SARS-CoV-2 RNA-binding protein suppresses extracellular miRNA release
et al., RNA Biology, doi:10.1080/15476286.2025.2527494, Jul 2025
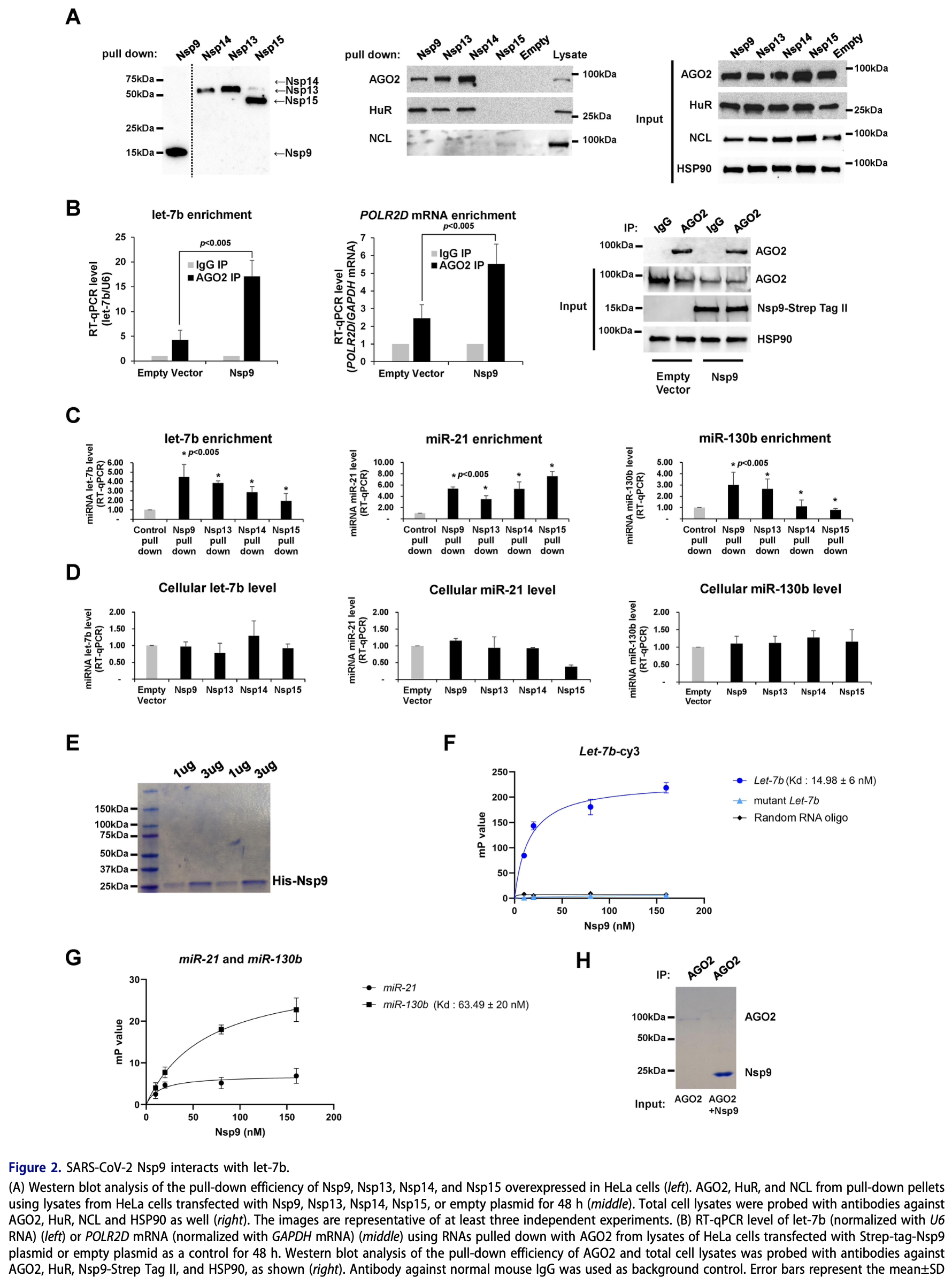
In vitro and animal study showing that SARS-CoV-2 RNA-binding protein Nsp9 suppresses extracellular release of microRNA let-7b, potentially inhibiting antiviral responses mediated by Toll-like Receptor 7 (TLR7). Authors identify dual mechanism inhibition of let-7b-mediated TLR7 activation, which would normally trigger antiviral inflammatory responses. Authors validated these findings in a Drosophila model, where Nsp9 expression induced an innate immune response that was suppressed by human let-7b co-expression. The study reveals a novel mechanism by which SARS-CoV-2 evades host immune responses and suggests that targeting the Nsp9-let-7b interaction could be a potential therapeutic strategy for COVID-19.
Mun et al., 7 Jul 2025, USA, peer-reviewed, 24 authors.
Contact: skang34@jhmi.edu, jehyun-yoon@ouhsc.edu, woonghee.lee@ucdenver.edu.
In vitro studies are an important part of preclinical research, however results may be very different in vivo.
SARS-CoV-2 RNA-binding protein suppresses extracellular miRNA release
RNA Biology, doi:10.1080/15476286.2025.2527494
SARS-CoV-2 is the betacoronavirus causing the COVID-19 pandemic. Although the SARS-CoV-2 genome and transcriptome were reported previously, the function of individual viral proteins is largely unknown. Utilizing biochemical and molecular biology methods, we identified that four SARS-CoV-2 RNA-binding proteins (RBPs) regulate the host RNA metabolism by direct interaction with mature miRNA let-7b revealed by Nuclear Magnetic Resonance spectroscopy (NMR). SARS-CoV-2 RBP Nsp9 primarily binds mature miRNA let-7b, a direct ligand of the Toll-like Receptor 7 (TLR7), one of the potential SARS-CoV-2 therapeutics. Nsp9 suppresses host gene expression possibly by promoting let-7b-mediated silencing of a cellular RNA polymerase, POLR2D. In addition, Nsp9 inhibits extracellular release of let-7b and subsequent antiviral activity via TLR7. These results demonstrate that SARS-CoV-2 hijacks the host RNA metabolism to suppress antiviral responses and to shut down cellular transcription. Our findings of how a natural ligand of TLR7, miRNA let-7b, is suppressed by SARS-CoV-2 RBPs will advance our understanding of COVID-19 and SARS-CoV-2 therapeutics.
Disclosure statement No potential conflict of interest was reported by the author(s).
Author contributions
References
Arunachalam, Wimmers, Mok, Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans, Science, doi:10.1126/science.abc6261
Biegel, Henderson, Cox, Cellular DEAD-box RNA helicase DDX6 modulates interaction of miR-122 with the 5′ untranslated region of hepatitis C virus RNA, Virology, doi:10.1016/j.virol.2017.04.014
Chatterjee, Ade, Nurik, Phosphorylation induces conformational rigidity at the C-terminal domain of AMPA receptors, J Phys Chem B, doi:10.1021/acs.jpcb.8b10749
Chen, Xiong, Bao, Convalescent plasma as a potential therapy for COVID-19, Lancet Infect Dis, doi:10.1016/S1473-3099(20)30141-9
Croce, Causes and consequences of microRNA dysregulation in cancer, Nat Rev Genet, doi:10.1038/nrg2634
Cullen, Viral and cellular messenger RNA targets of viral microRNAs, Nature, doi:10.1038/nature07757
Delaglio, Grzesiek, Vuister, Nmrpipe: a multidimensional spectral processing system based on UNIX pipes, J Biomol NMR, doi:10.1007/bf00197809
Diebold, Kaisho, Hemmi, Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA, Science, doi:10.1126/science.1093616
Dudás, Puglisi, Korn, Backbone chemical shift spectral assignments of SARS coronavirus-2 non-structural protein nsp9, Biomol NMR Assign, doi:10.1007/s12104-021-10011-0
Dunckley, Parker, The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif, Embo J, doi:10.1093/emboj/18.19.5411
Eastman, Swails, Chodera, OpenMM 7: rapid development of high performance algorithms for molecular dynamics, PLOS Comput Biol, doi:10.1371/journal.pcbi.1005659
Egloff, Ferron, Campanacci, The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world, Proc Natl Acad Sci, doi:10.1073/pnas.0307877101
Furuichi, Lafiandra, Shatkin, 5′-terminal structure and mRNA stability, Nature, doi:10.1038/266235a0
Gao, Tian, Yang, Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies, Biosci Trends, doi:10.5582/bst.2020.01047
Gautret, Lagier, Parola, Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial, Int J Antimicrob Agents, doi:10.1016/j.ijantimicag.2020.105949
Gomez-Carballa, Pardo-Seco, Pischedda, Sex-biased expression of the TLR7 gene in severe COVID-19 patients: insights from transcriptomics and epigenomics, Environ Res, doi:10.1016/j.envres.2022.114288
Gordon, Jang, Bouhaddou, A SARS-CoV-2 protein interaction map reveals targets for drug repurposing, Nature, doi:10.1038/s41586-020-2286-9
Groban, Narayanan, Jacobson, Conformational changes in protein loops and helices induced by post-translational phosphorylation, PLOS Comput Biol, doi:10.1371/journal.pcbi.0020032
Guichard, Lu, Kanca, A comprehensive Drosophila resource to identify key functional interactions between SARS-CoV-2 factors and host proteins, Cell Rep, doi:10.1016/j.celrep.2023.112842
Guo, Price, RNA polymerase II transcription elongation control, Chem Rev, doi:10.1021/cr400105n
Gómez-Carballa, Pardo-Seco, Pischedda, Sex-biased expression of the TLR7 gene in severe COVID-19 patients: insights from transcriptomics and epigenomics, Environ Res, doi:10.1016/j.envres.2022.114288
Hossein-Khannazer, Shokoohian, Shpichka, Novel therapeutic approaches for treatment of COVID-19, J Mol Med, doi:10.1007/s00109-020-01927-6
Hu, Xie, Onishi, Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling, Cell, doi:10.1016/j.cell.2009.08.037
Huang Da, Sherman, Lempicki, Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists, Nucleic Acids Res, doi:10.1093/nar/gkn923
Huang Da, Sherman, Lempicki, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources, Nat Protoc, doi:10.1038/nprot.2008.211
Jeong, Jiang, Albino, Rapid identification of monospecific monoclonal antibodies using a human proteome microarray, Mol Cell Proteomics, doi:10.1074/mcp.O111.016253
Khushman, Bhardwaj, Patel, Exosomal markers (CD63 and CD9) expression pattern using immunohistochemistry in resected malignant and nonmalignant pancreatic specimens, Pancrease, doi:10.1097/MPA.0000000000000847
Kim, Chung, Jo, Identification of coronavirus isolated from a patient in Korea with COVID-19, Osong Public Health Res Perspect, doi:10.24171/j.phrp.2020.11.1.02
Kim, Kuwano, Srikantan, HuR recruits let-7/RISC to repress c-Myc expression, Genes Dev, doi:10.1101/gad.1812509
Kim, Lee, Yang, The architecture of SARS-CoV-2 transcriptome, Cell, doi:10.1016/j.cell.2020.04.011
Kincaid, Sullivan, Hobman, Virus-encoded microRNAs: an overview and a look to the future, PLoS Pathog, doi:10.1371/journal.ppat.1003018
Kwon, Liu, Mostov, Intercellular transfer of GPRC5B via exosomes drives HGF-mediated outward growth, Curr Biol, doi:10.1016/j.cub.2013.12.010
Lee, Cornilescu, Dashti, Integrative NMR for biomolecular research, J Biomol NMR, doi:10.1007/s10858-016-0029-x
Lee, Rahimi, Lee, POKY: a software suite for multidimensional NMR and 3D structure calculation of biomolecules, Bioinformatics, doi:10.1093/bioinformatics/btab180
Lehmann, Krüger, Park, An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration, Nat Neurosci, doi:10.1038/nn.3113
Li, Lian, Moser, Identification of GW182 and its novel isoform TNGW1 as translational repressors in Ago2-mediated silencing, J Cell Sci, doi:10.1242/jcs.036905
Lin, Zhang, Xiang, Exosomes derived from HeLa cells break down vascular integrity by triggering endoplasmic reticulum stress in endothelial cells, J Extracell Vesicles, doi:10.1080/20013078.2020.1722385
Littler, Gully, Colson, Crystal structure of the SARS-CoV-2 non-structural protein 9, Nsp9, Iscience, doi:10.1016/j.isci.2020.101258
Maeta, Kataoka, Nishiya, RNA polymerase II subunit D is essential for zebrafish development, Sci Rep, doi:10.1038/s41598-020-70110-1
Mantovani, Daga, Fallerini, Rare variants in Toll-like receptor 7 results in functional impairment and downregulation of cytokine-mediated signaling in COVID-19 patients, Genes Immun, doi:10.1038/s41435-021-00157-1
Mcnab, Mayer-Barber, Sher, Type I interferons in infectious disease, Nat Rev Immunol, doi:10.1038/nri3787
Menachery, Graham, Baric, Jumping species-a mechanism for coronavirus persistence and survival, Curr Opin Virol, doi:10.1016/j.coviro.2017.01.002
Merino, Raad, Bugnon, Novel SARS-CoV-2 encoded small RNAs in the passage to humans, Bioinformatics, doi:10.1093/bioinformatics/btaa1002
Miknis, Donaldson, Umland, Severe acute respiratory syndrome coronavirus nsp9 dimerization is essential for efficient viral growth, J Virol, doi:10.1128/JVI.01505-08
Min, Choi, Mun, Mature microRNA-binding protein QKI suppresses extracellular microRNA let-7b release, J Cell Sci, doi:10.1242/jcs.261575
Min, Davila, Zealy, eIF4E phosphorylation by MST1 reduces translation of a subset of mRNAs, but increases lncRNA translation, Biochim Biophys Acta Gene Regul Mech, doi:10.1016/j.bbagrm.2017.05.002
Min, Jo, Shin, AUF1 facilitates microRNA-mediated gene silencing, Nucleic Acids Res, doi:10.1093/nar/gkx149
Min, Zealy, Davila, Profiling of m6A RNA modifications identified an age-associated regulation of AGO 2 mRNA stability, Aging Cell, doi:10.1111/acel.12753
Minakhina, Steward, Melanotic mutants in Drosophila: pathways and phenotypes, Genetics, doi:10.1534/genetics.106.061978
Mizrahi, Nachshon, Shitrit, Virus-induced changes in mRNA secondary structure uncover cis-regulatory elements that directly control gene expression, Mol Cell, doi:10.1016/j.molcel.2018.09.003
Mun, Lee, Choi, Targeting of CYP2E1 by miRnas in alcohol-induced intestine injury, Mol Cells, doi:10.1016/j.mocell.2024.100074
Nikolov, Burley, RNA polymerase II transcription initiation: a structural view, Proc Natl Acad Sci, doi:10.1073/pnas.94.1.15
O'boyle, Banck, James, Open babel: an open chemical toolbox, J Cheminform, doi:10.1186/1758-2946-3-33
O'connell, Kahn, Gibson, MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development, Immunity, doi:10.1016/j.immuni.2010.09.009
Park, Osinski, Hernandez, The mechanism of RNA capping by SARS-CoV-2, Nature, doi:10.1038/s41586-022-05185-z
Rahimi, Chiu, Lopez Giraldo, REDEN: interactive multi-fitting decomposition-based NMR peak picking assistant, J Magn Reson, doi:10.1016/j.jmr.2023.107600
Rüdel, Wang, Lenobel, Phosphorylation of human argonaute proteins affects small RNA binding, Nucleic Acids Res, doi:10.1093/nar/gkq1032
Shen, Bax, Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks, J Biomol NMR, doi:10.1007/s10858-013-9741-y
Snijder, Decroly, Ziebuhr, The nonstructural proteins directing coronavirus RNA synthesis and processing, Adv Virus Res
Sola, Almazán, Zúñiga, Continuous and discontinuous RNA synthesis in coronaviruses, Annu Rev Virol, doi:10.1146/annurev-virology-100114-055218
Sonoda, Lee, Park, miRNA profiling of urinary exosomes to assess the progression of acute kidney injury, Sci Rep, doi:10.1038/s41598-019-40747-8
Stevens, mRNA-decapping enzyme from Saccharomyces cerevisiae: purification and unique specificity for long RNA chains, Mol Cell Biol, doi:10.1128/MCB.8.5.2005
Thapar, Structural basis for regulation of RNA-binding proteins by phosphorylation, ACS Chem Biol, doi:10.1021/cb500860x
Titos, Juginović, Vaccaro, A gut-secreted peptide suppresses arousability from sleep, Cell, doi:10.1016/j.cell.2023.02.022
Trott, Olson, AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J Comput Chem, doi:10.1002/jcc.21334
Yoon, Abdelmohsen, Kim, Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination, Nat Commun, doi:10.1038/ncomms3939
Yoon, Abdelmohsen, Srikantan, LincRNA-p21 suppresses target mRNA translation, Mol Cell, doi:10.1016/j.molcel.2012.06.027
Yoon, Jo, White, AUF1 promotes let-7b loading on argonaute 2, Genes Dev, doi:10.1101/gad.263749.115
Zealy, Wrenn, Davila, microRNA-binding proteins: specificity and function, Wiley Interdiscip Rev: RNA, doi:10.1002/wrna.1414
Zhang, Amahong, Sun, The miRNA: a small but powerful RNA for COVID-19, Brief Bioinform, doi:10.1093/bib/bbab062
Zhou, Yang, Wang, A pneumonia outbreak associated with a new coronavirus of probable bat origin, Nature, doi:10.1038/s41586-020-2012-7
Zhu, Zhang, Song, SARS-CoV-2-encoded MiRNAs inhibit host type I interferon pathway and mediate allelic differential expression of susceptible gene, Front Immunol, doi:10.3389/fimmu.2021.767726
Zhu, Zhang, Wang, A novel coronavirus from patients with pneumonia in China, N Engl J Med, doi:10.1056/NEJMoa2001017
DOI record:
{
"DOI": "10.1080/15476286.2025.2527494",
"ISSN": [
"1547-6286",
"1555-8584"
],
"URL": "http://dx.doi.org/10.1080/15476286.2025.2527494",
"alternative-id": [
"10.1080/15476286.2025.2527494"
],
"assertion": [
{
"label": "Peer Review Statement",
"name": "peerreview_statement",
"order": 1,
"value": "The publishing and review policy for this title is described in its Aims & Scope."
},
{
"URL": "http://www.tandfonline.com/action/journalInformation?show=aimsScope&journalCode=krnb20",
"label": "Aim & Scope",
"name": "aims_and_scope_url",
"order": 2,
"value": "http://www.tandfonline.com/action/journalInformation?show=aimsScope&journalCode=krnb20"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Revised",
"name": "revised",
"order": 1,
"value": "2025-05-19"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "2025-06-17"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Published",
"name": "published",
"order": 3,
"value": "2025-07-07"
}
],
"author": [
{
"affiliation": [
{
"name": "University of Oklahoma",
"place": [
"Oklahoma City, USA"
]
}
],
"family": "Mun",
"given": "Hyejin",
"sequence": "first"
},
{
"affiliation": [
{
"name": "University of Oklahoma",
"place": [
"Oklahoma City, USA"
]
}
],
"family": "Shin",
"given": "Chang Hoon",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "University of Colorado",
"place": [
"Denver, USA"
]
}
],
"family": "Fei",
"given": "Qingxuan",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "University of Colorado",
"place": [
"Denver, USA"
]
}
],
"family": "Giraldo",
"given": "Andrea Estefania Lopez",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "University of Oklahoma",
"place": [
"Oklahoma City, USA"
]
}
],
"family": "Choi",
"given": "Kyoung-Min",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Gangneung-Wonju National University",
"place": [
"Gangneung-si, Republic of Korea"
]
}
],
"family": "Lee",
"given": "Ji Won",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Gangneung-Wonju National University",
"place": [
"Gangneung-si, Republic of Korea"
]
}
],
"family": "Kim",
"given": "Kyungmin",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Gangneung-Wonju National University",
"place": [
"Gangneung-si, Republic of Korea"
]
}
],
"family": "Min",
"given": "Kyung-Won",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "The University of Texas MD Anderson Cancer Center",
"place": [
"Houston, USA"
]
}
],
"family": "Shi",
"given": "Leilei",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "The University of Texas MD Anderson Cancer Center",
"place": [
"Houston, USA"
]
}
],
"family": "Bedford",
"given": "Mark T.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "NOSQUEST Inc",
"place": [
"Seongnam, Republic of Korea"
]
}
],
"family": "Kim",
"given": "Dong-Chan",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Medical University of South Carolina",
"place": [
"Charleston, USA"
]
}
],
"family": "Chun",
"given": "Yoo Lim",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "University of Oklahoma Health Sciences Center",
"place": [
"Oklahoma City, USA"
]
}
],
"family": "Ryu",
"given": "Seonghyun",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "University of Oklahoma Health Sciences Center",
"place": [
"Oklahoma City, USA"
]
}
],
"family": "Kim",
"given": "Dongin",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Kyungpook National University",
"place": [
"Daegu, Republic of Korea"
]
}
],
"family": "Chang",
"given": "Jeong Ho",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "University of Oklahoma",
"place": [
"Oklahoma City, USA"
]
}
],
"family": "Westrope",
"given": "Ryan T.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "University of Oklahoma",
"place": [
"Oklahoma City, USA"
]
}
],
"family": "Shay",
"given": "Michelle",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "University of Oklahoma",
"place": [
"Oklahoma City, USA"
]
}
],
"family": "Nguyen",
"given": "Edward",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Hanyang University",
"place": [
"Seoul, Republic of Korea"
]
}
],
"family": "Hur",
"given": "Junho K.",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "University of Minnesota",
"place": [
"Duluth, USA"
]
}
],
"family": "Agyenda",
"given": "Abigail",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "University of Minnesota",
"place": [
"Duluth, USA"
]
}
],
"family": "Kim",
"given": "Nam Chul",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Johns Hopkins University School of Medicine",
"place": [
"Baltimore, USA"
]
},
{
"name": "Johns Hopkins University School of Medicine",
"place": [
"Baltimore, USA"
]
}
],
"family": "Kang",
"given": "Sung-Ung",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "University of Colorado",
"place": [
"Denver, USA"
]
}
],
"family": "Lee",
"given": "Woonghee",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "University of Oklahoma",
"place": [
"Oklahoma City, USA"
]
},
{
"name": "University of Oklahoma",
"place": [
"Oklahoma City, USA"
]
}
],
"family": "Yoon",
"given": "Je-Hyun",
"sequence": "additional"
}
],
"container-title": "RNA Biology",
"container-title-short": "RNA Biology",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"www.tandfonline.com"
]
},
"created": {
"date-parts": [
[
2025,
7,
1
]
],
"date-time": "2025-07-01T11:28:20Z",
"timestamp": 1751369300000
},
"deposited": {
"date-parts": [
[
2025,
7,
7
]
],
"date-time": "2025-07-07T08:25:54Z",
"timestamp": 1751876754000
},
"funder": [
{
"DOI": "10.13039/501100014188",
"award": [
"NRF-2019R1A2C4069796"
],
"doi-asserted-by": "crossref",
"id": [
{
"asserted-by": "crossref",
"id": "10.13039/501100014188",
"id-type": "DOI"
}
],
"name": "Ministry of Science and ICT"
},
{
"DOI": "10.13039/100010346",
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/100010346",
"id-type": "DOI"
}
],
"name": "University of Colorado Denver"
},
{
"award": [
"RP180804"
],
"name": "CPRIT PAAC"
},
{
"DOI": "10.13039/100008901",
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/100008901",
"id-type": "DOI"
}
],
"name": "College of Pharmacy"
}
],
"indexed": {
"date-parts": [
[
2025,
7,
8
]
],
"date-time": "2025-07-08T04:05:28Z",
"timestamp": 1751947528489,
"version": "3.41.2"
},
"is-referenced-by-count": 0,
"issue": "1",
"issued": {
"date-parts": [
[
2025,
7,
7
]
]
},
"journal-issue": {
"issue": "1",
"published-print": {
"date-parts": [
[
2025,
12,
31
]
]
}
},
"language": "en",
"license": [
{
"URL": "http://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2025,
7,
7
]
],
"date-time": "2025-07-07T00:00:00Z",
"timestamp": 1751846400000
}
}
],
"link": [
{
"URL": "https://www.tandfonline.com/doi/pdf/10.1080/15476286.2025.2527494",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "301",
"original-title": [],
"page": "1-17",
"prefix": "10.1080",
"published": {
"date-parts": [
[
2025,
7,
7
]
]
},
"published-online": {
"date-parts": [
[
2025,
7,
7
]
]
},
"published-print": {
"date-parts": [
[
2025,
12,
31
]
]
},
"publisher": "Informa UK Limited",
"reference": [
{
"DOI": "10.1056/NEJMoa2001017",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_2_1"
},
{
"DOI": "10.1038/s41586-020-2012-7",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_3_1"
},
{
"DOI": "10.24171/j.phrp.2020.11.1.02",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_4_1"
},
{
"DOI": "10.1016/j.coviro.2017.01.002",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_5_1"
},
{
"DOI": "10.1016/bs.aivir.2016.08.008",
"article-title": "The nonstructural proteins directing coronavirus RNA synthesis and processing",
"author": "Snijder E",
"doi-asserted-by": "crossref",
"first-page": "59",
"journal-title": "Adv Virus Res",
"key": "e_1_3_5_6_1",
"unstructured": "Snijder E, Decroly E, Ziebuhr J. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv Virus Res. 2016;96:59–126.",
"volume": "96",
"year": "2016"
},
{
"DOI": "10.1146/annurev-virology-100114-055218",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_7_1"
},
{
"DOI": "10.1038/s41586-022-05185-z",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_8_1"
},
{
"DOI": "10.1016/j.cell.2020.04.011",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_9_1"
},
{
"DOI": "10.1016/j.ijantimicag.2020.105949",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_10_1"
},
{
"DOI": "10.5582/bst.2020.01047",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_11_1"
},
{
"DOI": "10.1016/S1473-3099(20)30141-9",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_12_1"
},
{
"DOI": "10.1007/s00109-020-01927-6",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_13_1"
},
{
"DOI": "10.1016/j.envres.2022.114288",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_14_1"
},
{
"DOI": "10.1038/s41435-021-00157-1",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_15_1"
},
{
"DOI": "10.1038/nn.3113",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_16_1"
},
{
"DOI": "10.1002/wrna.1414",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_17_1"
},
{
"DOI": "10.1111/acel.12753",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_18_1"
},
{
"DOI": "10.1093/nar/gkx149",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_19_1"
},
{
"DOI": "10.1016/j.isci.2020.101258",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_20_1"
},
{
"DOI": "10.1038/s41598-020-70110-1",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_21_1"
},
{
"DOI": "10.1101/gad.263749.115",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_22_1"
},
{
"DOI": "10.1038/ncomms3939",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_23_1"
},
{
"DOI": "10.1101/gad.1812509",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_24_1"
},
{
"DOI": "10.1038/s41586-020-2286-9",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_25_1"
},
{
"DOI": "10.1074/mcp.O111.016253",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_26_1"
},
{
"DOI": "10.1242/jcs.261575",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_27_1"
},
{
"DOI": "10.1016/j.mocell.2024.100074",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_28_1"
},
{
"DOI": "10.1093/nar/gkq1032",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_29_1"
},
{
"DOI": "10.1016/j.bbagrm.2017.05.002",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_30_1"
},
{
"DOI": "10.1128/JVI.01505-08",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_31_1"
},
{
"DOI": "10.1371/journal.pcbi.0020032",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_32_1"
},
{
"DOI": "10.1021/acs.jpcb.8b10749",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_33_1"
},
{
"DOI": "10.1097/MPA.0000000000000847",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_34_1"
},
{
"DOI": "10.1080/20013078.2020.1722385",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_35_1"
},
{
"DOI": "10.1016/j.celrep.2023.112842",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_36_1"
},
{
"DOI": "10.1534/genetics.106.061978",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_37_1"
},
{
"DOI": "10.1073/pnas.0307877101",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_38_1"
},
{
"DOI": "10.1073/pnas.94.1.15",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_39_1"
},
{
"DOI": "10.1021/cr400105n",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_40_1"
},
{
"DOI": "10.1242/jcs.036905",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_41_1"
},
{
"DOI": "10.1016/j.virol.2017.04.014",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_42_1"
},
{
"DOI": "10.1038/nrg2634",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_43_1"
},
{
"DOI": "10.1016/j.immuni.2010.09.009",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_44_1"
},
{
"DOI": "10.1038/nature07757",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_45_1"
},
{
"DOI": "10.1371/journal.ppat.1003018",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_46_1"
},
{
"DOI": "10.3389/fimmu.2021.767726",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_47_1"
},
{
"DOI": "10.1093/bib/bbab062",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_48_1"
},
{
"DOI": "10.1093/bioinformatics/btaa1002",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_49_1"
},
{
"DOI": "10.1021/cb500860x",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_50_1"
},
{
"DOI": "10.1038/266235a0",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_51_1"
},
{
"DOI": "10.1093/emboj/18.19.5411",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_52_1"
},
{
"DOI": "10.1128/MCB.8.5.2005",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_53_1"
},
{
"DOI": "10.1016/j.molcel.2018.09.003",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_54_1"
},
{
"DOI": "10.1038/nri3787",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_55_1"
},
{
"DOI": "10.1126/science.abc6261",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_56_1"
},
{
"DOI": "10.1016/j.envres.2022.114288",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_57_1"
},
{
"DOI": "10.1126/science.1093616",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_58_1"
},
{
"DOI": "10.1016/j.cell.2023.02.022",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_59_1"
},
{
"DOI": "10.1016/j.molcel.2012.06.027",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_60_1"
},
{
"DOI": "10.1016/j.cub.2013.12.010",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_61_1"
},
{
"DOI": "10.1038/s41598-019-40747-8",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_62_1"
},
{
"DOI": "10.1002/jcc.21334",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_63_1"
},
{
"DOI": "10.1186/1758-2946-3-33",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_64_1"
},
{
"DOI": "10.1016/j.cell.2009.08.037",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_65_1"
},
{
"DOI": "10.1093/nar/gkn923",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_66_1"
},
{
"DOI": "10.1038/nprot.2008.211",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_67_1"
},
{
"DOI": "10.1007/bf00197809",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_68_1"
},
{
"DOI": "10.1093/bioinformatics/btab180",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_69_1"
},
{
"DOI": "10.1007/s12104-021-10011-0",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_70_1"
},
{
"DOI": "10.1007/s10858-016-0029-x",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_71_1"
},
{
"DOI": "10.1016/j.jmr.2023.107600",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_72_1"
},
{
"DOI": "10.1371/journal.pcbi.1005659",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_73_1"
},
{
"DOI": "10.1007/s10858-013-9741-y",
"doi-asserted-by": "publisher",
"key": "e_1_3_5_74_1"
}
],
"reference-count": 73,
"references-count": 73,
"relation": {},
"resource": {
"primary": {
"URL": "https://www.tandfonline.com/doi/full/10.1080/15476286.2025.2527494"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "SARS-CoV-2 RNA-binding protein suppresses extracellular miRNA release",
"type": "journal-article",
"update-policy": "https://doi.org/10.1080/tandf_crossmark_01",
"volume": "22"
}