yong",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Chen",
"given": "Xian",
"sequence": "additional"
}
],
"container-title": "iScience",
"container-title-short": "iScience",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"cell.com",
"elsevier.com",
"sciencedirect.com"
]
},
"created": {
"date-parts": [
[
2025,
5,
11
]
],
"date-time": "2025-05-11T22:49:11Z",
"timestamp": 1747003751000
},
"deposited": {
"date-parts": [
[
2025,
6,
9
]
],
"date-time": "2025-06-09T18:52:06Z",
"timestamp": 1749495126000
},
"funder": [
{
"DOI": "10.13039/100000062",
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/100000062",
"id-type": "DOI"
}
],
"name": "National Institute of Diabetes and Digestive and Kidney Diseases"
}
],
"indexed": {
"date-parts": [
[
2025,
6,
9
]
],
"date-time": "2025-06-09T19:10:03Z",
"timestamp": 1749496203430,
"version": "3.41.0"
},
"is-referenced-by-count": 0,
"issue": "6",
"issued": {
"date-parts": [
[
2025,
6
]
]
},
"journal-issue": {
"issue": "6",
"published-print": {
"date-parts": [
[
2025,
6
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://www.elsevier.com/tdm/userlicense/1.0/",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
6,
1
]
],
"date-time": "2025-06-01T00:00:00Z",
"timestamp": 1748736000000
}
},
{
"URL": "https://www.elsevier.com/legal/tdmrep-license",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
6,
1
]
],
"date-time": "2025-06-01T00:00:00Z",
"timestamp": 1748736000000
}
},
{
"URL": "http://creativecommons.org/licenses/by-nc-nd/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
5,
7
]
],
"date-time": "2025-05-07T00:00:00Z",
"timestamp": 1746576000000
}
}
],
"link": [
{
"URL": "https://api.elsevier.com/content/article/PII:S2589004225008934?httpAccept=text/xml",
"content-type": "text/xml",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://api.elsevier.com/content/article/PII:S2589004225008934?httpAccept=text/plain",
"content-type": "text/plain",
"content-version": "vor",
"intended-application": "text-mining"
}
],
"member": "78",
"original-title": [],
"page": "112632",
"prefix": "10.1016",
"published": {
"date-parts": [
[
2025,
6
]
]
},
"published-print": {
"date-parts": [
[
2025,
6
]
]
},
"publisher": "Elsevier BV",
"reference": [
{
"DOI": "10.1016/S1473-3099(20)30120-1",
"article-title": "An interactive web-based dashboard to track COVID-19 in real time",
"author": "Dong",
"doi-asserted-by": "crossref",
"first-page": "533",
"journal-title": "Lancet Infect. Dis.",
"key": "10.1016/j.isci.2025.112632_bib1",
"volume": "20",
"year": "2020"
},
{
"article-title": "Clinical Characteristics of Hospitalized Covid-19 Patients in New York City",
"author": "Paranjpe",
"journal-title": "medRxiv",
"key": "10.1016/j.isci.2025.112632_bib2",
"year": "2020"
},
{
"article-title": "SARS-CoV-2 receptor networks in diabetic kidney disease, BK-Virus nephropathy and COVID-19 associated acute kidney injury",
"author": "Menon",
"journal-title": "medRxiv",
"key": "10.1016/j.isci.2025.112632_bib3",
"year": "2020"
},
{
"article-title": "Long-term observation of antibody titers against SARS-CoV-2 following vaccination",
"author": "Sugiyama",
"journal-title": "Public Health Pract.",
"key": "10.1016/j.isci.2025.112632_bib4",
"volume": "4",
"year": "2022"
},
{
"DOI": "10.1371/journal.pone.0273712",
"article-title": "Neutralizing-antibody response to SARS-CoV-2 for 12 months after the COVID-19 workplace outbreaks in Japan",
"author": "Ishizaki",
"doi-asserted-by": "crossref",
"journal-title": "PLoS One",
"key": "10.1016/j.isci.2025.112632_bib5",
"volume": "17",
"year": "2022"
},
{
"DOI": "10.1093/emph/eoac010",
"article-title": "Mutation rate of SARS-CoV-2 and emergence of mutators during experimental evolution",
"author": "Amicone",
"doi-asserted-by": "crossref",
"first-page": "142",
"journal-title": "Evol. Med. Public Health",
"key": "10.1016/j.isci.2025.112632_bib6",
"volume": "10",
"year": "2022"
},
{
"DOI": "10.1186/s12985-022-01951-7",
"article-title": "Mutations in SARS-CoV-2 structural proteins: a global analysis",
"author": "Abavisani",
"doi-asserted-by": "crossref",
"first-page": "220",
"journal-title": "Virol. J.",
"key": "10.1016/j.isci.2025.112632_bib7",
"volume": "19",
"year": "2022"
},
{
"DOI": "10.1016/j.celrep.2023.112075",
"article-title": "Durability of neutralization against Omicron subvariants after vaccination and breakthrough infection",
"author": "Zhu",
"doi-asserted-by": "crossref",
"journal-title": "Cell Rep.",
"key": "10.1016/j.isci.2025.112632_bib8",
"volume": "42",
"year": "2023"
},
{
"DOI": "10.1038/s41579-022-00809-7",
"article-title": "SARS-CoV-2 variant evasion of monoclonal antibodies based on in vitro studies",
"author": "Cox",
"doi-asserted-by": "crossref",
"first-page": "112",
"journal-title": "Nat. Rev. Microbiol.",
"key": "10.1016/j.isci.2025.112632_bib9",
"volume": "21",
"year": "2023"
},
{
"article-title": "Exploring the Potential of Broadly Neutralizing Antibodies for Treating SARS-CoV-2 Variants of Global Concern in 2023: A Comprehensive Clinical Review",
"author": "Gutlapalli",
"journal-title": "Cureus",
"key": "10.1016/j.isci.2025.112632_bib10",
"volume": "15",
"year": "2023"
},
{
"DOI": "10.3390/vaccines11030711",
"article-title": "Deep Mutational Scanning to Predict Escape from Bebtelovimab in SARS-CoV-2 Omicron Subvariants",
"author": "Alcantara",
"doi-asserted-by": "crossref",
"first-page": "711",
"journal-title": "Vaccines",
"key": "10.1016/j.isci.2025.112632_bib11",
"volume": "11",
"year": "2023"
},
{
"article-title": "SARS-CoV-2 3CLpro mutations selected in a VSV-based system confer resistance to nirmatrelvir, ensitrelvir, and GC376",
"author": "Heilmann",
"journal-title": "Sci. Transl. Med.",
"key": "10.1016/j.isci.2025.112632_bib12",
"volume": "15",
"year": "2022"
},
{
"DOI": "10.1038/s41467-022-29104-y",
"article-title": "De novo emergence of a remdesivir resistance mutation during treatment of persistent SARS-CoV-2 infection in an immunocompromised patient: a case report",
"author": "Gandhi",
"doi-asserted-by": "crossref",
"first-page": "1547",
"journal-title": "Nat. Commun.",
"key": "10.1016/j.isci.2025.112632_bib13",
"volume": "13",
"year": "2022"
},
{
"article-title": "Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir",
"author": "Hu",
"journal-title": "Preprint at bioRxiv",
"key": "10.1016/j.isci.2025.112632_bib14",
"year": "2022"
},
{
"DOI": "10.1016/S1473-3099(22)00529-1",
"article-title": "COVID-19 sequelae: can long-term effects be predicted?",
"author": "Gavriilaki",
"doi-asserted-by": "crossref",
"first-page": "1651",
"journal-title": "Lancet Infect. Dis.",
"key": "10.1016/j.isci.2025.112632_bib15",
"volume": "22",
"year": "2022"
},
{
"DOI": "10.1056/EVIDe2300165",
"article-title": "The Persistence of SARS-CoV-2 and Its Role in Long Covid",
"author": "Lineburg",
"doi-asserted-by": "crossref",
"journal-title": "NEJM Evid.",
"key": "10.1016/j.isci.2025.112632_bib16",
"volume": "2",
"year": "2023"
},
{
"DOI": "10.3389/fimmu.2020.00827",
"article-title": "Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19)",
"author": "Diao",
"doi-asserted-by": "crossref",
"first-page": "827",
"journal-title": "Front. Immunol.",
"key": "10.1016/j.isci.2025.112632_bib17",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1038/s41591-020-0901-9",
"article-title": "Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19",
"author": "Liao",
"doi-asserted-by": "crossref",
"first-page": "842",
"journal-title": "Nat. Med.",
"key": "10.1016/j.isci.2025.112632_bib18",
"volume": "26",
"year": "2020"
},
{
"DOI": "10.1016/j.medidd.2020.100057",
"article-title": "Viral sepsis is a complication in patients with Novel Corona Virus Disease (COVID-19)",
"author": "Liu",
"doi-asserted-by": "crossref",
"journal-title": "Med. Drug Discov.",
"key": "10.1016/j.isci.2025.112632_bib19",
"volume": "8",
"year": "2020"
},
{
"DOI": "10.1016/j.ccc.2021.05.003",
"article-title": "Pathophysiology of acute respiratory distress syndrome and COVID-19 lung injury",
"author": "Swenson",
"doi-asserted-by": "crossref",
"first-page": "749",
"journal-title": "Crit. Care Clin.",
"key": "10.1016/j.isci.2025.112632_bib20",
"volume": "37",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2020.10.028",
"article-title": "Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection",
"author": "Wei",
"doi-asserted-by": "crossref",
"first-page": "76",
"journal-title": "Cell",
"key": "10.1016/j.isci.2025.112632_bib21",
"volume": "184",
"year": "2020"
},
{
"DOI": "10.1038/s41591-020-1021-2",
"article-title": "Immune complement and coagulation dysfunction in adverse outcomes of SARS-CoV-2 infection",
"author": "Ramlall",
"doi-asserted-by": "crossref",
"first-page": "1609",
"journal-title": "Nat. Med.",
"key": "10.1016/j.isci.2025.112632_bib22",
"volume": "26",
"year": "2020"
},
{
"article-title": "Novel gene-specific translation mechanism of dysregulated, chronic inflammation reveals promising, multifaceted COVID-19 therapeutics",
"author": "Wang",
"journal-title": "bioRxiv",
"key": "10.1016/j.isci.2025.112632_bib23",
"year": "2020"
},
{
"author": "Muneer",
"key": "10.1016/j.isci.2025.112632_bib24",
"series-title": "Non-Canonical Translation Regulatory Function of G9a in Chronic Inflammation Associated Diseases",
"year": "2023"
},
{
"article-title": "Genome-scale CRISPR‒Cas9 screen identifies novel host factors as potential therapeutic targets for SARS-CoV-2 infection",
"author": "Sakai",
"journal-title": "bioRxiv",
"key": "10.1016/j.isci.2025.112632_bib25",
"year": "2023"
},
{
"DOI": "10.1016/j.cell.2011.05.006",
"article-title": "Analysis of the human endogenous coregulator complexome",
"author": "Malovannaya",
"doi-asserted-by": "crossref",
"first-page": "787",
"journal-title": "Cell",
"key": "10.1016/j.isci.2025.112632_bib26",
"volume": "145",
"year": "2011"
},
{
"DOI": "10.1016/j.cell.2015.06.043",
"article-title": "The BioPlex Network: A Systematic Exploration of the Human Interactome",
"author": "Huttlin",
"doi-asserted-by": "crossref",
"first-page": "425",
"journal-title": "Cell",
"key": "10.1016/j.isci.2025.112632_bib27",
"volume": "162",
"year": "2015"
},
{
"DOI": "10.1038/s41577-020-0331-4",
"article-title": "Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages",
"author": "Merad",
"doi-asserted-by": "crossref",
"first-page": "355",
"journal-title": "Nat. Rev. Immunol.",
"key": "10.1016/j.isci.2025.112632_bib28",
"volume": "20",
"year": "2020"
},
{
"DOI": "10.1016/j.isci.2019.07.001",
"article-title": "Multi-omic Dissection of Oncogenically Active Epiproteomes Identifies Drivers of Proliferative and Invasive Breast Tumors",
"author": "Wrobel",
"doi-asserted-by": "crossref",
"first-page": "359",
"journal-title": "iScience",
"key": "10.1016/j.isci.2025.112632_bib29",
"volume": "17",
"year": "2019"
},
{
"DOI": "10.1016/j.chembiol.2023.09.012",
"article-title": "Non-canonical function of histone methyltransferase G9a in the translational regulation of chronic inflammation",
"author": "Muneer",
"doi-asserted-by": "crossref",
"first-page": "1525",
"journal-title": "Cell Chem. Biol.",
"key": "10.1016/j.isci.2025.112632_bib30",
"volume": "30",
"year": "2023"
},
{
"DOI": "10.1016/j.cell.2021.01.004",
"article-title": "Multi-organ proteomic landscape of COVID-19 autopsies",
"author": "Nie",
"doi-asserted-by": "crossref",
"first-page": "775",
"journal-title": "Cell",
"key": "10.1016/j.isci.2025.112632_bib31",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1038/s41587-022-01474-0",
"article-title": "A comprehensive SARS-CoV-2-human protein-protein interactome reveals COVID-19 pathobiology and potential host therapeutic targets",
"author": "Zhou",
"doi-asserted-by": "crossref",
"first-page": "128",
"journal-title": "Nat. Biotechnol.",
"key": "10.1016/j.isci.2025.112632_bib32",
"volume": "41",
"year": "2023"
},
{
"DOI": "10.1038/nrm.2016.132",
"article-title": "Post-transcriptional gene regulation by mRNA modifications",
"author": "Zhao",
"doi-asserted-by": "crossref",
"first-page": "31",
"journal-title": "Nat. Rev. Mol. Cell Biol.",
"key": "10.1016/j.isci.2025.112632_bib33",
"volume": "18",
"year": "2017"
},
{
"DOI": "10.1016/j.cell.2017.05.045",
"article-title": "Dynamic RNA Modifications in Gene Expression Regulation",
"author": "Roundtree",
"doi-asserted-by": "crossref",
"first-page": "1187",
"journal-title": "Cell",
"key": "10.1016/j.isci.2025.112632_bib34",
"volume": "169",
"year": "2017"
},
{
"DOI": "10.1038/s41419-021-04012-z",
"article-title": "RBM15-mediated N6-methyladenosine modification affects COVID-19 severity by regulating the expression of multitarget genes",
"author": "Meng",
"doi-asserted-by": "crossref",
"first-page": "732",
"journal-title": "Cell Death Dis.",
"key": "10.1016/j.isci.2025.112632_bib35",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2021.02.008",
"article-title": "In vivo structural characterization of the SARS-CoV-2 RNA genome identifies host proteins vulnerable to repurposed drugs",
"author": "Sun",
"doi-asserted-by": "crossref",
"first-page": "1865",
"journal-title": "Cell",
"key": "10.1016/j.isci.2025.112632_bib36",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1128/mBio.01067-21",
"article-title": "Methyltransferase-like 3 modulates severe acute respiratory syndrome coronavirus-2 RNA N6-methyladenosine modification and replication",
"author": "Zhang",
"doi-asserted-by": "crossref",
"journal-title": "mBio",
"key": "10.1016/j.isci.2025.112632_bib37",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1016/j.antiviral.2021.105232",
"article-title": "S-adenosylmethionine-dependent methyltransferase inhibitor DZNep blocks transcription and translation of SARS-CoV-2 genome with a low tendency to select for drug-resistant viral variants",
"author": "Kumar",
"doi-asserted-by": "crossref",
"journal-title": "Antiviral Res.",
"key": "10.1016/j.isci.2025.112632_bib38",
"volume": "197",
"year": "2022"
},
{
"DOI": "10.3389/fimmu.2021.774776",
"article-title": "m6A Regulator-Mediated Methylation Modification Patterns and Characteristics of Immunity in Blood Leukocytes of COVID-19 Patients",
"author": "Qiu",
"doi-asserted-by": "crossref",
"journal-title": "Front. Immunol.",
"key": "10.1016/j.isci.2025.112632_bib39",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.3389/fpubh.2022.914193",
"article-title": "m6A regulator-mediated methylation modification patterns and characteristics in COVID-19 patients",
"author": "Qing",
"doi-asserted-by": "crossref",
"journal-title": "Front. Public Health",
"key": "10.1016/j.isci.2025.112632_bib40",
"volume": "10",
"year": "2022"
},
{
"DOI": "10.1016/j.meegid.2022.105389",
"article-title": "The risk of COVID-19 can be predicted by a nomogram based on m6A-related genes",
"author": "Lu",
"doi-asserted-by": "crossref",
"journal-title": "Infect. Genet. Evol.",
"key": "10.1016/j.isci.2025.112632_bib41",
"volume": "106",
"year": "2022"
},
{
"DOI": "10.1016/j.gendis.2021.12.005",
"article-title": "Systematic analysis of clinical relevance and molecular characterization of m6A in COVID-19 patients",
"author": "An",
"doi-asserted-by": "crossref",
"first-page": "1170",
"journal-title": "Genes Dis.",
"key": "10.1016/j.isci.2025.112632_bib42",
"volume": "9",
"year": "2022"
},
{
"DOI": "10.1038/s41556-021-00653-6",
"article-title": "A PRC2-independent function for EZH2 in regulating rRNA 2'-O methylation and IRES-dependent translation",
"author": "Yi",
"doi-asserted-by": "crossref",
"first-page": "341",
"journal-title": "Nat. Cell Biol.",
"key": "10.1016/j.isci.2025.112632_bib43",
"volume": "23",
"year": "2021"
},
{
"DOI": "10.1038/s41586-020-2332-7",
"article-title": "Proteomics of SARS-CoV-2-infected host cells reveals therapy targets",
"author": "Bojkova",
"doi-asserted-by": "crossref",
"first-page": "469",
"journal-title": "Nature",
"key": "10.1016/j.isci.2025.112632_bib44",
"volume": "583",
"year": "2020"
},
{
"DOI": "10.1038/s41422-020-00465-7",
"article-title": "The m(6)A methylome of SARS-CoV-2 in host cells",
"author": "Liu",
"doi-asserted-by": "crossref",
"first-page": "404",
"journal-title": "Cell Res.",
"key": "10.1016/j.isci.2025.112632_bib45",
"volume": "31",
"year": "2021"
},
{
"DOI": "10.1101/gad.348320.121",
"article-title": "Targeting the m(6)A RNA modification pathway blocks SARS-CoV-2 and HCoV-OC43 replication",
"author": "Burgess",
"doi-asserted-by": "crossref",
"first-page": "1005",
"journal-title": "Genes Dev.",
"key": "10.1016/j.isci.2025.112632_bib46",
"volume": "35",
"year": "2021"
},
{
"article-title": "Novel brain-penetrant inhibitor of G9a methylase blocks Alzheimer disease proteopathology for precision medication",
"author": "Xie",
"journal-title": "Preprint at medRxiv.",
"key": "10.1016/j.isci.2025.112632_bib47",
"volume": "23297491",
"year": "2023"
},
{
"DOI": "10.1146/annurev-anchem-061516-045357",
"article-title": "Relative and absolute quantitation in mass spectrometry–based proteomics",
"author": "Ankney",
"doi-asserted-by": "crossref",
"first-page": "49",
"journal-title": "Annu. Rev. Anal. Chem.",
"key": "10.1016/j.isci.2025.112632_bib48",
"volume": "11",
"year": "2018"
},
{
"DOI": "10.1021/cb400133j",
"article-title": "An orally bioavailable chemical probe of the Lysine Methyltransferases EZH2 and EZH1",
"author": "Konze",
"doi-asserted-by": "crossref",
"first-page": "1324",
"journal-title": "ACS Chem. Biol.",
"key": "10.1016/j.isci.2025.112632_bib49",
"volume": "8",
"year": "2013"
},
{
"DOI": "10.1038/s41586-020-2286-9",
"article-title": "A SARS-CoV-2 protein interaction map reveals targets for drug repurposing",
"author": "Gordon",
"doi-asserted-by": "crossref",
"first-page": "459",
"journal-title": "Nature",
"key": "10.1016/j.isci.2025.112632_bib50",
"volume": "583",
"year": "2020"
},
{
"article-title": "A SARS-CoV-2-host proximity interactome",
"author": "Samavarchi-Tehrani",
"journal-title": "Preprint at bioRxiv",
"key": "10.1016/j.isci.2025.112632_bib51",
"year": "2020"
},
{
"article-title": "Global BioID-based SARS-CoV-2 proteins proximal interactome unveils novel ties between viral polypeptides and host factors involved in multiple COVID19-associated mechanisms",
"author": "Laurent",
"journal-title": "Preprint at bioRxiv",
"key": "10.1016/j.isci.2025.112632_bib52",
"year": "2020"
},
{
"DOI": "10.1126/science.abc6261",
"article-title": "Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans",
"author": "Arunachalam",
"doi-asserted-by": "crossref",
"first-page": "1210",
"journal-title": "Science",
"key": "10.1016/j.isci.2025.112632_bib53",
"volume": "369",
"year": "2020"
},
{
"article-title": "Association of COVID-19 inflammation with activation of the C5a–C5aR1 axis",
"author": "Carvelli",
"first-page": "1",
"journal-title": "Nature",
"key": "10.1016/j.isci.2025.112632_bib54",
"year": "2020"
},
{
"article-title": "The trinity of COVID-19: immunity, inflammation and intervention",
"author": "Tay",
"first-page": "1",
"journal-title": "Nat. Rev. Immunol.",
"key": "10.1016/j.isci.2025.112632_bib55",
"year": "2020"
},
{
"DOI": "10.1038/s41586-022-05282-z",
"article-title": "SARS-CoV-2 disrupts host epigenetic regulation via histone mimicry",
"author": "Kee",
"doi-asserted-by": "crossref",
"first-page": "381",
"journal-title": "Nature",
"key": "10.1016/j.isci.2025.112632_bib56",
"volume": "610",
"year": "2022"
},
{
"DOI": "10.1038/s41564-023-01344-8",
"article-title": "SARS-CoV-2 restructures host chromatin architecture",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "679",
"journal-title": "Nat. Microbiol.",
"key": "10.1016/j.isci.2025.112632_bib57",
"volume": "8",
"year": "2023"
},
{
"DOI": "10.1155/2022/5794422",
"article-title": "METTL3 Accelerates Breast Cancer Progression via Regulating EZH2 m6A Modification",
"author": "Hu",
"doi-asserted-by": "crossref",
"journal-title": "J. Healthc. Eng.",
"key": "10.1016/j.isci.2025.112632_bib58",
"volume": "2022",
"year": "2022"
},
{
"DOI": "10.1016/j.medj.2020.07.002",
"article-title": "Virus-Host Interactome and Proteomic Survey Reveal Potential Virulence Factors Influencing SARS-CoV-2 Pathogenesis",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "99",
"journal-title": "Med",
"key": "10.1016/j.isci.2025.112632_bib59",
"volume": "2",
"year": "2020"
},
{
"DOI": "10.1126/science.abe9403",
"article-title": "Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms",
"author": "Gordon",
"doi-asserted-by": "crossref",
"journal-title": "Science",
"key": "10.1016/j.isci.2025.112632_bib60",
"volume": "370",
"year": "2020"
},
{
"DOI": "10.6004/jadpro.2022.13.2.7",
"article-title": "Tazemetostat: EZH2 Inhibitor",
"author": "Straining",
"doi-asserted-by": "crossref",
"first-page": "158",
"journal-title": "J. Adv. Pract. Oncol.",
"key": "10.1016/j.isci.2025.112632_bib61",
"volume": "13",
"year": "2022"
},
{
"DOI": "10.1515/cclm-2020-0188",
"article-title": "Prominent changes in blood coagulation of patients with SARS-CoV-2 infection",
"author": "Han",
"doi-asserted-by": "crossref",
"first-page": "1116",
"journal-title": "Clin. Chem. Lab. Med.",
"key": "10.1016/j.isci.2025.112632_bib62",
"volume": "58",
"year": "2020"
},
{
"DOI": "10.1038/s41421-021-00249-2",
"article-title": "SARS-CoV-2 cell tropism and multiorgan infection",
"author": "Liu",
"doi-asserted-by": "crossref",
"first-page": "17",
"journal-title": "Cell Discov.",
"key": "10.1016/j.isci.2025.112632_bib63",
"volume": "7",
"year": "2021"
},
{
"DOI": "10.1016/j.molcel.2020.08.006",
"article-title": "Growth factor receptor signaling inhibition prevents SARS-CoV-2 replication",
"author": "Klann",
"doi-asserted-by": "crossref",
"first-page": "164",
"journal-title": "Mol. Cell",
"key": "10.1016/j.isci.2025.112632_bib64",
"volume": "80",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.06.034",
"article-title": "The Global Phosphorylation Landscape of SARS-CoV-2 Infection",
"author": "Bouhaddou",
"doi-asserted-by": "crossref",
"first-page": "685",
"journal-title": "Cell",
"key": "10.1016/j.isci.2025.112632_bib65",
"volume": "182",
"year": "2020"
},
{
"article-title": "Multi-level proteomics reveals host-perturbation strategies of SARS-CoV-2 and SARS-CoV",
"author": "Stukalov",
"journal-title": "Preprint at bioRxiv",
"key": "10.1016/j.isci.2025.112632_bib66",
"year": "2020"
},
{
"article-title": "System-wide hematopoietic and immune signaling aberrations in COVID-19 revealed by deep proteome and phosphoproteome analysis",
"author": "Kaneko",
"journal-title": "Preprint at medRxiv",
"key": "10.1016/j.isci.2025.112632_bib67",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2020.10.037",
"article-title": "Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19",
"author": "Su",
"doi-asserted-by": "crossref",
"first-page": "1479",
"journal-title": "Cell",
"key": "10.1016/j.isci.2025.112632_bib68",
"volume": "183",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2022.01.014",
"article-title": "Multiple early factors anticipate post-acute COVID-19 sequelae",
"author": "Su",
"doi-asserted-by": "crossref",
"first-page": "881",
"journal-title": "Cell",
"key": "10.1016/j.isci.2025.112632_bib69",
"volume": "185",
"year": "2022"
},
{
"article-title": "The FDA-approved drug Alectinib compromises SARS-CoV-2 nucleocapsid phosphorylation and inhibits viral infection in vitro",
"author": "Yaron",
"journal-title": "Preprint at bioRxiv",
"key": "10.1016/j.isci.2025.112632_bib70",
"year": "2020"
},
{
"DOI": "10.1186/s13073-020-00763-0",
"article-title": "Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein",
"author": "Davidson",
"doi-asserted-by": "crossref",
"first-page": "15",
"journal-title": "Genome Med.",
"key": "10.1016/j.isci.2025.112632_bib71",
"volume": "12",
"year": "2020"
},
{
"article-title": "METTL3 plays multiple functions in biological processes",
"author": "Liu",
"first-page": "1631",
"journal-title": "Am. J. Cancer Res.",
"key": "10.1016/j.isci.2025.112632_bib72",
"volume": "10",
"year": "2020"
},
{
"DOI": "10.1016/j.molcel.2016.03.021",
"article-title": "The m6A methyltransferase METTL3 promotes translation in human cancer cells",
"author": "Lin",
"doi-asserted-by": "crossref",
"first-page": "335",
"journal-title": "Mol. Cell",
"key": "10.1016/j.isci.2025.112632_bib73",
"volume": "62",
"year": "2016"
},
{
"DOI": "10.1016/j.celrep.2021.109091",
"article-title": "METTL3 regulates viral m6A RNA modification and host cell innate immune responses during SARS-CoV-2 infection",
"author": "Li",
"doi-asserted-by": "crossref",
"journal-title": "Cell Rep.",
"key": "10.1016/j.isci.2025.112632_bib74",
"volume": "35",
"year": "2021"
},
{
"article-title": "The RNA demethylase FTO controls m6A marking on SARS-CoV-2 and classifies COVID-19 severity in patients",
"author": "Malbec",
"first-page": "497749",
"journal-title": "Preprint at bioRxiv",
"key": "10.1016/j.isci.2025.112632_bib75",
"year": "2022"
},
{
"DOI": "10.1016/j.ebiom.2023.104851",
"article-title": "Probing long COVID through a proteomic lens: a comprehensive two-year longitudinal cohort study of hospitalised survivors",
"author": "Gu",
"doi-asserted-by": "crossref",
"journal-title": "EBioMedicine",
"key": "10.1016/j.isci.2025.112632_bib76",
"volume": "98",
"year": "2023"
},
{
"DOI": "10.1038/s41598-020-80120-8",
"article-title": "In-depth blood proteome profiling analysis revealed distinct functional characteristics of plasma proteins between severe and non-severe COVID-19 patients",
"author": "Park",
"doi-asserted-by": "crossref",
"journal-title": "Sci. Rep.",
"key": "10.1016/j.isci.2025.112632_bib77",
"volume": "10",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.05.032",
"article-title": "Proteomic and Metabolomic Characterization of COVID-19 Patient Sera",
"author": "Shen",
"doi-asserted-by": "crossref",
"first-page": "59",
"journal-title": "Cell",
"key": "10.1016/j.isci.2025.112632_bib78",
"volume": "182",
"year": "2020"
},
{
"DOI": "10.1016/j.cell.2020.12.004",
"article-title": "Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "106",
"journal-title": "Cell",
"key": "10.1016/j.isci.2025.112632_bib79",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1038/s41467-021-21213-4",
"article-title": "A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry",
"author": "Zhu",
"doi-asserted-by": "crossref",
"first-page": "961",
"journal-title": "Nat. Commun.",
"key": "10.1016/j.isci.2025.112632_bib80",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2020.10.030",
"article-title": "Identification of required host factors for SARS-CoV-2 infection in human cells",
"author": "Daniloski",
"doi-asserted-by": "crossref",
"first-page": "92",
"journal-title": "Cell",
"key": "10.1016/j.isci.2025.112632_bib81",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1016/j.cell.2020.12.006",
"article-title": "Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks",
"author": "Schneider",
"doi-asserted-by": "crossref",
"first-page": "120",
"journal-title": "Cell",
"key": "10.1016/j.isci.2025.112632_bib82",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1038/s41591-021-01282-0",
"article-title": "Developing therapeutic approaches for twenty-first-century emerging infectious viral diseases",
"author": "Meganck",
"doi-asserted-by": "crossref",
"first-page": "401",
"journal-title": "Nat. Med.",
"key": "10.1016/j.isci.2025.112632_bib83",
"volume": "27",
"year": "2021"
},
{
"DOI": "10.1056/NEJMsr2105280",
"article-title": "SARS-CoV-2 variants and vaccines",
"author": "Krause",
"doi-asserted-by": "crossref",
"first-page": "179",
"journal-title": "N. Engl. J. Med.",
"key": "10.1016/j.isci.2025.112632_bib84",
"volume": "385",
"year": "2021"
},
{
"DOI": "10.1038/s44161-022-00096-8",
"article-title": "Hyperactivity of the CD155 immune checkpoint suppresses anti-viral immunity in patients with coronary artery disease",
"author": "Zhao",
"doi-asserted-by": "crossref",
"first-page": "634",
"journal-title": "Nat. Cardiovasc. Res.",
"key": "10.1016/j.isci.2025.112632_bib85",
"volume": "1",
"year": "2022"
},
{
"DOI": "10.1128/mBio.01174-14",
"article-title": "Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses",
"author": "Menachery",
"doi-asserted-by": "crossref",
"journal-title": "mBio",
"key": "10.1016/j.isci.2025.112632_bib86",
"volume": "5",
"year": "2014"
},
{
"DOI": "10.3389/fcell.2022.849298",
"article-title": "Epitranscriptomics of SARS-CoV-2 infection",
"author": "Izadpanah",
"doi-asserted-by": "crossref",
"journal-title": "Front. Cell Dev. Biol.",
"key": "10.1016/j.isci.2025.112632_bib87",
"volume": "10",
"year": "2022"
},
{
"article-title": "Targeting SARS-CoV-2 using polycomb inhibitors as antiviral agents",
"author": "Ayaz",
"journal-title": "Future Medicine",
"key": "10.1016/j.isci.2025.112632_bib88",
"year": "2020"
},
{
"DOI": "10.15252/embj.2022111608",
"article-title": "Attenuation of SARS-CoV-2 replication and associated inflammation by concomitant targeting of viral and host cap 2'-O-ribose methyltransferases",
"author": "Bergant",
"doi-asserted-by": "crossref",
"journal-title": "EMBO J.",
"key": "10.1016/j.isci.2025.112632_bib89",
"volume": "41",
"year": "2022"
},
{
"DOI": "10.1016/j.molcel.2017.12.024",
"article-title": "Phosphorylation of EZH2 by AMPK suppresses PRC2 methyltransferase activity and oncogenic function",
"author": "Wan",
"doi-asserted-by": "crossref",
"first-page": "279",
"journal-title": "Mol. Cell",
"key": "10.1016/j.isci.2025.112632_bib90",
"volume": "69",
"year": "2018"
},
{
"article-title": "USP7-Based Deubiquitinase-Targeting Chimeras Stabilize AMPK",
"author": "Liu",
"first-page": "11507",
"journal-title": "J. Am. Chem. Soc.",
"key": "10.1016/j.isci.2025.112632_bib91",
"volume": "146",
"year": "2024"
},
{
"DOI": "10.1038/nature12711",
"article-title": "Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor",
"author": "Ge",
"doi-asserted-by": "crossref",
"first-page": "535",
"journal-title": "Nature",
"key": "10.1016/j.isci.2025.112632_bib92",
"volume": "503",
"year": "2013"
},
{
"DOI": "10.1016/j.cell.2021.06.008",
"article-title": "Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses",
"author": "Zhou",
"doi-asserted-by": "crossref",
"first-page": "4380",
"journal-title": "Cell",
"key": "10.1016/j.isci.2025.112632_bib93",
"volume": "184",
"year": "2021"
},
{
"DOI": "10.1007/s12250-018-0012-7",
"article-title": "Serological evidence of bat SARS-related coronavirus infection in humans, China",
"author": "Wang",
"doi-asserted-by": "crossref",
"first-page": "104",
"journal-title": "Virol. Sin.",
"key": "10.1016/j.isci.2025.112632_bib94",
"volume": "33",
"year": "2018"
},
{
"DOI": "10.1128/jvi.00997-22",
"article-title": "WTAP targets the METTL3 m6A-methyltransferase complex to cytoplasmic hepatitis C virus RNA to regulate infection",
"author": "Sacco",
"doi-asserted-by": "crossref",
"journal-title": "J. Virol.",
"key": "10.1016/j.isci.2025.112632_bib95",
"volume": "96",
"year": "2022"
},
{
"article-title": "Hepatitis B virus X protein recruits methyltransferases to affect cotranscriptional N6-methyladenosine modification of viral/host RNAs",
"author": "Kim",
"journal-title": "Proc. Natl. Acad. Sci. USA",
"key": "10.1016/j.isci.2025.112632_bib96",
"volume": "118",
"year": "2021"
},
{
"DOI": "10.3390/ph14111135",
"article-title": "Regulation of m6A methylation as a new therapeutic option against COVID-19",
"author": "Zannella",
"doi-asserted-by": "crossref",
"first-page": "1135",
"journal-title": "Pharmaceuticals",
"key": "10.1016/j.isci.2025.112632_bib97",
"volume": "14",
"year": "2021"
},
{
"DOI": "10.1093/nar/gky1007",
"article-title": "N 6-methyladenosine modification and METTL3 modulate enterovirus 71 replication",
"author": "Hao",
"doi-asserted-by": "crossref",
"first-page": "362",
"journal-title": "Nucleic Acids Res.",
"key": "10.1016/j.isci.2025.112632_bib98",
"volume": "47",
"year": "2019"
},
{
"DOI": "10.3390/epigenomes6030016",
"article-title": "Global m6A RNA Methylation in SARS-CoV-2 Positive Nasopharyngeal Samples in a Mexican Population: A First Approximation Study",
"author": "Batista-Roche",
"doi-asserted-by": "crossref",
"first-page": "16",
"journal-title": "Epigenomes",
"key": "10.1016/j.isci.2025.112632_bib99",
"volume": "6",
"year": "2022"
},
{
"DOI": "10.3233/ADR-220090",
"article-title": "The Effects of SARS-CoV-2 Infection on the Cognitive Functioning of Patients with Pre-Existing Dementia",
"author": "Dubey",
"doi-asserted-by": "crossref",
"first-page": "119",
"journal-title": "J. Alzheimers Dis. Rep.",
"key": "10.1016/j.isci.2025.112632_bib100",
"volume": "7",
"year": "2023"
},
{
"DOI": "10.1002/alz.057892",
"article-title": "Plasma biomarkers of neurodegeneration and neuroinflammation in hospitalized COVID-19 patients with and without new neurological symptoms",
"author": "Boutajangout",
"doi-asserted-by": "crossref",
"journal-title": "Alzheimer's Dement.",
"key": "10.1016/j.isci.2025.112632_bib101",
"volume": "17",
"year": "2021"
},
{
"DOI": "10.1038/s41467-020-19055-7",
"article-title": "A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19",
"author": "Xie",
"doi-asserted-by": "crossref",
"first-page": "5214",
"journal-title": "Nat. Commun.",
"key": "10.1016/j.isci.2025.112632_bib102",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1002/cmdc.201300450",
"article-title": "A chemical tool for in vitro and in vivo precipitation of lysine methyltransferase G9a",
"author": "Konze",
"doi-asserted-by": "crossref",
"first-page": "549",
"journal-title": "ChemMedChem",
"key": "10.1016/j.isci.2025.112632_bib103",
"volume": "9",
"year": "2014"
},
{
"DOI": "10.1021/jm401480r",
"article-title": "Discovery of an in vivo chemical probe of the lysine methyltransferases G9a and GLP",
"author": "Liu",
"doi-asserted-by": "crossref",
"first-page": "8931",
"journal-title": "J. Med. Chem.",
"key": "10.1016/j.isci.2025.112632_bib104",
"volume": "56",
"year": "2013"
},
{
"DOI": "10.1093/bioinformatics/btu170",
"article-title": "Trimmomatic: a flexible trimmer for Illumina sequence data",
"author": "Bolger",
"doi-asserted-by": "crossref",
"first-page": "2114",
"journal-title": "Bioinformatics",
"key": "10.1016/j.isci.2025.112632_bib105",
"volume": "30",
"year": "2014"
},
{
"DOI": "10.1093/bioinformatics/bts635",
"article-title": "STAR: ultrafast universal RNA-seq aligner",
"author": "Dobin",
"doi-asserted-by": "crossref",
"first-page": "15",
"journal-title": "Bioinformatics",
"key": "10.1016/j.isci.2025.112632_bib106",
"volume": "29",
"year": "2013"
},
{
"DOI": "10.1093/bioinformatics/btp352",
"article-title": "The sequence alignment/map format and SAMtools",
"author": "Li",
"doi-asserted-by": "crossref",
"first-page": "2078",
"journal-title": "Bioinformatics",
"key": "10.1016/j.isci.2025.112632_bib107",
"volume": "25",
"year": "2009"
},
{
"DOI": "10.1038/s41467-019-09234-6",
"article-title": "Metascape provides a biologist-oriented resource for the analysis of systems-level datasets",
"author": "Zhou",
"doi-asserted-by": "crossref",
"first-page": "1523",
"journal-title": "Nat. Commun.",
"key": "10.1016/j.isci.2025.112632_bib108",
"volume": "10",
"year": "2019"
},
{
"DOI": "10.1038/s41467-021-21361-7",
"article-title": "Shotgun transcriptome, spatial omics, and isothermal profiling of SARS-CoV-2 infection reveals unique host responses, viral diversification, and drug interactions",
"author": "Butler",
"doi-asserted-by": "crossref",
"first-page": "1660",
"journal-title": "Nat. Commun.",
"key": "10.1016/j.isci.2025.112632_bib109",
"volume": "12",
"year": "2021"
},
{
"article-title": "Systemic tissue and cellular disruption from SARS-CoV-2 infection revealed in COVID-19 autopsies and spatial omics tissue maps",
"author": "Park",
"journal-title": "Preprint at bioRxiv",
"key": "10.1016/j.isci.2025.112632_bib110",
"year": "2021"
},
{
"DOI": "10.1016/j.neuron.2017.12.036",
"article-title": "Epitranscriptomic m6A regulation of axon regeneration in the adult mammalian nervous system",
"author": "Weng",
"doi-asserted-by": "crossref",
"first-page": "313",
"journal-title": "Neuron",
"key": "10.1016/j.isci.2025.112632_bib111",
"volume": "97",
"year": "2018"
},
{
"DOI": "10.1038/nprot.2014.006",
"article-title": "Full-length RNA-seq from single cells using Smart-seq2",
"author": "Picelli",
"doi-asserted-by": "crossref",
"first-page": "171",
"journal-title": "Nat. Protoc.",
"key": "10.1016/j.isci.2025.112632_bib112",
"volume": "9",
"year": "2014"
},
{
"DOI": "10.1016/0003-2697(84)90782-6",
"article-title": "A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids",
"author": "Wessel",
"doi-asserted-by": "crossref",
"first-page": "141",
"journal-title": "Anal. Biochem.",
"key": "10.1016/j.isci.2025.112632_bib113",
"volume": "138",
"year": "1984"
}
],
"reference-count": 113,
"references-count": 113,
"relation": {},
"resource": {
"primary": {
"URL": "https://linkinghub.elsevier.com/retrieve/pii/S2589004225008934"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Targeting G9a-m6A translational mechanism of SARS-CoV-2 pathogenesis for multifaceted therapeutics of COVID-19 and its sequalae",
"type": "journal-article",
"update-policy": "https://doi.org/10.1016/elsevier_cm_policy",
"volume": "28"
}