Upper airway gene expression reveals suppressed immune responses to SARS-CoV-2 compared with other respiratory viruses
et al., Nature Communications, doi:10.1038/s41467-020-19587-y, Nov 2020
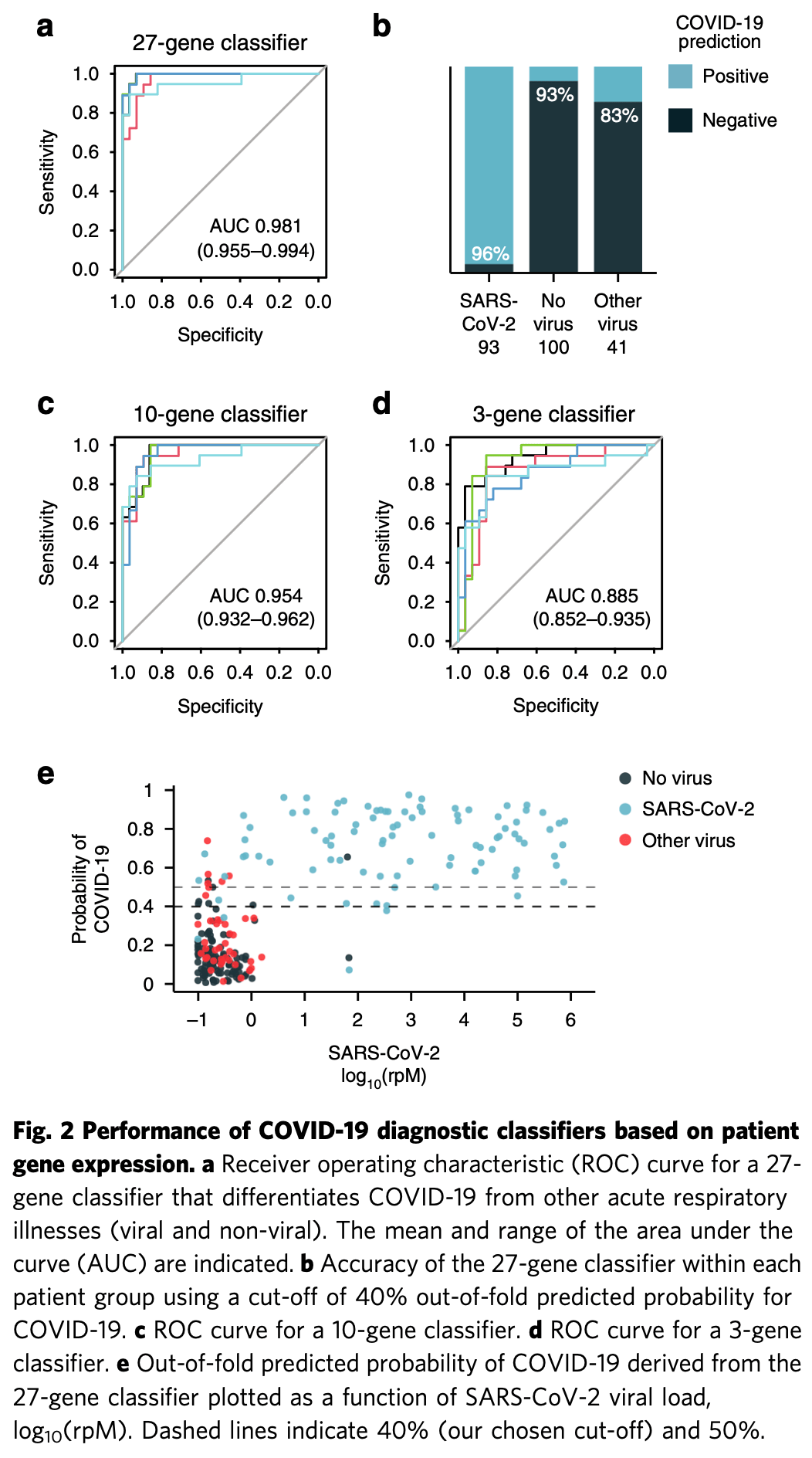
Upper airway gene expression analysis showing attenuated immune responses in COVID-19 patients compared to other respiratory viral infections. Authors analyzed nasopharyngeal/oropharyngeal swab specimens from 234 patients (93 with COVID-19, 41 with other viral respiratory infections, 100 with non-viral respiratory illnesses) using metagenomic RNA sequencing. COVID-19 patients showed robust interferon responses but markedly suppressed activation of toll-like receptor, interleukin, and chemokine signaling pathways compared to other viral infections. The IL-1 and NLRP3 inflammasome pathways were particularly non-responsive to SARS-CoV-2, with reduced neutrophil and macrophage recruitment to the upper airway. Authors developed machine learning classifiers using 27, 10, and 3 host genes that differentiated COVID-19 from other respiratory illnesses with AUROCs of 0.981, 0.954, and 0.885 respectively.
Mick et al., 17 Nov 2020, peer-reviewed, 23 authors.
Contact: chaz.langelier@ucsf.edu.
Upper airway gene expression reveals suppressed immune responses to SARS-CoV-2 compared with other respiratory viruses
Nature Communications, doi:10.1038/s41467-020-19587-y
SARS-CoV-2 infection is characterized by peak viral load in the upper airway prior to or at the time of symptom onset, an unusual feature that has enabled widespread transmission of the virus and precipitated a global pandemic. How SARS-CoV-2 is able to achieve high titer in the absence of symptoms remains unclear. Here, we examine the upper airway host transcriptional response in patients with COVID-19 (n = 93), other viral (n = 41) or non-viral (n = 100) acute respiratory illnesses (ARIs). Compared with other viral ARIs, COVID-19 is characterized by a pronounced interferon response but attenuated activation of other innate immune pathways, including toll-like receptor, interleukin and chemokine signaling. The IL-1 and NLRP3 inflammasome pathways are markedly less responsive to SARS-CoV-2, commensurate with a signature of diminished neutrophil and macrophage recruitment. This pattern resembles previously described distinctions between symptomatic and asymptomatic viral infections and may partly explain the propensity for pre-symptomatic transmission in COVID-19. We further use machine learning to build 27-, 10-and 3-gene classifiers that differentiate COVID-19 from other ARIs with AUROCs of 0.981, 0.954 and 0.885, respectively. Classifier performance is stable across a wide range of viral load, suggesting utility in mitigating false positive or false negative results of direct SARS-CoV-2 tests.
Author contributions C.L., A.K., E.M., and J.K. conceived and designed the study. A.K., N.N., G.C., A.M.D., G.R.K., P.H.S., M.T., and S.A.M. oversaw or performed sample processing, library preparation, and sequencing. C.L., S.J.S., K.N.K., P.A.P., K.R., L.M.L., and N.S. performed metadata collation or clinical chart review. E.M., J.K., A.O.P., C.L., K.R., and S.L.H. performed data analysis. S.A.C. contributed to data interpretation. E.M., J.K., and A.O.P. generated data visualizations. C.S.C., J.L.D., and J.M.B. provided guidance, advice, and comments on the study design and manuscript. C.L., E.M., J.K., and S.A.C. wrote the manuscript with input from all authors.
Competing interests The authors declare no competing interests.
Additional information Supplementary information is available for this paper at https://doi.org/10.1038/s41467- 020-19587-y . Correspondence and requests for materials should be addressed to C.L. Peer review information Nature Communications thanks Klaus Jung, Klaus Schughart and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available. Reprints and permission information is available at http://www.nature.com/reprints Publisher's note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Arons, Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility, N. Engl. J. Med
Blanco-Melo, Imbalanced host response to SARS-CoV-2 drives development of COVID-19, Cell, doi:10.1016/j.cell.2020.04.026
Bray, Pimentel, Melsted, Pachter, Near-optimal probabilistic RNA-seq quantification, Nat. Biotechnol
Butler, Shotgun transcriptome and isothermal profiling of SARS-CoV-2 infection reveals unique host responses, viral diversification, and drug interactions, doi:10.1101/2020.04.20.048066
Cao, COVID-19: immunopathology and its implications for therapy, Nat. Rev. Immunol
Chun, Baek, Kim, Transmission onset distribution of COVID-19, Int. J. Infect. Dis, doi:10.1016/j.ijid.2020.07.075
Croft, The Reactome pathway knowledgebase, Nucleic Acids Res
Deng, Metagenomic sequencing with spiked primer enrichment for viral diagnostics and genomic surveillance, Nat. Microbiol
Dong, Du, Gardner, An interactive web-based dashboard to track COVID-19 in real time, Lancet Infect. Dis
Friedman, Hastie, Tibshirani, Regularization paths for generalized linear models via coordinate descent, J. Stat. Softw., Artic
Gandhi, Yokoe, Havlir, Asymptomatic Transmission, the Achilles' Heel of Current Strategies to Control Covid-19, N. Engl. J. Med
Giacomelli, Self-reported olfactory and taste disorders in patients with severe acute respiratory coronavirus 2 infection: a cross-sectional study, Clin. Infect. Dis
He, Temporal dynamics in viral shedding and transmissibility of COVID-19, Nat. Med
Hiraishi, Ishida, Sudo, Nagahama, WDR74 participates in an early cleavage of the pre-rRNA processing pathway in cooperation with the nucleolar AAA-ATPase NVL2, Biochem. Biophys. Res. Commun
Huang, Blood single cell immune profiling reveals the interferon-MAPK pathway mediated adaptive immune response for COVID-19, doi:10.1101/2020.03.15.20033472
Huang, Temporal dynamics of host molecular responses differentiate symptomatic and asymptomatic influenza A infection, PLOS Genet
Kalantar, IDseqan open source cloud-based pipeline and analysis service for metagenomic pathogen detection and monitoring, doi:10.1101/2020.04.07.030551
Koller, Stahel, Sharpening Wald-type inference in robust regression for small samples, Comput. Stat. Data Anal
Korotkevich, Sukhov, Sergushichev, Fast gene set enrichment analysis, doi:10.1101/060012
Langelier, Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults, Proc. Natl Acad. Sci
Lavezzo, Suppression of a SARS-CoV-2 outbreak in the Italian municipality of Vo, Nature, doi:10.1038/s41586-020-2488-1
Lee, FAM83A confers EGFR-TKI resistance in breast cancer cells and in mice, J. Clin. Invest
Li, Minimap2: pairwise alignment for nucleotide sequences, Bioinformatics
Liao, Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19, Nat. Med
Liaw, Wiener, Classification and regression by random forest, R. N
Lopez-Castejon, Brough, Understanding the mechanism of IL-1β secretion, Cytokine Growth Factor Rev
Love, Huber, Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol
Maechler, robustbase: Basic robust statistics
Mayday, Khan, Chow, Zinter, Derisi, Miniaturization and optimization of 384-well compatible RNA sequencing library preparation, PLoS ONE
Menu, Vince, The NLRP3 inflammasome in health and disease: the good, the bad and the ugly, Clin. Exp. Immunol
Moein, Smell dysfunction: a biomarker for COVID-19, Int. Forum Allergy Rhinol
Moghadas, The implications of silent transmission for the control of COVID-19 outbreaks, Proc. Natl Acad. Sci
Ndzinu, Takeuchi, Saito, Yoshida, Yamaoka, eIF4A2 is a host factor required for efficient HIV-1 replication, Microbes Infect
Newman, Robust enumeration of cell subsets from tissue expression profiles, Nat. Methods
Ng, Attig, Bolland, Tissue-specific and interferon-inducible expression of nonfunctional ACE2 through endogenous retroelement cooption, Nat Genet, doi:10.1038/s41588-020-00732-8
Nienhold, Ciani, Koelzer, Two distinct immunopathological profiles in autopsy lungs of COVID-19, Nat Commun, doi:10.1038/s41467-020-18854-2
Onabajo, Banday, Stanifer, Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor, Nat Genet, doi:10.1038/s41588-020-00731-9
Peiris, Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study, Lancet
Pine, Evaluation of the External RNA Controls Consortium (ERCC) reference material using a modified Latin square design, BMC Biotechnol
Pirhonen, Sareneva, Kurimoto, Julkunen, Matikainen, Virus infection activates IL-1 beta and IL-18 production in human macrophages by a caspase-1-dependent pathway, J. Immunol
Ramesh, Metagenomic next-generation sequencing of samples from pediatric febrile illness in Tororo, Uganda, PLoS ONE
Ramlall, Immune complement and coagulation dysfunction in adverse outcomes of SARS-CoV-2 infection, Nat. Med
Renaud, Victoria-Feser, A robust coefficient of determination for regression, J. Stat. Plan. Inference
Ritchie, limma powers differential expression analyses for RNAsequencing and microarray studies, Nucleic Acids Res
Rothe, Transmission of 2019-nCoV infection from an asymptomatic contact in Germany, N. Engl. J. Med
Soneson, Love, Robinson, Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences, F1000Research
Song, EIF4A2 interacts with the membrane protein of transmissible gastroenteritis coronavirus and plays a role in virus replication, Res. Vet. Sci
Subramanian, Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles, Proc. Natl Acad. Sci
To, Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study, Lancet Infect. Dis
Travaglini, A molecular cell atlas of the human lung from single cell RNA sequencing, doi:10.1101/742320
Tsalik, Host gene expression classifiers diagnose acute respiratory illness etiology, Sci. Transl. Med
Venables, Ripley, Modern Applied Statistics with S
Wang, Detection of SARS-CoV-2 in different types of clinical specimens, JAMA
Wood, Lu, Langmead, Improved metagenomic analysis with Kraken 2, Genome Biol
Wu, Risk Factors Associated With Acute Respiratory Distress Syndrome and Death in Patients With Coronavirus Disease 2019 Pneumonia in Wuhan, China, JAMA Intern. Med, doi:10.1001/jamainternmed.2020.0994
Wölfel, Virological assessment of hospitalized patients with COVID, doi:10.1038/s41586-020-2196-x
Yang, Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections, doi:10.1101/2020.02.11.20021493
Yohai, High Breakdown-Point and High Efficiency Robust Estimates for Regression, Ann. Stat
Zhou, Heightened innate immune responses in the respiratory tract of COVID-19 patients, Cell Host Microbe
Ziegler, SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues, Cell, doi:10.1016/j.cell.2020.04.035
Zou, SARS-CoV-2 viral load in upper respiratory specimens of infected patients, N. Engl. J. Med
DOI record:
{
"DOI": "10.1038/s41467-020-19587-y",
"ISSN": [
"2041-1723"
],
"URL": "http://dx.doi.org/10.1038/s41467-020-19587-y",
"abstract": "<jats:title>Abstract</jats:title><jats:p>SARS-CoV-2 infection is characterized by peak viral load in the upper airway prior to or at the time of symptom onset, an unusual feature that has enabled widespread transmission of the virus and precipitated a global pandemic. How SARS-CoV-2 is able to achieve high titer in the absence of symptoms remains unclear. Here, we examine the upper airway host transcriptional response in patients with COVID-19 (<jats:italic>n</jats:italic> = 93), other viral (<jats:italic>n</jats:italic> = 41) or non-viral (<jats:italic>n</jats:italic> = 100) acute respiratory illnesses (ARIs). Compared with other viral ARIs, COVID-19 is characterized by a pronounced interferon response but attenuated activation of other innate immune pathways, including toll-like receptor, interleukin and chemokine signaling. The IL-1 and NLRP3 inflammasome pathways are markedly less responsive to SARS-CoV-2, commensurate with a signature of diminished neutrophil and macrophage recruitment. This pattern resembles previously described distinctions between symptomatic and asymptomatic viral infections and may partly explain the propensity for pre-symptomatic transmission in COVID-19. We further use machine learning to build 27-, 10- and 3-gene classifiers that differentiate COVID-19 from other ARIs with AUROCs of 0.981, 0.954 and 0.885, respectively. Classifier performance is stable across a wide range of viral load, suggesting utility in mitigating false positive or false negative results of direct SARS-CoV-2 tests.</jats:p>",
"alternative-id": [
"19587"
],
"article-number": "5854",
"assertion": [
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Received",
"name": "received",
"order": 1,
"value": "22 May 2020"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "Accepted",
"name": "accepted",
"order": 2,
"value": "16 October 2020"
},
{
"group": {
"label": "Article History",
"name": "ArticleHistory"
},
"label": "First Online",
"name": "first_online",
"order": 3,
"value": "17 November 2020"
},
{
"group": {
"label": "Competing interests",
"name": "EthicsHeading"
},
"name": "Ethics",
"order": 1,
"value": "The authors declare no competing interests."
}
],
"author": [
{
"ORCID": "https://orcid.org/0000-0002-7299-808X",
"affiliation": [],
"authenticated-orcid": false,
"family": "Mick",
"given": "Eran",
"sequence": "first"
},
{
"ORCID": "https://orcid.org/0000-0003-2412-756X",
"affiliation": [],
"authenticated-orcid": false,
"family": "Kamm",
"given": "Jack",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Pisco",
"given": "Angela Oliveira",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-5953-0004",
"affiliation": [],
"authenticated-orcid": false,
"family": "Ratnasiri",
"given": "Kalani",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Babik",
"given": "Jennifer M.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Castañeda",
"given": "Gloria",
"sequence": "additional"
},
{
"affiliation": [],
"family": "DeRisi",
"given": "Joseph L.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Detweiler",
"given": "Angela M.",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-4349-8131",
"affiliation": [],
"authenticated-orcid": false,
"family": "Hao",
"given": "Samantha L.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Kangelaris",
"given": "Kirsten N.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Kumar",
"given": "G. Renuka",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-6562-4004",
"affiliation": [],
"authenticated-orcid": false,
"family": "Li",
"given": "Lucy M.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Mann",
"given": "Sabrina A.",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0001-7141-5420",
"affiliation": [],
"authenticated-orcid": false,
"family": "Neff",
"given": "Norma",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Prasad",
"given": "Priya A.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Serpa",
"given": "Paula Hayakawa",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Shah",
"given": "Sachin J.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Spottiswoode",
"given": "Natasha",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Tan",
"given": "Michelle",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Calfee",
"given": "Carolyn S.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Christenson",
"given": "Stephanie A.",
"sequence": "additional"
},
{
"affiliation": [],
"family": "Kistler",
"given": "Amy",
"sequence": "additional"
},
{
"ORCID": "https://orcid.org/0000-0002-6708-4646",
"affiliation": [],
"authenticated-orcid": false,
"family": "Langelier",
"given": "Charles",
"sequence": "additional"
}
],
"container-title": "Nature Communications",
"container-title-short": "Nat Commun",
"content-domain": {
"crossmark-restriction": false,
"domain": [
"link.springer.com"
]
},
"created": {
"date-parts": [
[
2020,
11,
17
]
],
"date-time": "2020-11-17T11:02:56Z",
"timestamp": 1605610976000
},
"deposited": {
"date-parts": [
[
2023,
10,
12
]
],
"date-time": "2023-10-12T14:03:22Z",
"timestamp": 1697119402000
},
"funder": [
{
"DOI": "10.13039/100000050",
"award": [
"1K23HL138461-01A1"
],
"doi-asserted-by": "publisher",
"id": [
{
"asserted-by": "publisher",
"id": "10.13039/100000050",
"id-type": "DOI"
}
],
"name": "U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute"
},
{
"name": "Chan Zuckerberg Biohub Chan Zuckerberg Initiative"
}
],
"indexed": {
"date-parts": [
[
2025,
8,
3
]
],
"date-time": "2025-08-03T04:08:20Z",
"timestamp": 1754194100840,
"version": "3.37.3"
},
"is-referenced-by-count": 135,
"issue": "1",
"issued": {
"date-parts": [
[
2020,
11,
17
]
]
},
"journal-issue": {
"issue": "1",
"published-online": {
"date-parts": [
[
2020,
12
]
]
}
},
"language": "en",
"license": [
{
"URL": "https://creativecommons.org/licenses/by/4.0",
"content-version": "tdm",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2020,
11,
17
]
],
"date-time": "2020-11-17T00:00:00Z",
"timestamp": 1605571200000
}
},
{
"URL": "https://creativecommons.org/licenses/by/4.0",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2020,
11,
17
]
],
"date-time": "2020-11-17T00:00:00Z",
"timestamp": 1605571200000
}
}
],
"link": [
{
"URL": "https://www.nature.com/articles/s41467-020-19587-y.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.nature.com/articles/s41467-020-19587-y",
"content-type": "text/html",
"content-version": "vor",
"intended-application": "text-mining"
},
{
"URL": "https://www.nature.com/articles/s41467-020-19587-y.pdf",
"content-type": "application/pdf",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "297",
"original-title": [],
"prefix": "10.1038",
"published": {
"date-parts": [
[
2020,
11,
17
]
]
},
"published-online": {
"date-parts": [
[
2020,
11,
17
]
]
},
"publisher": "Springer Science and Business Media LLC",
"reference": [
{
"DOI": "10.1016/S1473-3099(20)30120-1",
"author": "E Dong",
"doi-asserted-by": "publisher",
"first-page": "533",
"journal-title": "Lancet Infect. Dis.",
"key": "19587_CR1",
"unstructured": "Dong, E., Du, H. & Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 20, 533–534 (2020).",
"volume": "20",
"year": "2020"
},
{
"DOI": "10.1038/s41591-020-0869-5",
"author": "X He",
"doi-asserted-by": "publisher",
"first-page": "672",
"journal-title": "Nat. Med.",
"key": "19587_CR2",
"unstructured": "He, X. et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 26, 672–675 (2020).",
"volume": "26",
"year": "2020"
},
{
"DOI": "10.1038/s41586-020-2488-1",
"doi-asserted-by": "publisher",
"key": "19587_CR3",
"unstructured": "Lavezzo, E. et al. Suppression of a SARS-CoV-2 outbreak in the Italian municipality of Vo’. Nature https://doi.org/10.1038/s41586-020-2488-1 (2020)."
},
{
"DOI": "10.1056/NEJMoa2008457",
"author": "MM Arons",
"doi-asserted-by": "publisher",
"first-page": "2081",
"journal-title": "N. Engl. J. Med.",
"key": "19587_CR4",
"unstructured": "Arons, M. M. et al. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N. Engl. J. Med. 382, 2081–2090 (2020).",
"volume": "382",
"year": "2020"
},
{
"DOI": "10.1056/NEJMc2001468",
"author": "C Rothe",
"doi-asserted-by": "publisher",
"first-page": "970",
"journal-title": "N. Engl. J. Med.",
"key": "19587_CR5",
"unstructured": "Rothe, C. et al. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 382, 970–971 (2020).",
"volume": "382",
"year": "2020"
},
{
"DOI": "10.1056/NEJMc2001737",
"author": "L Zou",
"doi-asserted-by": "publisher",
"first-page": "1177",
"journal-title": "N. Engl. J. Med.",
"key": "19587_CR6",
"unstructured": "Zou, L. et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 382, 1177–1179 (2020).",
"volume": "382",
"year": "2020"
},
{
"DOI": "10.1016/j.ijid.2020.07.075",
"doi-asserted-by": "publisher",
"key": "19587_CR7",
"unstructured": "Chun, J. Y., Baek, G. & Kim, Y. Transmission onset distribution of COVID-19. Int. J. Infect. Dis. https://doi.org/10.1016/j.ijid.2020.07.075 (2020)."
},
{
"DOI": "10.1016/S1473-3099(20)30196-1",
"author": "KK-W To",
"doi-asserted-by": "publisher",
"first-page": "565",
"journal-title": "Lancet Infect. Dis.",
"key": "19587_CR8",
"unstructured": "To, K. K.-W. et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 20, 565–574 (2020).",
"volume": "20",
"year": "2020"
},
{
"DOI": "10.1001/jamainternmed.2020.0994",
"doi-asserted-by": "publisher",
"key": "19587_CR9",
"unstructured": "Wu, C. et al. Risk Factors Associated With Acute Respiratory Distress Syndrome and Death in Patients With Coronavirus Disease 2019 Pneumonia in Wuhan, China. JAMA Intern. Med. https://doi.org/10.1001/jamainternmed.2020.0994 (2020)."
},
{
"DOI": "10.1073/pnas.1809700115",
"author": "C Langelier",
"doi-asserted-by": "publisher",
"first-page": "E12353 LP",
"journal-title": "Proc. Natl Acad. Sci. USA",
"key": "19587_CR10",
"unstructured": "Langelier, C. et al. Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults. Proc. Natl Acad. Sci. USA 115, E12353 LP–E12312362 (2018).",
"volume": "115",
"year": "2018"
},
{
"DOI": "10.1126/scitranslmed.aad6873",
"author": "EL Tsalik",
"doi-asserted-by": "publisher",
"first-page": "322ra11 LP",
"journal-title": "Sci. Transl. Med.",
"key": "19587_CR11",
"unstructured": "Tsalik, E. L. et al. Host gene expression classifiers diagnose acute respiratory illness etiology. Sci. Transl. Med. 8, 322ra11 LP–322ra11ra11 LP (2016).",
"volume": "8",
"year": "2016"
},
{
"DOI": "10.1101/2020.02.11.20021493",
"doi-asserted-by": "publisher",
"key": "19587_CR12",
"unstructured": "Yang, Y. et al. Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. Preprint at https://doi.org/10.1101/2020.02.11.20021493 (2020)."
},
{
"DOI": "10.1038/s41586-020-2196-x",
"doi-asserted-by": "publisher",
"key": "19587_CR13",
"unstructured": "Wölfel, R. et al. Virological assessment of hospitalized patients with COVID-2019. Nature https://doi.org/10.1038/s41586-020-2196-x (2020)."
},
{
"author": "W Wang",
"first-page": "1843",
"journal-title": "JAMA",
"key": "19587_CR14",
"unstructured": "Wang, W. et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 323, 1843–1844 (2020).",
"volume": "323",
"year": "2020"
},
{
"DOI": "10.1073/pnas.0506580102",
"author": "A Subramanian",
"doi-asserted-by": "publisher",
"first-page": "15545 LP",
"journal-title": "Proc. Natl Acad. Sci. USA",
"key": "19587_CR15",
"unstructured": "Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545 LP–15515550 (2005).",
"volume": "102",
"year": "2005"
},
{
"DOI": "10.1101/2020.04.20.048066",
"doi-asserted-by": "publisher",
"key": "19587_CR16",
"unstructured": "Butler, D. J. et al. Shotgun transcriptome and isothermal profiling of SARS-CoV-2 infection reveals unique host responses, viral diversification, and drug interactions. Preprint at https://doi.org/10.1101/2020.04.20.048066 (2020)."
},
{
"DOI": "10.1101/2020.03.15.20033472",
"doi-asserted-by": "publisher",
"key": "19587_CR17",
"unstructured": "Huang, L. et al. Blood single cell immune profiling reveals the interferon-MAPK pathway mediated adaptive immune response for COVID-19. Preprint at https://doi.org/10.1101/2020.03.15.20033472 (2020)."
},
{
"DOI": "10.1016/j.cell.2020.04.035",
"doi-asserted-by": "publisher",
"key": "19587_CR18",
"unstructured": "Ziegler, C. G. K. et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell https://doi.org/10.1016/j.cell.2020.04.035 (2020)."
},
{
"DOI": "10.1038/s41588-020-00731-9",
"doi-asserted-by": "publisher",
"key": "19587_CR19",
"unstructured": "Onabajo, O. O., Banday, A. R., Stanifer, M. L. et al. Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor. Nat Genet https://doi.org/10.1038/s41588-020-00731-9 (2020)."
},
{
"DOI": "10.1038/s41588-020-00732-8",
"doi-asserted-by": "publisher",
"key": "19587_CR20",
"unstructured": "Ng, K. W., Attig, J., Bolland, W. et al. Tissue-specific and interferon-inducible expression of nonfunctional ACE2 through endogenous retroelement co-option. Nat Genet https://doi.org/10.1038/s41588-020-00732-8 (2020)."
},
{
"DOI": "10.1016/j.cell.2020.04.026",
"doi-asserted-by": "publisher",
"key": "19587_CR21",
"unstructured": "Blanco-Melo, D. et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell https://doi.org/10.1016/j.cell.2020.04.026 (2020)."
},
{
"DOI": "10.1093/cid/ciaa330",
"author": "A Giacomelli",
"doi-asserted-by": "publisher",
"first-page": "889",
"journal-title": "Clin. Infect. Dis.",
"key": "19587_CR22",
"unstructured": "Giacomelli, A. et al. Self-reported olfactory and taste disorders in patients with severe acute respiratory coronavirus 2 infection: a cross-sectional study. Clin. Infect. Dis. 71, 889–890 (2020).",
"volume": "71",
"year": "2020"
},
{
"DOI": "10.1002/alr.22587",
"author": "ST Moein",
"doi-asserted-by": "publisher",
"first-page": "944",
"journal-title": "Int. Forum Allergy Rhinol.",
"key": "19587_CR23",
"unstructured": "Moein, S. T. et al. Smell dysfunction: a biomarker for COVID-19. Int. Forum Allergy Rhinol. 10, 944–950 (2020).",
"volume": "10",
"year": "2020"
},
{
"DOI": "10.1016/j.cytogfr.2011.10.001",
"author": "G Lopez-Castejon",
"doi-asserted-by": "publisher",
"first-page": "189",
"journal-title": "Cytokine Growth Factor Rev.",
"key": "19587_CR24",
"unstructured": "Lopez-Castejon, G. & Brough, D. Understanding the mechanism of IL-1β secretion. Cytokine Growth Factor Rev. 22, 189–195 (2011).",
"volume": "22",
"year": "2011"
},
{
"DOI": "10.4049/jimmunol.162.12.7322",
"author": "J Pirhonen",
"doi-asserted-by": "crossref",
"first-page": "7322",
"journal-title": "J. Immunol.",
"key": "19587_CR25",
"unstructured": "Pirhonen, J., Sareneva, T., Kurimoto, M., Julkunen, I. & Matikainen, S. Virus infection activates IL-1 beta and IL-18 production in human macrophages by a caspase-1-dependent pathway. J. Immunol. 162, 7322–7329 (1999).",
"volume": "162",
"year": "1999"
},
{
"DOI": "10.1111/j.1365-2249.2011.04440.x",
"author": "P Menu",
"doi-asserted-by": "publisher",
"first-page": "1",
"journal-title": "Clin. Exp. Immunol.",
"key": "19587_CR26",
"unstructured": "Menu, P. & Vince, J. E. The NLRP3 inflammasome in health and disease: the good, the bad and the ugly. Clin. Exp. Immunol. 166, 1–15 (2011).",
"volume": "166",
"year": "2011"
},
{
"DOI": "10.1038/s41591-020-1021-2",
"author": "V Ramlall",
"doi-asserted-by": "publisher",
"first-page": "1609",
"journal-title": "Nat. Med.",
"key": "19587_CR27",
"unstructured": "Ramlall, V. et al. Immune complement and coagulation dysfunction in adverse outcomes of SARS-CoV-2 infection. Nat. Med. 26, 1609–1615 (2020).",
"volume": "26",
"year": "2020"
},
{
"DOI": "10.1016/j.bbrc.2017.10.148",
"author": "N Hiraishi",
"doi-asserted-by": "publisher",
"first-page": "116",
"journal-title": "Biochem. Biophys. Res. Commun.",
"key": "19587_CR28",
"unstructured": "Hiraishi, N., Ishida, Y.-I., Sudo, H. & Nagahama, M. WDR74 participates in an early cleavage of the pre-rRNA processing pathway in cooperation with the nucleolar AAA-ATPase NVL2. Biochem. Biophys. Res. Commun. 495, 116–123 (2018).",
"volume": "495",
"year": "2018"
},
{
"DOI": "10.1016/j.rvsc.2018.12.005",
"author": "Z Song",
"doi-asserted-by": "publisher",
"first-page": "39",
"journal-title": "Res. Vet. Sci.",
"key": "19587_CR29",
"unstructured": "Song, Z. et al. EIF4A2 interacts with the membrane protein of transmissible gastroenteritis coronavirus and plays a role in virus replication. Res. Vet. Sci. 123, 39–46 (2019).",
"volume": "123",
"year": "2019"
},
{
"DOI": "10.1016/j.micinf.2018.05.001",
"author": "JK Ndzinu",
"doi-asserted-by": "publisher",
"first-page": "346",
"journal-title": "Microbes Infect.",
"key": "19587_CR30",
"unstructured": "Ndzinu, J. K., Takeuchi, H., Saito, H., Yoshida, T. & Yamaoka, S. eIF4A2 is a host factor required for efficient HIV-1 replication. Microbes Infect. 20, 346–352 (2018).",
"volume": "20",
"year": "2018"
},
{
"DOI": "10.1172/JCI60498",
"author": "S-Y Lee",
"doi-asserted-by": "publisher",
"first-page": "3211",
"journal-title": "J. Clin. Invest.",
"key": "19587_CR31",
"unstructured": "Lee, S.-Y. et al. FAM83A confers EGFR-TKI resistance in breast cancer cells and in mice. J. Clin. Invest. 122, 3211–3220 (2012).",
"volume": "122",
"year": "2012"
},
{
"DOI": "10.1056/NEJMe2009758",
"author": "M Gandhi",
"doi-asserted-by": "publisher",
"first-page": "2158",
"journal-title": "N. Engl. J. Med.",
"key": "19587_CR32",
"unstructured": "Gandhi, M., Yokoe, D. S. & Havlir, D. V. Asymptomatic Transmission, the Achilles’ Heel of Current Strategies to Control Covid-19. N. Engl. J. Med. 382, 2158–2160 (2020).",
"volume": "382",
"year": "2020"
},
{
"DOI": "10.1073/pnas.2008373117",
"author": "SM Moghadas",
"doi-asserted-by": "publisher",
"first-page": "17513 LP",
"journal-title": "Proc. Natl Acad. Sci. USA",
"key": "19587_CR33",
"unstructured": "Moghadas, S. M. et al. The implications of silent transmission for the control of COVID-19 outbreaks. Proc. Natl Acad. Sci. USA 117, 17513 LP–17517515 (2020).",
"volume": "117",
"year": "2020"
},
{
"DOI": "10.1016/S0140-6736(03)13412-5",
"author": "JSM Peiris",
"doi-asserted-by": "publisher",
"first-page": "1767",
"journal-title": "Lancet",
"key": "19587_CR34",
"unstructured": "Peiris, J. S. M. et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361, 1767–1772 (2003).",
"volume": "361",
"year": "2003"
},
{
"DOI": "10.1371/journal.pgen.1002234",
"author": "Y Huang",
"doi-asserted-by": "publisher",
"first-page": "e1002234",
"journal-title": "PLOS Genet.",
"key": "19587_CR35",
"unstructured": "Huang, Y. et al. Temporal dynamics of host molecular responses differentiate symptomatic and asymptomatic influenza A infection. PLOS Genet. 7, e1002234 (2011).",
"volume": "7",
"year": "2011"
},
{
"DOI": "10.1038/s41577-020-0308-3",
"author": "X Cao",
"doi-asserted-by": "publisher",
"first-page": "269",
"journal-title": "Nat. Rev. Immunol.",
"key": "19587_CR36",
"unstructured": "Cao, X. COVID-19: immunopathology and its implications for therapy. Nat. Rev. Immunol. 20, 269–270 (2020).",
"volume": "20",
"year": "2020"
},
{
"DOI": "10.1016/j.chom.2020.04.017",
"author": "Z Zhou",
"doi-asserted-by": "publisher",
"first-page": "883",
"journal-title": "Cell Host Microbe",
"key": "19587_CR37",
"unstructured": "Zhou, Z. et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe 27, 883–890 (2020). e2.",
"volume": "27",
"year": "2020"
},
{
"DOI": "10.1038/s41467-020-18854-2",
"author": "R Nienhold",
"doi-asserted-by": "publisher",
"first-page": "5086",
"journal-title": "Nat Commun",
"key": "19587_CR38",
"unstructured": "Nienhold, R., Ciani, Y., Koelzer, V. H. et al. Two distinct immunopathological profiles in autopsy lungs of COVID-19. Nat Commun 11, 5086 https://doi.org/10.1038/s41467-020-18854-2 (2020).",
"volume": "11",
"year": "2020"
},
{
"DOI": "10.1038/s41591-020-0901-9",
"author": "M Liao",
"doi-asserted-by": "publisher",
"first-page": "842",
"journal-title": "Nat. Med.",
"key": "19587_CR39",
"unstructured": "Liao, M. et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 26, 842–844 (2020).",
"volume": "26",
"year": "2020"
},
{
"DOI": "10.1186/s12896-016-0281-x",
"author": "PS Pine",
"doi-asserted-by": "publisher",
"journal-title": "BMC Biotechnol.",
"key": "19587_CR40",
"unstructured": "Pine, P. S. et al. Evaluation of the External RNA Controls Consortium (ERCC) reference material using a modified Latin square design. BMC Biotechnol. 16, 54 (2016).",
"volume": "16",
"year": "2016"
},
{
"DOI": "10.1038/s41564-019-0637-9",
"author": "X Deng",
"doi-asserted-by": "publisher",
"first-page": "443",
"journal-title": "Nat. Microbiol.",
"key": "19587_CR41",
"unstructured": "Deng, X. et al. Metagenomic sequencing with spiked primer enrichment for viral diagnostics and genomic surveillance. Nat. Microbiol. 5, 443–454 (2020).",
"volume": "5",
"year": "2020"
},
{
"DOI": "10.1186/s13059-019-1891-0",
"author": "DE Wood",
"doi-asserted-by": "publisher",
"journal-title": "Genome Biol.",
"key": "19587_CR42",
"unstructured": "Wood, D. E., Lu, J. & Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 20, 257 (2019).",
"volume": "20",
"year": "2019"
},
{
"DOI": "10.1093/bioinformatics/bty191",
"author": "H Li",
"doi-asserted-by": "publisher",
"first-page": "3094",
"journal-title": "Bioinformatics",
"key": "19587_CR43",
"unstructured": "Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).",
"volume": "34",
"year": "2018"
},
{
"DOI": "10.1101/2020.04.07.030551",
"doi-asserted-by": "publisher",
"key": "19587_CR44",
"unstructured": "Kalantar, K. L. et al. IDseq – an open source cloud-based pipeline and analysis service for metagenomic pathogen detection and monitoring. Preprint at https://doi.org/10.1101/2020.04.07.030551 (2020)."
},
{
"DOI": "10.1371/journal.pone.0218318",
"author": "A Ramesh",
"doi-asserted-by": "publisher",
"first-page": "e0218318",
"journal-title": "PLoS ONE",
"key": "19587_CR45",
"unstructured": "Ramesh, A. et al. Metagenomic next-generation sequencing of samples from pediatric febrile illness in Tororo, Uganda. PLoS ONE 14, e0218318 (2019).",
"volume": "14",
"year": "2019"
},
{
"DOI": "10.1371/journal.pone.0206194",
"author": "MY Mayday",
"doi-asserted-by": "publisher",
"first-page": "e0206194",
"journal-title": "PLoS ONE",
"key": "19587_CR46",
"unstructured": "Mayday, M. Y., Khan, L. M., Chow, E. D., Zinter, M. S. & DeRisi, J. L. Miniaturization and optimization of 384-well compatible RNA sequencing library preparation. PLoS ONE 14, e0206194 (2019).",
"volume": "14",
"year": "2019"
},
{
"DOI": "10.1007/978-0-387-21706-2",
"doi-asserted-by": "crossref",
"key": "19587_CR47",
"unstructured": "Venables, W. N. & Ripley, B. D. Modern Applied Statistics with S. (Springer, 2002)."
},
{
"DOI": "10.1038/nbt.3519",
"author": "NL Bray",
"doi-asserted-by": "publisher",
"first-page": "525",
"journal-title": "Nat. Biotechnol.",
"key": "19587_CR48",
"unstructured": "Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525 (2016).",
"volume": "34",
"year": "2016"
},
{
"DOI": "10.12688/f1000research.7563.1",
"author": "C Soneson",
"doi-asserted-by": "publisher",
"first-page": "1521",
"journal-title": "F1000Research",
"key": "19587_CR49",
"unstructured": "Soneson, C., Love, M. I. & Robinson, M. D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research 4, 1521 (2015).",
"volume": "4",
"year": "2015"
},
{
"DOI": "10.1093/nar/gkv007",
"author": "ME Ritchie",
"doi-asserted-by": "publisher",
"first-page": "e47",
"journal-title": "Nucleic Acids Res.",
"key": "19587_CR50",
"unstructured": "Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47–e47 (2015).",
"volume": "43",
"year": "2015"
},
{
"author": "MI Love",
"journal-title": "Genome Biol.",
"key": "19587_CR51",
"unstructured": "Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).",
"volume": "15",
"year": "2014"
},
{
"DOI": "10.1093/nar/gkt1102",
"author": "D Croft",
"doi-asserted-by": "publisher",
"first-page": "D472",
"journal-title": "Nucleic Acids Res.",
"key": "19587_CR52",
"unstructured": "Croft, D. et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 42, D472–D477 (2014).",
"volume": "42",
"year": "2014"
},
{
"DOI": "10.1101/060012",
"doi-asserted-by": "publisher",
"key": "19587_CR53",
"unstructured": "Korotkevich, G., Sukhov, V. & Sergushichev, A. Fast gene set enrichment analysis. Preprint at https://doi.org/10.1101/060012 (2019)."
},
{
"key": "19587_CR54",
"unstructured": "Maechler, M. et al. robustbase: Basic robust statistics. (2020)."
},
{
"DOI": "10.1016/j.csda.2011.02.014",
"author": "M Koller",
"doi-asserted-by": "publisher",
"first-page": "2504",
"journal-title": "Comput. Stat. Data Anal.",
"key": "19587_CR55",
"unstructured": "Koller, M. & Stahel, W. A. Sharpening Wald-type inference in robust regression for small samples. Comput. Stat. Data Anal. 55, 2504–2515 (2011).",
"volume": "55",
"year": "2011"
},
{
"DOI": "10.1214/aos/1176350366",
"author": "VJ Yohai",
"doi-asserted-by": "publisher",
"first-page": "642",
"journal-title": "Ann. Stat.",
"key": "19587_CR56",
"unstructured": "Yohai, V. J. High Breakdown-Point and High Efficiency Robust Estimates for Regression. Ann. Stat. 15, 642–656 (1987).",
"volume": "15",
"year": "1987"
},
{
"DOI": "10.1016/j.jspi.2010.01.008",
"author": "O Renaud",
"doi-asserted-by": "publisher",
"first-page": "1852",
"journal-title": "J. Stat. Plan. Inference",
"key": "19587_CR57",
"unstructured": "Renaud, O. & Victoria-Feser, M.-P. A robust coefficient of determination for regression. J. Stat. Plan. Inference 140, 1852–1862 (2010).",
"volume": "140",
"year": "2010"
},
{
"DOI": "10.1038/nmeth.3337",
"author": "AM Newman",
"doi-asserted-by": "publisher",
"first-page": "453",
"journal-title": "Nat. Methods",
"key": "19587_CR58",
"unstructured": "Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015).",
"volume": "12",
"year": "2015"
},
{
"DOI": "10.1101/742320",
"doi-asserted-by": "publisher",
"key": "19587_CR59",
"unstructured": "Travaglini, K. J. et al. A molecular cell atlas of the human lung from single cell RNA sequencing. Preprint at https://doi.org/10.1101/742320 (2020)."
},
{
"author": "J Friedman",
"first-page": "1",
"journal-title": "J. Stat. Softw., Artic.",
"key": "19587_CR60",
"unstructured": "Friedman, J., Hastie, T. & Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw., Artic. 33, 1–22 (2010).",
"volume": "33",
"year": "2010"
},
{
"author": "A Liaw",
"first-page": "18",
"journal-title": "R. N.",
"key": "19587_CR61",
"unstructured": "Liaw, A. & Wiener, M. Classification and regression by random forest. R. N. 2, 18–22 (2002).",
"volume": "2",
"year": "2002"
}
],
"reference-count": 61,
"references-count": 61,
"relation": {
"has-preprint": [
{
"asserted-by": "object",
"id": "10.1101/2020.05.18.20105171",
"id-type": "doi"
}
]
},
"resource": {
"primary": {
"URL": "https://www.nature.com/articles/s41467-020-19587-y"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [],
"subtitle": [],
"title": "Upper airway gene expression reveals suppressed immune responses to SARS-CoV-2 compared with other respiratory viruses",
"type": "journal-article",
"update-policy": "https://doi.org/10.1007/springer_crossmark_policy",
"volume": "11"
}