Integration of human organoids single‐cell transcriptomic profiles and human genetics repurposes critical cell type‐specific drug targets for severe COVID‐19
et al., Cell Proliferation, doi:10.1111/cpr.13558, Oct 2023
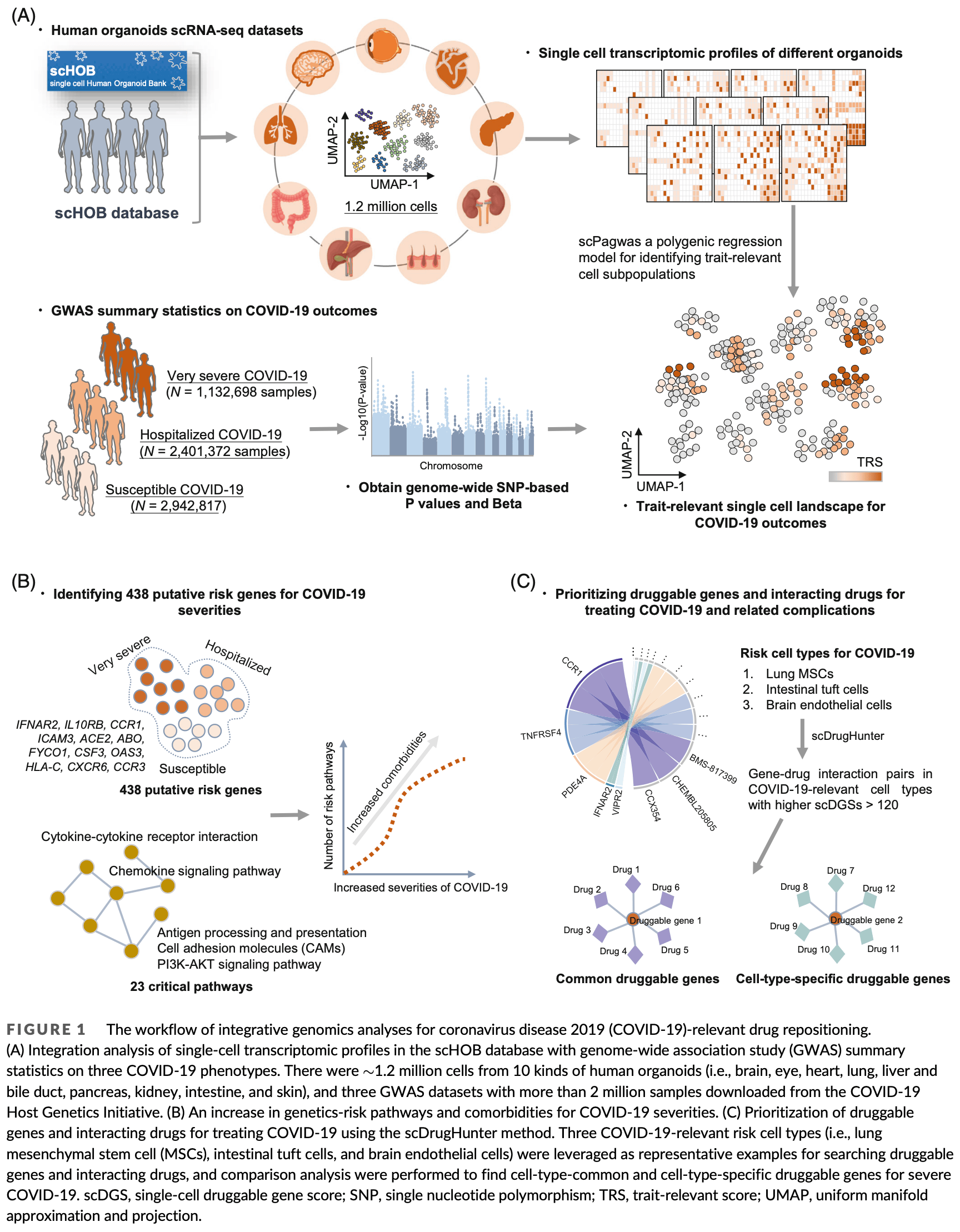
In silico study integrating human organoid data with single-cell transcriptomic profiles and GWAS to identify cell type-specific drug targets for severe COVID-19. The authors found 39 cell types across eight organoid types associated with COVID-19 severity. Lung mesenchymal stem cells (MSCs) showed increased proximity to fibroblasts, suggesting a role in lung repair. Brain endothelial cells were significantly associated with severe COVID-19, particularly due to increased interactions with microglia. The study repurposed 33 druggable genes, including IFNAR2, TYK2, and VIPR2, and identified their interacting drugs as potential COVID-19 therapeutics. Detailed single-cell analyses and genetic associations revealed pathways like cytokine signaling and chemokine signaling as potential therapeutic targets.
Ma et al., 8 Oct 2023, peer-reviewed, 17 authors.
Contact: yaoyinghao@ojlab.ac.cn, sujz@wmu.edu.cn.
In silico studies are an important part of preclinical research, however results may be very different in vivo.
Integration of human organoids single‐cell transcriptomic profiles and human genetics repurposes critical cell type‐specific drug targets for severe COVID‐19
Cell Proliferation, doi:10.1111/cpr.13558
Human organoids recapitulate the cell type diversity and function of their primary organs holding tremendous potentials for basic and translational research. Advances in single-cell RNA sequencing (scRNA-seq) technology and genome-wide association study (GWAS) have accelerated the biological and therapeutic interpretation of traitrelevant cell types or states. Here, we constructed a computational framework to integrate atlas-level organoid scRNA-seq data, GWAS summary statistics, expression quantitative trait loci, and gene-drug interaction data for distinguishing critical cell populations and drug targets relevant to coronavirus disease 2019 (COVID-19) severity. We found that 39 cell types across eight kinds of organoids were significantly associated with COVID-19 outcomes. Notably, subset of lung mesenchymal stem cells increased proximity with fibroblasts predisposed to repair COVID-19-damaged lung tissue. Brain endothelial cell subset exhibited significant associations with severe COVID-19, and this cell subset showed a notable increase in cell-to-cell interactions with other brain cell types, including microglia. We repurposed 33 druggable genes, including IFNAR2, TYK2, and VIPR2, and their interacting drugs for COVID-19 in a cell-type-specific manner. Overall, our results showcase that host genetic
AUTHOR CONTRIBUTIONS Yunlong
CONFLICT OF INTEREST STATEMENT The authors declare no competing interests.
SUPPORTING INFORMATION Additional supporting information can be found online in the Supporting Information section at the end of this article.
References
Abdo-Cuza, Castellanos-Gutiérrez, Treto-Ramirez, Safety and efficacy of intranasal recombinant human interferon alfa 2b as prophylaxis for COVID-19 in patients on a hemodialysis program, J Renal Endocrinol
Anderson, Rogers, Loupe, Single nucleus multiomics identifies ZEB1 and MAFB as candidate regulators of Alzheimer's disease-specific cis-regulatory elements, Cell. Genomics
Athar, Füllgrabe, George, ArrayExpress update-from bulk to single-cell expression data, Nucleic Acids Res
Auton, Brooks, Durbin, Garrison, Kang, A global reference for human genetic variation, Nature
Auwul, Rahman, Gov, Shahjaman, Moni, Bioinformatics and machine learning approach identifies potential drug targets and pathways in COVID-19, Brief Bioinform
Baldridge, Nice, Mccune, Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection, Science
Barbeira, Bonazzola, Gamazon, Exploiting the GTEx resources to decipher the mechanisms at GWAS loci, Genome Biol
Barbeira, Dickinson, Bonazzola, Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics, Nat Commun
Barbeira, Pividori, Zheng, Wheeler, Nicolae et al., Integrating predicted transcriptome from multiple tissues improves association detection, PLoS Genet
Boldrini, Canoll, Klein, How COVID-19 affects the brain, JAMA Psychiatry
Borkowski, Wili Nski, Szczesny, Binderman, Mathematical analysis of synthetic measures based on radar charts, Math Model Anal
Bulik-Sullivan, Loh, Finucane, LD score regression distinguishes confounding from polygenicity in genome-wide association studies, Nat Genet
Butler, Hoffman, Smibert, Papalexi, Satija, Integrating single-cell transcriptomic data across different conditions, technologies, and species, Nat Biotechnol
Cantuti-Castelvetri, Ojha, Pedro, Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity, Science
Chen, Park, Spence, Studying SARS-CoV-2 infectivity and therapeutic responses with complex organoids, Nat Cell Biol
Chung, Miasojedow, Startek, Gambin, Jaccard/Tanimoto similarity test and estimation methods for biological presenceabsence data, BMC Bioinformatics
Clough, Barrett, The Gene Expression Omnibus database, Methods Mol Biol
Cruceanu, Dony, Krontira, Cell-type-specific impact of glucocorticoid receptor activation on the developing brain: a cerebral organoid study, Am J Psychiatry
Daly, Simonetti, Klein, Neuropilin-1 is a host factor for SARS-CoV-2 infection, Science
De Leeuw, Mooij, Heskes, Posthuma, MAGMA: generalized gene-set analysis of GWAS data, PLoS Comput Biol
Delorey, Ziegler, Heimberg, COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets, Nature
Detomaso, Jones, Subramaniam, Ashuach, Ye et al., Functional interpretation of single cell similarity maps, Nat Commun
Dey, Gazal, Van De Geijn, SNP-to-gene linking strategies reveal contributions of enhancer-related and candidate master-regulator genes to autoimmune disease, Cell Genomics
Ding, Liang, Is SARS-CoV-2 also an enteric pathogen with potential fecal-oral transmission? A COVID-19 virological and clinical review, Gastroenterology
Dobin, Davis, Schlesinger, STAR: ultrafast universal RNA-seq aligner, Bioinformatics
Dong, Ma, Zhou, Integrated genomics analysis highlights important SNPs and genes implicated in moderate-to-severe asthma based on GWAS and eQTL datasets, BMC Pulm Med
Downes, Cross, Hua, Identification of LZTFL1 as a candidate effector gene at a COVID-19 risk locus, Nat Genet
Ellinghaus, Degenhardt, Bujanda, Genomewide association study of severe Covid-19 with respiratory failure, N Engl J Med
Fang, Wolf, Knezevic, A genetics-led approach defines the drug target landscape of 30 immune-related traits, Nat Genet
Finucane, Reshef, Anttila, Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types, Nat Genet
Freshour, Kiwala, Cotto, Integration of the drug-gene interaction database (DGIdb 4.0) with open crowdsource efforts, Nucleic Acids Res
Fullard, Lee, Voloudakis, Single-nucleus transcriptome analysis of human brain immune response in patients with severe COVID-19, Genome Med
Gaziano, Giambartolomei, Pereira, Actionable druggable genome-wide mendelian randomization identifies repurposing opportunities for COVID-19, Nat Med
Geary, The contiguity ratio and statistical mapping, Incorporated Stat
Guan, Liang, -H, Zhao, Comorbidity and its impact on 1590 patients with COVID-19 in China: a nationwide analysis, Eur Respir J
Gulati, Sikandar, Wesche, Single-cell transcriptional diversity is a hallmark of developmental potential, Science
Han, Duan, Yang, Identification of SARS-CoV-2 inhibitors using lung and colonic organoids, Nature
Hirt, Booij, Grob, Drug screening and genome editing in human pancreatic cancer organoids identifies drug-gene interactions and candidates for off-label therapy, Cell Genomics
Holloway, Wu, Czerwinski, Differentiation of human intestinal organoids with endogenous vascular endothelial cells, Dev Cell
Horowitz, Kosmicki, Damask, Genome-wide analysis provides genetic evidence that ACE2 influences COVID-19 risk and yields risk scores associated with severe disease, Nat Genet
Hung, Lung, Tso, Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial, Lancet
Jagadeesh, Dey, Montoro, Identifying diseasecritical cell types and cellular processes by integrating single-cell RNA-sequencing and human genetics, Nat Genet
Jeong, Bulyk, Blood cell traits' GWAS loci colocalization with variation in PU. 1 genomic occupancy prioritizes causal noncoding regulatory variants, Cell Genomics
Jin, Guerrero-Juarez, Zhang, Inference and analysis of cell-cell communication using CellChat, Nat Commun
Jyothula, Peters, Liang, Fulminant lung fibrosis in nonresolvable COVID-19 requiring transplantation, EBioMedicine
Kanehisa, Goto, KEGG: Kyoto encyclopedia of genes and genomes, Nucleic Acids Res
Kathiriya, Wang, Zhou, Human alveolar type 2 epithelium transdifferentiates into metaplastic KRT5+ basal cells, Nat Cell Biol
Korsunsky, Millard, Fan, Fast, sensitive and accurate integration of single-cell data with harmony, Nat Methods
Kousathanas, Pairo-Castineira, Rawlik, Whole-genome sequencing reveals host factors underlying critical COVID-19, Nature
Leinonen, Sugawara, Shumway, Collaboration INSD. The sequence read archive, Nucleic Acids Res
Leng, Ma, Zhang, Spatial region-resolved proteome map reveals mechanism of COVID-19-associated heart injury, Cell Rep
Li, Li, Wu, Plasma proteomic and metabolomic characterization of COVID-19 survivors 6 months after discharge, Cell Death Dis
Li, Ouyang, Zhan, Tian, CRISPR-based functional genomics screening in human-pluripotent-stem-cell-derived cell types, Cell. Genomics
Li, Wei, He, Sun, Liu, Interferon-induced transmembrane protein 3 gene polymorphisms are associated with COVID-19 susceptibility and severity: a meta-analysis, J Infect
Liao, Smyth, Shi, featureCounts: an efficient general purpose program for assigning sequence reads to genomic features, Bioinformatics
Liddelow, Guttenplan, Clarke, Neurotoxic reactive astrocytes are induced by activated microglia, Nature
Lotfollahi, Naghipourfar, Luecken, Mapping singlecell data to reference atlases by transfer learning, Nat Biotechnol
Luo, Liu, Xie, Single nucleus multi-omics identifies human cortical cell regulatory genome diversity, Cell Genomics
Ma, Deng, Zhou, Polygenic regression uncovers trait-relevant cellular contexts through pathway activation transformation of single-cell RNA sequencing data, Cell Genom
Ma, Huang, Zhao, Integrative genomics analysis reveals a 21q22. 11 locus contributing risk to COVID-19, Hum Mol Genet
Ma, Li, Establishment of a strong link between smoking and cancer pathogenesis through DNA methylation analysis, Sci Rep
Ma, Qiu, Deng, Integrating single-cell sequencing data with GWAS summary statistics reveals CD16+ monocytes and memory CD8+ T cells involved in severe COVID-19, Genome Med
Ma, Wang, Xu, Yu, Ma, Integrative genomics analysis of various omics data and networks identify risk genes and variants vulnerable to childhood-onset asthma, BMC Med Genomics
Ma, Zhou, Su, None
Mallard, Linnér, Grotzinger, Multivariate GWAS of psychiatric disorders and their cardinal symptoms reveal two dimensions of cross-cutting genetic liabilities, Cell Genomics
Meinhardt, Radke, Dittmayer, Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19, Nat Neurosci
Melms, Biermann, Huang, A molecular single-cell lung atlas of lethal COVID-19, Nature
Menche, Sharma, Kitsak, Uncovering disease-disease relationships through the incomplete interactome, Science
Monteil, Kwon, Prado, Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2, Cell
Mountjoy, Schmidt, Carmona, An open approach to systematically prioritize causal variants and genes at all published human GWAS trait-associated loci, Nat Genet
Ng, Sun, Je, Tan, Unravelling pathophysiology of neurological and psychiatric complications of COVID-19 using brain organoids, Neuroscientist
Pairo-Castineira, Clohisey, Klaric, Genetic mechanisms of critical illness in COVID-19, Nature
Pathak, Singh, Miller-Fleming, Integrative genomic analyses identify susceptibility genes underlying COVID-19 hospitalization, Nat Commun
Qiu, Mao, Tang, Reversed graph embedding resolves complex single-cell trajectories, Nat Methods
Quinlan, Hall, BEDTools: a flexible suite of utilities for comparing genomic features, Bioinformatics
Reay, Cairns, Advancing the use of genome-wide association studies for drug repurposing, Nat Rev Genet
Rendeiro, Ravichandran, Bram, The spatial landscape of lung pathology during COVID-19 progression, Nature
Roberts, Partha, Rhead, Expanded COVID-19 phenotype definitions reveal distinct patterns of genetic association and protective effects, Nat Genet
Rofeal, El-Malek, Ribosomal proteins as a possible tool for blocking SARS-COV 2 virus replication for a potential prospective treatment, Med Hypotheses
Samuel, Majd, Richter, Androgen signaling regulates SARS-CoV-2 receptor levels and is associated with severe COVID-19 symptoms in men, Cell Stem Cell
Sashindranath, Nandurkar, Endothelial dysfunction in the brain: setting the stage for stroke and other cerebrovascular complications of COVID-19, Stroke
Sun, Liu, Zou, Acute gastrointestinal injury in critically ill patients with COVID-19 in Wuhan, China, World J Gastroenterol
Turner, qqman: an R package for visualizing GWAS results using QQ and Manhattan plots, Biorxiv
Varga, Flammer, Steiger, Endothelial cell infection and endotheliitis in COVID-19, The Lancet
Wang, Duncan, Shi, Zhang, WEB-based GEne SeT AnaLysis toolkit (WebGestalt): update 2013, Nucleic Acids Res
Wang, Lin, Jiang, EPIC: inferring relevant cell types for complex traits by integrating genome-wide association studies and single-cell RNA sequencing, PLoS Genet
Wang, Liu, Luo, Rational drug repositioning for coronavirus-associated diseases using directional mapping and sideeffect inference, Iscience
Wilen, Lee, Hsieh, Tropism for tuft cells determines immune promotion of norovirus pathogenesis, Science
Wolf, Angerer, Theis, SCANPY: large-scale single-cell gene expression data analysis, Genome Biol
Wu, Mcgoogan, Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention, Jama
Wu, Qi, Wray, Visscher, Zeng et al., Joint analysis of GWAS and multi-omics QTL summary statistics reveals a large fraction of GWAS signals shared with molecular phenotypes, Cell Genomics
Xiang, Deng, Qiu, Single cell sequencing analysis identifies genetics-modulated ORMDL3+ cholangiocytes having higher metabolic effects on primary biliary cholangitis, J Nanobiotechnol
Xu, Feng, Wang, Mesenchymal stem cell treatment for COVID-19, EBioMedicine
Xu, Ma, Yuan, COVID-19 quarantine reveals that behavioral changes have an effect on myopia progression, Ophthalmology
Xu, Xie, Al-Aly, Long-term gastrointestinal outcomes of COVID-19, Nat Commun
Yang, Fan, Huang, Clinical and molecular characteristics of COVID-19 patients with persistent SARS-CoV-2 infection, Nat Commun
Yuan, Fan, Xue, Qu, Su, Co-expression of mitochondrial genes and ACE2 in cornea involved in COVID-19, Invest Ophthalmol Vis Sci
Zang, Castro, Mccune, TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes, Sci Immunol
Zhang, Hou, Dey, Polygenic enrichment distinguishes disease associations of individual cells in single-cell RNA-seq data, Nat Genet
Zhang, Zhang, Koeken, Altered and allele-specific open chromatin landscape reveals epigenetic and genetic regulators of innate immunity in COVID-19, Cell Genomics
Zheng, Terry, Belgrader, Massively parallel digital transcriptional profiling of single cells, Nat Commun
Zheng, Zhang, Zhao, Multi-ancestry mendelian randomization of omics traits revealing drug targets of COVID-19 severity, EBioMedicine
Zhou, Butler-Laporte, Nakanishi, A neanderthal OAS1 isoform protects individuals of European ancestry against COVID-19 susceptibility and severity, Nat Med
Zhou, Yu, Du, Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study, The Lancet
Zhu, Yan, Feng, Mesenchymal stem cell treatment improves outcome of COVID-19 patients via multiple immunomodulatory mechanisms, Cell Res
DOI record:
{
"DOI": "10.1111/cpr.13558",
"ISSN": [
"0960-7722",
"1365-2184"
],
"URL": "http://dx.doi.org/10.1111/cpr.13558",
"abstract": "<jats:title>Abstract</jats:title><jats:p>Human organoids recapitulate the cell type diversity and function of their primary organs holding tremendous potentials for basic and translational research. Advances in single‐cell RNA sequencing (scRNA‐seq) technology and genome‐wide association study (GWAS) have accelerated the biological and therapeutic interpretation of trait‐relevant cell types or states. Here, we constructed a computational framework to integrate atlas‐level organoid scRNA‐seq data, GWAS summary statistics, expression quantitative trait loci, and gene–drug interaction data for distinguishing critical cell populations and drug targets relevant to coronavirus disease 2019 (COVID‐19) severity. We found that 39 cell types across eight kinds of organoids were significantly associated with COVID‐19 outcomes. Notably, subset of lung mesenchymal stem cells increased proximity with fibroblasts predisposed to repair COVID‐19‐damaged lung tissue. Brain endothelial cell subset exhibited significant associations with severe COVID‐19, and this cell subset showed a notable increase in cell‐to‐cell interactions with other brain cell types, including microglia. We repurposed 33 druggable genes, including <jats:italic>IFNAR2</jats:italic>, <jats:italic>TYK2</jats:italic>, and <jats:italic>VIPR2</jats:italic>, and their interacting drugs for COVID‐19 in a cell‐type‐specific manner. Overall, our results showcase that host genetic determinants have cellular‐specific contribution to COVID‐19 severity, and identification of cell type‐specific drug targets may facilitate to develop effective therapeutics for treating severe COVID‐19 and its complications.</jats:p>",
"alternative-id": [
"10.1111/cpr.13558"
],
"assertion": [
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Received",
"name": "received",
"order": 0,
"value": "2023-07-31"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Accepted",
"name": "accepted",
"order": 1,
"value": "2023-09-18"
},
{
"group": {
"label": "Publication History",
"name": "publication_history"
},
"label": "Published",
"name": "published",
"order": 2,
"value": "2023-10-08"
}
],
"author": [
{
"ORCID": "http://orcid.org/0000-0002-1299-4802",
"affiliation": [
{
"name": "National Engineering Research Center of Ophthalmology and Optometry Eye Hospital, Wenzhou Medical University Wenzhou China"
},
{
"name": "Department of Biomedical Informatics Institute of Biomedical Big Data, Wenzhou Medical University Wenzhou China"
},
{
"name": "Oujiang Laboratory, Zhejiang Lab for Regenerative Medicine, Vision and Brain Health Zhejiang China"
}
],
"authenticated-orcid": false,
"family": "Ma",
"given": "Yunlong",
"sequence": "first"
},
{
"affiliation": [
{
"name": "National Engineering Research Center of Ophthalmology and Optometry Eye Hospital, Wenzhou Medical University Wenzhou China"
},
{
"name": "Department of Biomedical Informatics Institute of Biomedical Big Data, Wenzhou Medical University Wenzhou China"
}
],
"family": "Zhou",
"given": "Yijun",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "National Engineering Research Center of Ophthalmology and Optometry Eye Hospital, Wenzhou Medical University Wenzhou China"
},
{
"name": "Oujiang Laboratory, Zhejiang Lab for Regenerative Medicine, Vision and Brain Health Zhejiang China"
}
],
"family": "Jiang",
"given": "Dingping",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Wenzhou Institute University of Chinese Academy of Sciences Wenzhou China"
}
],
"family": "Dai",
"given": "Wei",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "National Engineering Research Center of Ophthalmology and Optometry Eye Hospital, Wenzhou Medical University Wenzhou China"
},
{
"name": "Department of Biomedical Informatics Institute of Biomedical Big Data, Wenzhou Medical University Wenzhou China"
}
],
"family": "Li",
"given": "Jingjing",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "School of Life Science and Technology Harbin Institute of Technology Harbin China"
}
],
"family": "Deng",
"given": "Chunyu",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "National Engineering Research Center of Ophthalmology and Optometry Eye Hospital, Wenzhou Medical University Wenzhou China"
},
{
"name": "Department of Biomedical Informatics Institute of Biomedical Big Data, Wenzhou Medical University Wenzhou China"
}
],
"family": "Chen",
"given": "Cheng",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "National Engineering Research Center of Ophthalmology and Optometry Eye Hospital, Wenzhou Medical University Wenzhou China"
},
{
"name": "Department of Biomedical Informatics Institute of Biomedical Big Data, Wenzhou Medical University Wenzhou China"
}
],
"family": "Zheng",
"given": "Gongwei",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "National Engineering Research Center of Ophthalmology and Optometry Eye Hospital, Wenzhou Medical University Wenzhou China"
},
{
"name": "Department of Biomedical Informatics Institute of Biomedical Big Data, Wenzhou Medical University Wenzhou China"
},
{
"name": "Oujiang Laboratory, Zhejiang Lab for Regenerative Medicine, Vision and Brain Health Zhejiang China"
}
],
"family": "Zhang",
"given": "Yaru",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "National Engineering Research Center of Ophthalmology and Optometry Eye Hospital, Wenzhou Medical University Wenzhou China"
},
{
"name": "Department of Biomedical Informatics Institute of Biomedical Big Data, Wenzhou Medical University Wenzhou China"
}
],
"family": "Qiu",
"given": "Fei",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "National Engineering Research Center of Ophthalmology and Optometry Eye Hospital, Wenzhou Medical University Wenzhou China"
},
{
"name": "Department of Biomedical Informatics Institute of Biomedical Big Data, Wenzhou Medical University Wenzhou China"
}
],
"family": "Sun",
"given": "Haojun",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "National Engineering Research Center of Ophthalmology and Optometry Eye Hospital, Wenzhou Medical University Wenzhou China"
},
{
"name": "Department of Biomedical Informatics Institute of Biomedical Big Data, Wenzhou Medical University Wenzhou China"
}
],
"family": "Xing",
"given": "Shilai",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "School of Medicine Hangzhou City University Hangzhou China"
}
],
"family": "Han",
"given": "Haijun",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "National Engineering Research Center of Ophthalmology and Optometry Eye Hospital, Wenzhou Medical University Wenzhou China"
}
],
"family": "Qu",
"given": "Jia",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Beijing Key Laboratory for Genetic Research of Skeletal Deformity, Key Laboratory of Big Data for Spinal Deformities, Department of Orthopedic Surgery Peking Union Medical College Hospital, Peking Union Medical College and Chinese Academy of Medical Sciences Beijing China"
}
],
"family": "Wu",
"given": "Nan",
"sequence": "additional"
},
{
"affiliation": [
{
"name": "Oujiang Laboratory, Zhejiang Lab for Regenerative Medicine, Vision and Brain Health Zhejiang China"
}
],
"family": "Yao",
"given": "Yinghao",
"sequence": "additional"
},
{
"ORCID": "http://orcid.org/0000-0003-1054-6042",
"affiliation": [
{
"name": "National Engineering Research Center of Ophthalmology and Optometry Eye Hospital, Wenzhou Medical University Wenzhou China"
},
{
"name": "Department of Biomedical Informatics Institute of Biomedical Big Data, Wenzhou Medical University Wenzhou China"
},
{
"name": "Oujiang Laboratory, Zhejiang Lab for Regenerative Medicine, Vision and Brain Health Zhejiang China"
}
],
"authenticated-orcid": false,
"family": "Su",
"given": "Jianzhong",
"sequence": "additional"
}
],
"container-title": "Cell Proliferation",
"container-title-short": "Cell Proliferation",
"content-domain": {
"crossmark-restriction": true,
"domain": [
"onlinelibrary.wiley.com"
]
},
"created": {
"date-parts": [
[
2023,
10,
11
]
],
"date-time": "2023-10-11T23:47:41Z",
"timestamp": 1697068061000
},
"deposited": {
"date-parts": [
[
2024,
3,
1
]
],
"date-time": "2024-03-01T09:30:25Z",
"timestamp": 1709285425000
},
"funder": [
{
"DOI": "10.13039/501100001809",
"award": [
"82172882",
"61871294",
"32200535"
],
"doi-asserted-by": "publisher",
"name": "National Natural Science Foundation of China"
},
{
"DOI": "10.13039/501100004731",
"award": [
"LR19C060001"
],
"doi-asserted-by": "publisher",
"name": "Natural Science Foundation of Zhejiang Province"
}
],
"indexed": {
"date-parts": [
[
2024,
3,
2
]
],
"date-time": "2024-03-02T00:15:22Z",
"timestamp": 1709338522263
},
"is-referenced-by-count": 2,
"issue": "3",
"issued": {
"date-parts": [
[
2023,
10,
8
]
]
},
"journal-issue": {
"issue": "3",
"published-print": {
"date-parts": [
[
2024,
3
]
]
}
},
"language": "en",
"license": [
{
"URL": "http://creativecommons.org/licenses/by/4.0/",
"content-version": "vor",
"delay-in-days": 0,
"start": {
"date-parts": [
[
2023,
10,
8
]
],
"date-time": "2023-10-08T00:00:00Z",
"timestamp": 1696723200000
}
}
],
"link": [
{
"URL": "https://onlinelibrary.wiley.com/doi/pdf/10.1111/cpr.13558",
"content-type": "unspecified",
"content-version": "vor",
"intended-application": "similarity-checking"
}
],
"member": "311",
"original-title": [],
"prefix": "10.1111",
"published": {
"date-parts": [
[
2023,
10,
8
]
]
},
"published-online": {
"date-parts": [
[
2023,
10,
8
]
]
},
"published-print": {
"date-parts": [
[
2024,
3
]
]
},
"publisher": "Wiley",
"reference": [
{
"DOI": "10.1001/jama.2020.2648",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_2_1"
},
{
"DOI": "10.1016/j.ophtha.2021.04.001",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_3_1"
},
{
"DOI": "10.1016/j.xgen.2022.100232",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_4_1"
},
{
"DOI": "10.1038/s41591-021-01310-z",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_5_1"
},
{
"DOI": "10.1038/s41556-021-00721-x",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_6_1"
},
{
"DOI": "10.1016/j.xgen.2022.100095",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_7_1"
},
{
"DOI": "10.1186/s12951-021-01154-2",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_8_1"
},
{
"article-title": "CRISPR‐based functional genomics screening in human‐pluripotent‐stem‐cell‐derived cell types. Cell",
"author": "Li K",
"issue": "5",
"journal-title": "Genomics",
"key": "e_1_2_9_9_1",
"volume": "3",
"year": "2023"
},
{
"DOI": "10.1038/s41586-020-2901-9",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_10_1"
},
{
"DOI": "10.1016/j.xgen.2022.100107",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_11_1"
},
{
"DOI": "10.1016/j.cell.2020.04.004",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_12_1"
},
{
"DOI": "10.1016/j.xgen.2022.100140",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_13_1"
},
{
"DOI": "10.1038/s41586-020-03065-y",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_14_1"
},
{
"DOI": "10.1056/NEJMoa2020283",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_15_1"
},
{
"DOI": "10.1093/hmg/ddab125",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_16_1"
},
{
"DOI": "10.1016/j.ebiom.2022.104112",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_17_1"
},
{
"DOI": "10.1038/s41591-021-01281-1",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_18_1"
},
{
"DOI": "10.1038/s41588-022-01187-9",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_19_1"
},
{
"DOI": "10.1186/s13073-022-01021-1",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_20_1"
},
{
"DOI": "10.1038/s41586-021-03570-8",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_21_1"
},
{
"DOI": "10.1007/978-1-4939-3578-9_5",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_22_1"
},
{
"DOI": "10.1093/nar/gky964",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_23_1"
},
{
"article-title": "The sequence read archive",
"author": "Leinonen R",
"first-page": "D19",
"issue": "1",
"journal-title": "Nucleic Acids Res",
"key": "e_1_2_9_24_1",
"volume": "39",
"year": "2010"
},
{
"DOI": "10.1093/bioinformatics/btq033",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_25_1"
},
{
"DOI": "10.1038/ncomms14049",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_26_1"
},
{
"DOI": "10.1093/bioinformatics/bts635",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_27_1"
},
{
"DOI": "10.1038/nature15393",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_28_1"
},
{
"DOI": "10.1093/bioinformatics/btt656",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_29_1"
},
{
"DOI": "10.1186/s13059-017-1382-0",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_30_1"
},
{
"DOI": "10.1038/nbt.4096",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_31_1"
},
{
"DOI": "10.1038/s41592-019-0619-0",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_32_1"
},
{
"DOI": "10.1038/s41587-021-01001-7",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_33_1"
},
{
"DOI": "10.1038/s41431-020-0636-6",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_34_1"
},
{
"DOI": "10.1101/005165",
"doi-asserted-by": "crossref",
"key": "e_1_2_9_35_1",
"unstructured": "TurnerSD.qqman: an R package for visualizing GWAS results using QQ and Manhattan plots. Biorxiv2014:005165."
},
{
"DOI": "10.1016/j.xgen.2023.100383",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_36_1"
},
{
"DOI": "10.1093/nar/28.1.27",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_37_1"
},
{
"DOI": "10.1038/s41588-018-0081-4",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_38_1"
},
{
"DOI": "10.1038/s41467-019-12235-0",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_39_1"
},
{
"DOI": "10.2307/2986645",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_40_1"
},
{
"DOI": "10.1038/s41467-018-03621-1",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_41_1"
},
{
"DOI": "10.1371/journal.pgen.1007889",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_42_1"
},
{
"DOI": "10.1371/journal.pcbi.1004219",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_43_1"
},
{
"DOI": "10.1186/s12920-020-00768-z",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_44_1"
},
{
"DOI": "10.1186/s12890-020-01303-7",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_45_1"
},
{
"article-title": "Jaccard/Tanimoto similarity test and estimation methods for biological presence‐absence data",
"author": "Chung NC",
"first-page": "1",
"issue": "15",
"journal-title": "BMC Bioinformatics",
"key": "e_1_2_9_46_1",
"volume": "20",
"year": "2019"
},
{
"DOI": "10.1093/nar/gkt439",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_47_1"
},
{
"DOI": "10.1093/nar/gku1179",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_48_1"
},
{
"DOI": "10.1038/ng.3211",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_49_1"
},
{
"key": "e_1_2_9_50_1",
"unstructured": "MaY ZhouY SuJ.https://github.com/x-burner-ux/scDrugHunter. Github2023."
},
{
"DOI": "10.1186/s13059-020-02252-4",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_51_1"
},
{
"DOI": "10.1093/nar/gkaa1084",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_52_1"
},
{
"DOI": "10.1038/s41588-019-0456-1",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_53_1"
},
{
"DOI": "10.1038/s41588-021-00945-5",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_54_1"
},
{
"DOI": "10.3846/mma.2020.11223",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_55_1"
},
{
"DOI": "10.1038/s41467-021-21246-9",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_56_1"
},
{
"DOI": "10.1016/j.devcel.2020.07.023",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_57_1"
},
{
"DOI": "10.1176/appi.ajp.2021.21010095",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_58_1"
},
{
"DOI": "10.1038/s41588-022-01167-z",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_59_1"
},
{
"DOI": "10.1371/journal.pgen.1010251",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_60_1"
},
{
"DOI": "10.1038/s41422-021-00573-y",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_61_1"
},
{
"DOI": "10.1016/j.ebiom.2022.103920",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_62_1"
},
{
"DOI": "10.1053/j.gastro.2020.04.052",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_63_1"
},
{
"DOI": "10.1038/s41467-023-36223-7",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_64_1"
},
{
"DOI": "10.1161/STROKEAHA.120.032711",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_65_1"
},
{
"DOI": "10.1186/s13073-021-00933-8",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_66_1"
},
{
"DOI": "10.1167/iovs.61.12.13",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_67_1"
},
{
"DOI": "10.1038/s41588-021-01006-7",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_68_1"
},
{
"DOI": "10.1038/s41467-021-24824-z",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_69_1"
},
{
"DOI": "10.1038/s41588-022-01042-x",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_70_1"
},
{
"DOI": "10.1038/s41588-021-00955-3",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_71_1"
},
{
"DOI": "10.1038/s41586-022-04576-6",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_72_1"
},
{
"DOI": "10.1038/s41598-016-0028-x",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_73_1"
},
{
"DOI": "10.1126/science.1257601",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_74_1"
},
{
"DOI": "10.1093/bib/bbab120",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_75_1"
},
{
"DOI": "10.1183/13993003.00547-2020",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_76_1"
},
{
"DOI": "10.1016/S0140-6736(20)30566-3",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_77_1"
},
{
"DOI": "10.1016/j.stem.2020.11.009",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_78_1"
},
{
"DOI": "10.1038/s41556-021-00809-4",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_79_1"
},
{
"DOI": "10.1126/science.aax0249",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_80_1"
},
{
"DOI": "10.1038/nmeth.4402",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_81_1"
},
{
"DOI": "10.1038/s41586-021-03475-6",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_82_1"
},
{
"DOI": "10.1038/s41419-022-04674-3",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_83_1"
},
{
"DOI": "10.3748/wjg.v26.i39.6087",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_84_1"
},
{
"DOI": "10.1126/sciimmunol.abc3582",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_85_1"
},
{
"DOI": "10.1126/science.aar3799",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_86_1"
},
{
"DOI": "10.1126/science.1258025",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_87_1"
},
{
"DOI": "10.1016/j.ebiom.2022.104351",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_88_1"
},
{
"DOI": "10.1016/j.celrep.2022.110955",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_89_1"
},
{
"DOI": "10.1038/s41586-021-03569-1",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_90_1"
},
{
"DOI": "10.1016/j.jinf.2022.04.029",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_91_1"
},
{
"DOI": "10.1016/j.mehy.2020.109904",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_92_1"
},
{
"article-title": "Clinical and molecular characteristics of COVID‐19 patients with persistent SARS‐CoV‐2 infection",
"author": "Yang B",
"first-page": "1",
"issue": "1",
"journal-title": "Nat Commun",
"key": "e_1_2_9_93_1",
"volume": "12",
"year": "2021"
},
{
"DOI": "10.1001/jamapsychiatry.2021.0500",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_94_1"
},
{
"DOI": "10.1177/10738584211015136",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_95_1"
},
{
"DOI": "10.1038/s41593-020-00758-5",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_96_1"
},
{
"DOI": "10.1126/science.abd2985",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_97_1"
},
{
"DOI": "10.1126/science.abd3072",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_98_1"
},
{
"DOI": "10.1038/nature21029",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_99_1"
},
{
"DOI": "10.1016/S0140-6736(20)30937-5",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_100_1"
},
{
"DOI": "10.1016/j.isci.2022.105348",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_101_1"
},
{
"DOI": "10.1038/s41576-021-00387-z",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_102_1"
},
{
"article-title": "Single nucleus multiomics identifies ZEB1 and MAFB as candidate regulators of Alzheimer's disease‐specific cis‐regulatory elements. Cell",
"author": "Anderson AG",
"issue": "3",
"journal-title": "Genomics",
"key": "e_1_2_9_103_1",
"volume": "3",
"year": "2023"
},
{
"DOI": "10.1016/j.xgen.2023.100327",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_104_1"
},
{
"DOI": "10.1016/j.xgen.2023.100344",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_105_1"
},
{
"DOI": "10.1016/S0140-6736(20)31042-4",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_106_1"
},
{
"DOI": "10.34172/jre.2021.05",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_107_1"
},
{
"DOI": "10.1016/j.xgen.2022.100145",
"doi-asserted-by": "publisher",
"key": "e_1_2_9_108_1"
}
],
"reference-count": 107,
"references-count": 107,
"relation": {},
"resource": {
"primary": {
"URL": "https://onlinelibrary.wiley.com/doi/10.1111/cpr.13558"
}
},
"score": 1,
"short-title": [],
"source": "Crossref",
"subject": [
"Cell Biology",
"General Medicine"
],
"subtitle": [],
"title": "Integration of human organoids single‐cell transcriptomic profiles and human genetics repurposes critical cell type‐specific drug targets for severe <scp>COVID</scp>‐19",
"type": "journal-article",
"update-policy": "http://dx.doi.org/10.1002/crossmark_policy",
"volume": "57"
}